The New Human Babesia sp. FR1 Is a European Member of the Babesia sp. MO1 Clade
Abstract
1. Introduction
2. Results
2.1. Babesia sp. FR1: Report of the Clinical Case
2.2. Analysis of 18S rRNA Sequences and Position of Babesia sp. FR1 within the B. Divergens-Like Phylogenetic Group
2.3. Major Differences in Ama-1 and Rap-1a Genes within the B. divergens-Like Phylogenetic Group
2.4. Intraspecific Sequence Diversity of Rap-1a and Ama-1 within B. divergens and B. capreoli
2.5. Genetic Variability within the B. divergens-Like Phylogenetic Group
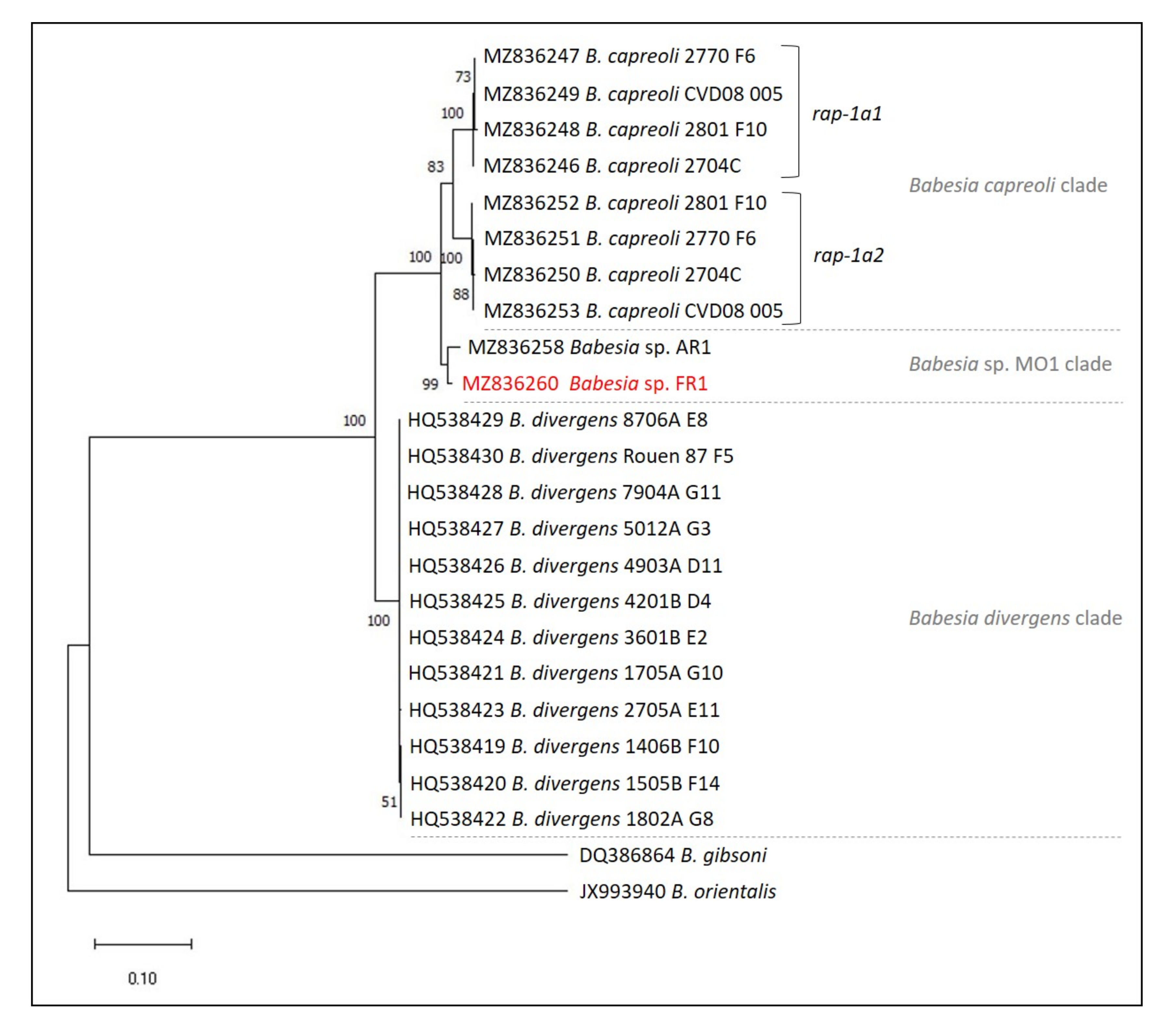
2.6. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Babesia Isolates and DNA Origins
4.2. Comparison of 18S rDNA Sequences within the B. divergens Taxonomic Group
4.3. Amplification of Ama-1 (Apical Membrane Antigen-1) and Rap-1a (Rhoptry Associated protein-1) Genes for B. capreoli, Babesia sp. AR1, and Babesia sp. FR1
4.4. Comparison of Ama-1 and Rap-1a DNA Sequences for B. divergens-Like Phylogenetic Group Members
4.5. Phylogenetic Analysis
4.6. Genbank Deposition
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Schnittger, L.; Rodriguez, A.E.; Florin-Christensen, M.; Morrison, D.A. Babesia: A world emerging. Infect. Genet. Evol. 2012, 12, 1788–1809. [Google Scholar] [CrossRef] [PubMed]
- Herwaldt, B.L.; de Bruyn, G.; Pieniazek, N.J.; Homer, M.; Lofy, K.H.; Slemenda, S.B.; Fritsche, T.R.; Persing, D.H.; Limaye, A.P. Babesia divergens-like infection, Washington State. Emerg. Infect. Dis. 2004, 10, 622–629. [Google Scholar] [CrossRef]
- Herwaldt, B.; Persing, D.H.; Précigout, E.A.; Goff, W.L.; Mathiesen, D.A.; Taylor, P.W.; Eberhard, M.L.; Gorenflot, A.F. A fatal case of babesiosis in Missouri: Identification of another piroplasm that infects humans. Ann. Intern. Med. 1996, 124, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Vannier, E.G.; Diuk-Wasser, M.A.; Ben Mamoun, C.; Krause, P.J. Babesiosis. Infect. Dis. Clin. N. Am. 2015, 29, 357–370. [Google Scholar] [CrossRef]
- Jiang, J.F.; Zheng, Y.C.; Jiang, R.R.; Li, H.; Huo, Q.B.; Jiang, B.G.; Sun, Y.; Jia, N.; Wang, Y.W.; Ma, L.; et al. Epidemiological, clinical, and laboratory characteristics of 48 cases of “Babesia venatorum” infection in China: A descriptive study. Lancet Infect. Dis. 2015, 15, 196–203. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, S.; Yang, J.; Liu, J.; Zhang, D.; Li, Y.; Luo, J.; Guan, G.; Yin, H. Babesia divergens in human in Gansu province, China. Emerg. Microbes Infect. 2019, 8, 959–961. [Google Scholar] [CrossRef] [PubMed]
