New Reference Genes for qRT-PCR Analysis as a Potential Target for Identification of Trichophyton verrucosum in Different Culture Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dermatophyte Strains and Growth Conditions
2.2. RNA Extraction and cDNA Synthesis
2.3. qRT-PCR Analysis
2.4. Data Analysis