- Jia, N.; Zheng, Y.C.; Jiang, J.F.; Jiang, R.R.; Jiang, B.G.; Wei, R.; Liu, H.B.; Huo, Q.B.; Sun, Y.; Chu, Y.L.; et al. Human babesiosis caused by a Babesia crassa-like pathogen: A case series. Clin. Infect. Dis. 2018, 67, 1110–1119. [Google Scholar] [CrossRef]
- Skrabalo, Z.; Deanovic, Z. Piroplasmosis in man; report of a case. Doc. Med. Geogr. Trop. 1957, 9, 11–16. [Google Scholar] [PubMed]
- Gray, J.S. Identity of the causal agents of human babesiosis in Europe. Int. J. Med. Microbiol. 2006, 296, 131–136. [Google Scholar] [CrossRef]
- Uguen, C.; Girard, L.; Brasseur, P.; Leblay, R. Human babesiosis in 1997. Rev. Med. Interne 1997, 18, 945–951. [Google Scholar] [CrossRef]
- Herwaldt, B.L.; Cacciò, S.; Gherlinzoni, F.; Aspöck, H.; Slemenda, S.B.; Piccaluga, P.; Martinelli, G.; Edelhofer, R.; Hollenstein, U.; Poletti, G.; et al. Molecular characterization of a non-Babesia divergens organism causing zoonotic babesiosis in Europe. Emerg. Infect. Dis. 2003, 9, 942–948. [Google Scholar] [CrossRef]
- Häselbarth, K.; Tenter, A.M.; Brade, V.; Krieger, G.; Hunfeld, K.P. First case of human babesiosis in Germany—Clinical presentation and molecular characterisation of the pathogen. Int. J. Med. Microbiol. 2007, 297, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Bläckberg, J.; Lazarevic, V.L.; Hunfeld, K.P.; Persson, K.E.M. Low-virulent Babesia venatorum infection masquerading as hemophagocytic syndrome. Ann. Hematol. 2018, 97, 731–733. [Google Scholar] [CrossRef] [PubMed]
- Stahl, P.; Poinsignon, Y.; Pouedras, P.; Ciubotaru, V.; Berry, L.; Emu, B.; Krause, P.J.; Ben Mamoun, C.; Cornillot, E. Case report of the patient source of the Babesia microti R1 reference strain and implications for travelers. J. Travel. Med. 2018, 25, tax073. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, A.; Hunfeld, K.P.; Baier, M.; Krumbholz, A.; Sachse, S.; Lorenzen, T.; Kiehntopf, M.; Fricke, H.J.; Straube, E. First confirmed autochthonous case of human Babesia microti infection in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 595–601. [Google Scholar] [CrossRef]
- Centeno-Lima, S.; do Rosário, V.; Parreira, R.; Maia, A.J.; Freudenthal, A.M.; Nijhof, A.M.; Jongejan, F. A fatal case of human babesiosis in Portugal: Molecular and phylogenetic analysis. Trop. Med. Int. Health. 2003, 8, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Corpelet, C.; Vacher, P.; Coudore, F.; Laurichesse, H.; Conort, N.; Souweine, B. Role of quinine in life-threatening Babesia divergens infection successfully treated with clindamycin. Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 74–75. [Google Scholar] [CrossRef]
- Haapasalo, K.; Suomalainen, P.; Sukura, A.; Siikamaki, H.; Jokiranta, T.S. Fatal babesiosis in man, Finland, 2004. Emerg. Infect. Dis. 2010, 16, 1116–1118. [Google Scholar] [CrossRef]
- Gonzalez, L.M.; Rojo, S.; Gonzalez-Camacho, F.; Luque, D.; Lobo, C.A.; Montero, E. Severe babesiosis in immunocompetent man, Spain, 2011. Emerg. Infect. Dis. 2014, 20, 724–726. [Google Scholar] [CrossRef]
- González, L.M.; Castro, E.; Lobo, C.A.; Richart, A.; Ramiro, R.; González-Camacho, F.; Luque, D.; Velasco, A.C.; Montero, E. First report of Babesia divergens infection in an HIV patient. Int. J. Infect. Dis. 2015, 33, 202–204. [Google Scholar] [CrossRef] [PubMed]
- Tanyel, E.; Guler, N.; Hokelek, M.; Ulger, F.; Sunbul, M. A case of severe babesiosis treated successfully with exchange transfusion. Int. J. Infect. Dis. 2015, 38, 83–85. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, S.; Lyons, C.; Abdou, M.; Patowary, R.; Aslam, S.; Kinsella, N.; Zintl, A.; Hunfeld, K.P.; Wormser, G.P.; Gray, J.; et al. Splenic dysfunction from celiac disease resulting in severe babesiosis. Ticks Tick Borne Dis. 2017, 8, 537–539. [Google Scholar] [CrossRef]
- Asensi, V.; González, L.M.; Fernández-Suárez, J.; Sevilla, E.; Navascués, R.Á.; Suárez, M.L.; Lauret, M.E.; Bernardo, A.; Carton, J.A.; Montero, E. A fatal case of Babesia divergens infection in Northwestern Spain. Ticks Tick Borne Dis. 2018, 9, 730–734. [Google Scholar] [CrossRef]
- Kukina, I.V.; Zelya, O.P.; Guzeeva, T.M.; Karan, L.S.; Perkovskaya, I.A.; Tymoshenko, N.I.; Guzeeva, M.V. Severe babesiosis caused by Babesia divergens in a host with intact spleen, Russia, 2018. Ticks Tick Borne Dis. 2019, 10, 101262. [Google Scholar] [CrossRef]
- Mørch, K.; Holmaas, G.; Frolander, P.S.; Kristoffersen, E.K. Severe human Babesia divergens infection in Norway. Int. J. Infect. Dis. 2015, 33, 37–38. [Google Scholar] [CrossRef] [PubMed]
- Kukina, I.V.; Guzeeva, T.M.; Zelya, O.P.; Ganushkina, L.A. Fatal human babesiosis caused by Babesia divergens in an asplenic host. IDCases 2018, 13, e00414. [Google Scholar] [CrossRef]
- Strizova, Z.; Havlova, K.; Patek, O.; Smrz, D.; Bartunkova, J. The first human case of babesiosis mimicking Reiter’s syndrome. Folia Parasitol. 2020, 67, 1–5. [Google Scholar] [CrossRef]
- Loutan, L.; Rossier, J.; Zufferey, G.; Cuénod, D.; Hatz, C.; Marti, H.P.; Gern, L. Human babesiosis: First case report in Switzerland. Rev. Med. Suisse Romande 1994, 114, 111–116. (In French) [Google Scholar]
- Martinot, M.; Zadeh, M.M.; Hansmann, Y.; Grawey, I.; Christmann, D.; Aguillon, S.; Jouglin, M.; Chauvin, A.; De Briel, D. Babesiosis in immunocompetent patients, Europe. Emerg. Infect. Dis. 2011, 17, 114–116. [Google Scholar] [CrossRef]
- Paleau, A.; Candolfi, E.; Souply, L.; De Briel, D.; Delarbre, J.M.; Lipsker, D.; Jouglin, M.; Malandrin, L.; Hansmann, Y.; Martinot, M. Human babesiosis in Alsace. Med. Mal. Infect. 2020, 50, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Lempereur, L.; Shiels, B.; Heyman, P.; Moreau, E.; Saegerman, C.; Losson, B.; Malandrin, L. A retrospective serological survey on human babesiosis in Belgium. Clin. Microbiol. Infect. 2015, 21, 96.e1–96.e7. [Google Scholar] [CrossRef]
- Duh, D.; Petrovec, M.; Bidovec, A.; Avsic-Zupanc, T. Cervids as Babesiae hosts, Slovenia. Emerg. Infect. Dis. 2005, 11, 1121–1123. [Google Scholar] [CrossRef]
- Malandrin, L.; Jouglin, M.; Sun, Y.; Brisseau, N.; Chauvin, A. Redescription of Babesia capreoli (Enigk and Friedhoff, 1962) from roe deer (Capreolus capreolus): Isolation, cultivation, host specificity, molecular characterisation and differentiation from Babesia divergens. Int. J. Parasitol. 2010, 40, 277–284. [Google Scholar] [CrossRef]
- Beattie, J.F.; Michelson, M.L.; Holman, P.J. Acute babesiosis caused by Babesia divergens in a resident of Kentucky. N. Engl. J. Med. 2002, 347, 697–698. [Google Scholar] [CrossRef]
- Burgess, M.J.; Rosenbaum, E.R.; Pritt, B.S.; Haselow, D.T.; Ferren, K.M.; Alzghoul, B.N.; Rico, J.C.; Sloan, L.M.; Ramanan, P.; Purushothaman, R.; et al. Possible transfusion-transmitted Babesia divergens-like/MO1 infection in an Arkansas patient. Clin. Infect. Dis. 2017, 64, 1622–1625. [Google Scholar] [CrossRef]
- Herc, E.; Pritt, B.; Huizenga, T.; Douce, R.; Hysell, M.; Newton, D.; Sidge, J.; Losman, E.; Sherbeck, J.; Kaul, D.R. Probable locally acquired Babesia divergens-like infection in woman, Michigan, USA. Emerg. Infect. Dis. 2018, 24, 1558–1560. [Google Scholar] [CrossRef] [PubMed]
- Goethert, H.K.; Telford, S.R., 3rd. Enzootic transmission of Babesia divergens among cottontail rabbits on Nantucket Island, Massachusetts. Am. J. Trop. Med. Hyg. 2003, 69, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Holman, P.J.; Spencer, A.M.; Droleskey, R.E.; Goethert, H.K.; Telford, S.R., 3rd. In vitro cultivation of a zoonotic Babesia sp. isolated from eastern cottontail rabbits (Sylvilagus floridanus) on Nantucket Island, Massachusetts. J. Clin. Microbiol. 2005, 43, 3995–4001. [Google Scholar] [CrossRef]
- Holman, P.J.; Spencer, A.M.; Telford, S.R., 3rd; Goethert, H.K.; Allen, A.J.; Knowles, D.P.; Goff, W.L. Comparative infectivity of Babesia divergens and a zoonotic Babesia divergens-like parasite in cattle. Am. J. Trop. Med. Hyg. 2005, 73, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Spencer, A.M.; Goethert, H.K.; Telford, S.R., 3rd; Holman, P.J. In vitro host erythrocyte specificity and differential morphology of Babesia divergens and a zoonotic Babesia sp. from eastern cottontail rabbits (Sylvilagus floridanus). J. Parasitol. 2006, 92, 333–340. [Google Scholar] [CrossRef]
- Holman, P.J. Phylogenetic and biologic evidence that Babesia divergens is not endemic in the United States. Ann. N. Y. Acad. Sci. 2006, 1081, 518–525. [Google Scholar] [CrossRef]
- Hildebrandt, A.; Gray, J.S.; Hunfeld, K.P. Human babesiosis in Europe: What clinicians need to know. Infection 2013, 41, 1057–1072. [Google Scholar] [CrossRef]
- Hildebrandt, A.; Zintl, A.; Montero, E.; Hunfeld, K.P.; Gray, J. Human Babesiosis in Europe. Pathogens 2021, 10, 1165. [Google Scholar] [CrossRef]
- Jahfari, S.; Hofhuis, A.; Fonville, M.; van der Giessen, J.; van Pelt, W.; Sprong, H. Molecular Detection of tick-borne pathogens in humans with tick bites and erythema migrans, in the Netherlands. PLoS Negl. Trop. Dis. 2016, 10, e0005042. [Google Scholar] [CrossRef]
- Moreau, E.; Bonsergent, C.; Al Dybiat, I.; Gonzalez, L.M.; Lobo, C.A.; Montero, E.; Malandrin, L. Babesia divergens apical membrane antigen-1 (BdAMA-1): A poorly polymorphic protein that induces a weak and late immune response. Exp. Parasitol. 2015, 155, 40–45. [Google Scholar] [CrossRef]
- Sun, Y. Caractérisation moléculaire, localisation cellulaire et conservation des protéines impliquées dans le processus d’invasion des érythrocytes par Babesia divergens. Ph.D. Thesis, Nantes University, Nantes, France, 2010; 202p. [Google Scholar]
- Bastian, S.; Jouglin, M.; Brisseau, N.; Malandrin, L.; Klegou, G.; L’Hostis, M.; Chauvin, A. Antibody prevalence and molecular identification of Babesia spp. in roe deer in France. J. Wildl. Dis. 2012, 48, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Krause, P.J. Human babesiosis. Int. J. Parasitol. 2019, 49, 165–174. [Google Scholar] [CrossRef]
- Gorenflot, A.; Moubri, K.; Precigout, E.; Carcy, B.; Schetters, T.P. Human babesiosis. Ann. Trop. Med. Parasitol. 1998, 92, 489–501. [Google Scholar] [CrossRef] [PubMed]
- L’Hostis, M.; Chauvin, A.; Valentin, A.; Marchand, A.; Gorenflot, A. Large scale survey of bovine babesiosis due to Babesia divergens in France. Vet. Rec. 1995, 136, 36–38. [Google Scholar] [CrossRef]
- Agoulon, A.; Malandrin, L.; Lepigeon, F.; Vénisse, M.; Bonnet, S.; Becker, C.A.; Hoch, T.; Bastian, S.; Plantard, O.; Beaudeau, F. A Vegetation Index qualifying pasture edges is related to Ixodes ricinus density and to Babesia divergens seroprevalence in dairy cattle herds. Vet. Parasitol. 2012, 185, 101–109. [Google Scholar] [CrossRef] [PubMed]
- González, L.M.; Estrada, K.; Grande, R.; Jiménez-Jacinto, V.; Vega-Alvarado, L.; Sevilla, E.; Barrera, J.; Cuesta, I.; Zaballos, Á.; Bautista, J.M.; et al. Comparative and functional genomics of the protozoan parasite Babesia divergens highlighting the invasion and egress processes. PLoS Negl. Trop. Dis. 2019, 13, e0007680. [Google Scholar] [CrossRef]
- Zamoto-Niikura, A.; Tsuji, M.; Imaoka, K.; Kimura, M.; Morikawa, S.; Holman, P.J.; Hirata, H.; Ishihara, C. Sika deer carrying Babesia parasites closely related to B. divergens, Japan. Emerg. Infect. Dis. 2014, 20, 1398–1400. [Google Scholar] [CrossRef]
- Zamoto-Niikura, A.; Tsuji, M.; Qiang, W.; Morikawa, S.; Hanaki, K.I.; Holman, P.J.; Ishihara, C. The Babesia divergens Asia lineage is maintained through enzootic cycles between Ixodes persulcatus and sika deer in Hokkaido, Japan. Appl. Environ. Microbiol. 2018, 84, e02491-17. [Google Scholar] [CrossRef] [PubMed]
- Malandrin, L.; L’Hostis, M.; Chauvin, A. Isolation of Babesia divergens from carrier cattle blood using in vitro culture. Vet. Res. 2004, 35, 131–139. [Google Scholar] [CrossRef]
- Dalrymple, B.P.; Casu, R.E.; Peters, J.M.; Dimmock, C.M.; Gale, K.R.; Böse, R.; Wright, I.G. Characterisation of a family of multi-copy genes encoding rhoptry protein homologues in Babesia bovis, Babesia ovis and Babesia canis. Mol. Biochem. Parasitol. 1993, 57, 181–192. [Google Scholar] [CrossRef]
- Suarez, C.E.; Palmer, G.H.; Hötzel, I.; McElwain, T.F. Structure, sequence, and transcriptional analysis of the Babesia bovis rap-1 multigene locus. Mol. Biochem. Parasitol. 1998, 93, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Suarez, C.E.; Palmer, G.H.; Florin-Christensen, M.; Hines, S.A.; Hötzel, I.; McElwain, T.F. Organization transcription, and expression of rhoptry associated protein genes in the Babesia bigemina rap-1 locus. Mol. Biochem. Parasitol. 2003, 127, 101–112. [Google Scholar] [CrossRef]
- Niu, Q.; Bonsergent, C.; Guan, G.; Yin, H.; Malandrin, L. Sequence and organization of the rhoptry-associated-protein-1 (rap-1) locus for the sheep hemoprotozoan Babesia sp. BQ1 Lintan (B. motasi phylogenetic group). Vet. Parasitol. 2013, 198, 24–38. [Google Scholar] [CrossRef]
- Niu, Q.; Valentin, C.; Bonsergent, C.; Malandrin, L. Strong conservation of rhoptry-associated-protein-1 (RAP-1) locus organization and sequence among Babesia isolates infecting sheep from China (Babesia motasi-like phylogenetic group). Infect. Genet. Evol. 2014, 28, 21–32. [Google Scholar] [CrossRef]
- Niu, Q.; Marchand, J.; Yang, C.; Bonsergent, C.; Guan, G.; Yin, H.; Malandrin, L. Rhoptry-associated protein (rap-1) genes in the sheep pathogen Babesia sp. Xinjiang: Multiple transcribed copies differing by 3′ end repeated sequences. Vet. Parasitol. 2015, 211, 158–169. [Google Scholar] [CrossRef]
- Skuce, P.J.; Mallon, T.R.; Taylor, S.M. Molecular cloning of a putative rhoptry associated protein homologue from Babesia divergens. Mol. Biochem. Parasitol. 1996, 77, 99–102. [Google Scholar] [CrossRef]
- Rodriguez, M.; Alhassan, A.; Ord, R.L.; Cursino-Santos, J.R.; Singh, M.; Gray, J.; Lobo, C.A. Identification and characterization of the RouenBd1987 Babesia divergens Rhopty-Associated Protein 1. PLoS ONE 2014, 9, e107727. [Google Scholar] [CrossRef][Green Version]
- Sun, Y.; Jouglin, M.; Bastian, S.; Chauvin, A.; Malandrin, L. Molecular cloning and genetic polymorphism of Babesia capreoli gene Bcp37/41, an ortholog of Babesia divergens merozoite surface antigen Bd37. Vet. Parasitol. 2011, 178, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
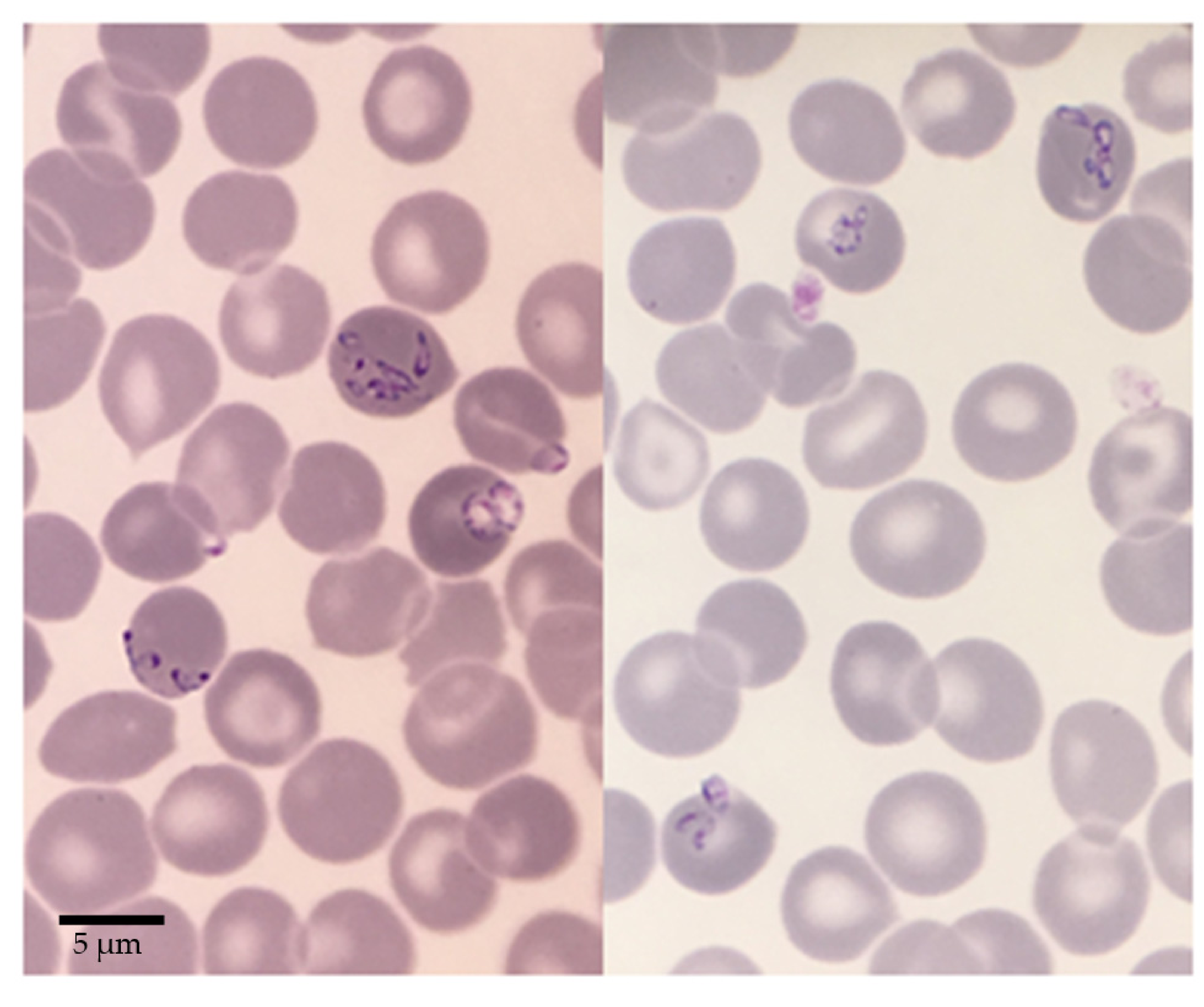
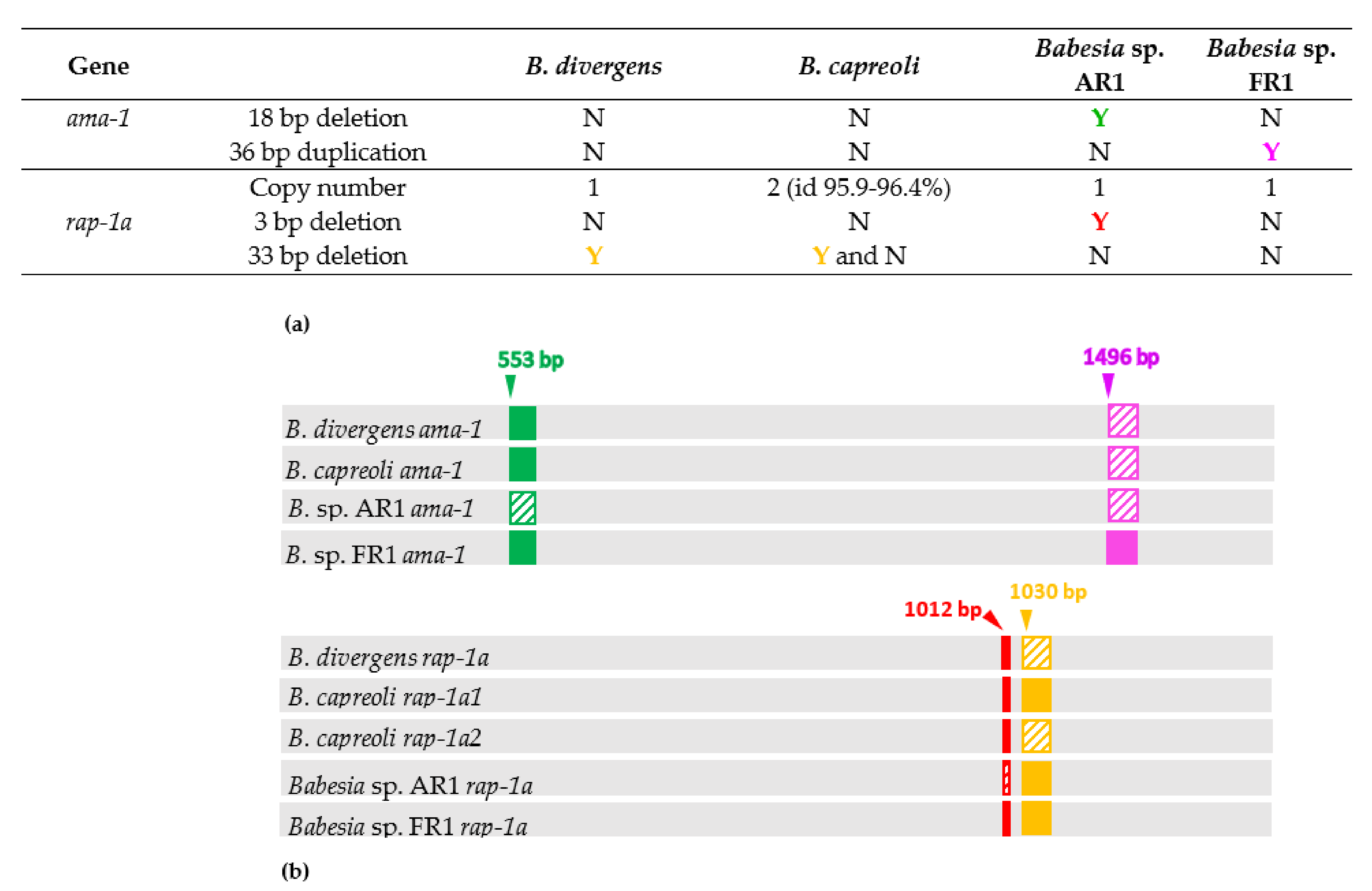
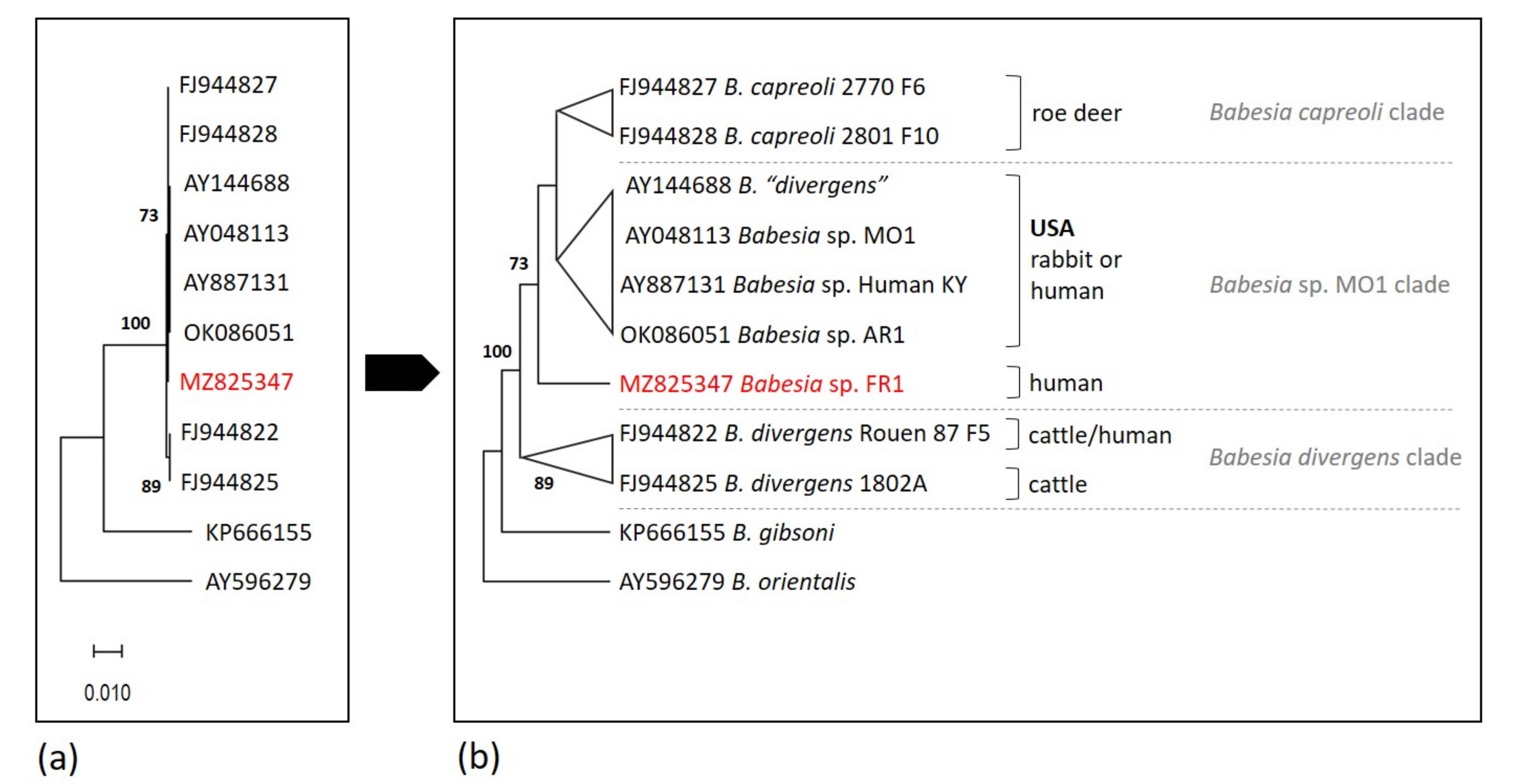
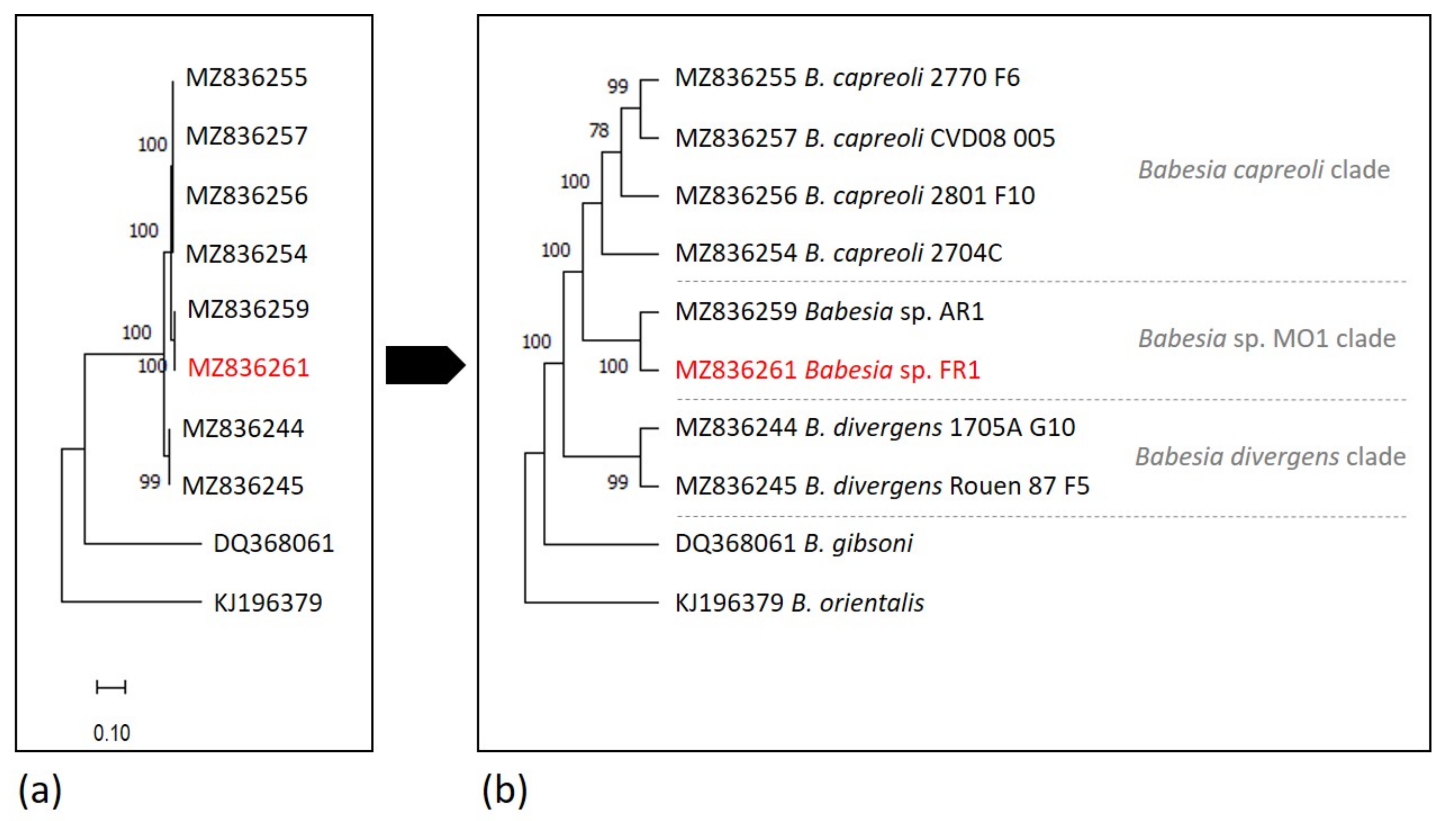
| Organism | Natural Host | Human Infection | Geographical Occurence | Vector | 18S rRNA Sequence Differences at Nucleotide Position b | |||
|---|---|---|---|---|---|---|---|---|
| 631 | 663 | 819 | 1637 | |||||
| B. divergens | Cattle | + | Europe | I. ricinus | A | A | T | C |
| B. capreoli | Roe deer | − | Europe | I. ricinus | G | T | T | T |
| Babesia sp. MO1/AR1 | Cottontail rabbit | + | USA | I. dentatus a | G | A | A | T |
| Babesia sp. FR1 | nd c | + | France | nd c | G | A | T | T |
| Babesia Species | Gene | Number of Isolates | Nucleotide Differences | Identities |
|---|---|---|---|---|
| B. divergens | 18S rRNA | 12 | None | 100% [33] |
| rap-1a | 12 | 0–6 nt/1242 bp | 99.6–100% [46] | |
| ama-1 | 9 | 0–2 nt/1821 bp | 99.9–100% [45] | |
| B. capreoli | 18S rRNA | 9 | None | 100% [33] |
| rap-1a1 | 4 | 0–7 nt/1236 bp | 99.5–100% | |
| rap-1a2 | 4 | 0–5 nt/1203 bp | 99.6–100% | |
| ama-1 | 4 | 0–6 nt/1821 bp | 99.7–100% |
| Organism | B. divergens | B. capreoli | Babesia sp. AR1 | Babesia sp. FR1 |
|---|---|---|---|---|
| B. divergens | 0 | |||
| 99.9–100% | ||||
| 99.6–100% | ||||
| B. capreoli | 3 | 0 | ||
| 95–95.3% | 99.7–100% | |||
| (rap-1a1) 86.6–89.1% | (rap-1a1) 99.5–100% | |||
| (rap1-a2) 88.6–89.1% | (rap1-a2) 99.6–100% | |||
| Babesia sp. AR1 | 3 | 2 | 0 | |
| 94.3% | 97.2–97.3% | 100% | ||
| 89.5–89.8% | (rap-1a1) 95.1–95.4% | 100% | ||
| (rap1-a2) 95.3–95.7% | ||||
| Babesia sp. FR1 | 2 | 1 | 1 | 0 |
| 94.5% | 97.3–97.4% | 98.7% | 100% | |
| 89.8–90% | (rap-1a1) 95.2–95.5% | 98.7% | 100% | |
| (rap1-a2) 95.7–96.1% |
| Target Gene | Primer Name | Sequence (5′–3′) | Tm (°C) | PCR | Amplicon Length (bp) | Sequencing | References |
|---|---|---|---|---|---|---|---|
| 18S rRNA | CRYPTOF | AACCTGGTTGATCCTGCCAGTAGTCAT | 63 | × | 1728 | × | [33] |
| CRYPTOR | TGATCCTTCTGCAGGTTCACCTA | × | × | ||||
| BAB-GF2 | GTCTTGTAATTGGAATGATGG | 61 | × | 560 | × | [11] | |
| BAB-GR2 | CCAAAGACTTTGATTTCTCT | × | × | ||||
| ama-1 | ama1-S1 | TGACTGCCATATCGACGAAG | 61 | × | ≈2000 | × | this study |
| ama1-R3 | CTCTAGTGAATTACGATAGC | × | × | [50] | |||
| ama1-As1 | GGCGGATATTCGGTTGAGG | × | this study | ||||
| ama1-S2 | CATGGCCAAGTTTGACCTTG | × | this study | ||||
| ama1-As2 | CTGCGTCACGCGTGAATTC | × | this study | ||||
| ama1-S3 | CTCCTGTGTATGGAGCCGA | × | this study | ||||
| ama1-As3 | GTGAAAGCGCGGTTGTGAC | × | this study | ||||
| ama1-S4 | AGCAGTTGGATCGCCTCTC | × | this study | ||||
| rap-1a | rap1-fw | AATGTCCTACTGGGAAACGC | 58 | × | ≈1300 | × | this study |
| rap1-rev | GCGGAGTCCATGCCTGTACC | × | × | this study | |||
| rap-1a1 (5′) | rap1-fw | see above | 58 | × | 1146 | × | |
| rap1-a1-rev | GCTTAGTAGCATGCATCTTC | × | × | this study | |||
| rap-1a1 (3′) | rap1-a1-fw | GGACTCCGAGAAAAAGGATG | 58 | × | 261 | × | this study |
| rap1-rev | see above | × | × | ||||
| rap-1a2 (5′) | rap1-fw | see above | 58 | × | 1123 | × | |
| rap1-a2-rev | TGGAACAACTTCTTCATAGG | × | × | this study | |||
| rap-1a2 (3′) | rap1-a2-fw | GGGCTTCTGGAAAAAGAAGG | 58 | × | 228 | × | this study |
| rap1-rev | see above | × | × |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonsergent, C.; de Carné, M.-C.; de la Cotte, N.; Moussel, F.; Perronne, V.; Malandrin, L. The New Human Babesia sp. FR1 Is a European Member of the Babesia sp. MO1 Clade. Pathogens 2021, 10, 1433. https://doi.org/10.3390/pathogens10111433
Bonsergent C, de Carné M-C, de la Cotte N, Moussel F, Perronne V, Malandrin L. The New Human Babesia sp. FR1 Is a European Member of the Babesia sp. MO1 Clade. Pathogens. 2021; 10(11):1433. https://doi.org/10.3390/pathogens10111433
Chicago/Turabian StyleBonsergent, Claire, Marie-Charlotte de Carné, Nathalie de la Cotte, François Moussel, Véronique Perronne, and Laurence Malandrin. 2021. "The New Human Babesia sp. FR1 Is a European Member of the Babesia sp. MO1 Clade" Pathogens 10, no. 11: 1433. https://doi.org/10.3390/pathogens10111433
APA StyleBonsergent, C., de Carné, M.-C., de la Cotte, N., Moussel, F., Perronne, V., & Malandrin, L. (2021). The New Human Babesia sp. FR1 Is a European Member of the Babesia sp. MO1 Clade. Pathogens, 10(11), 1433. https://doi.org/10.3390/pathogens10111433





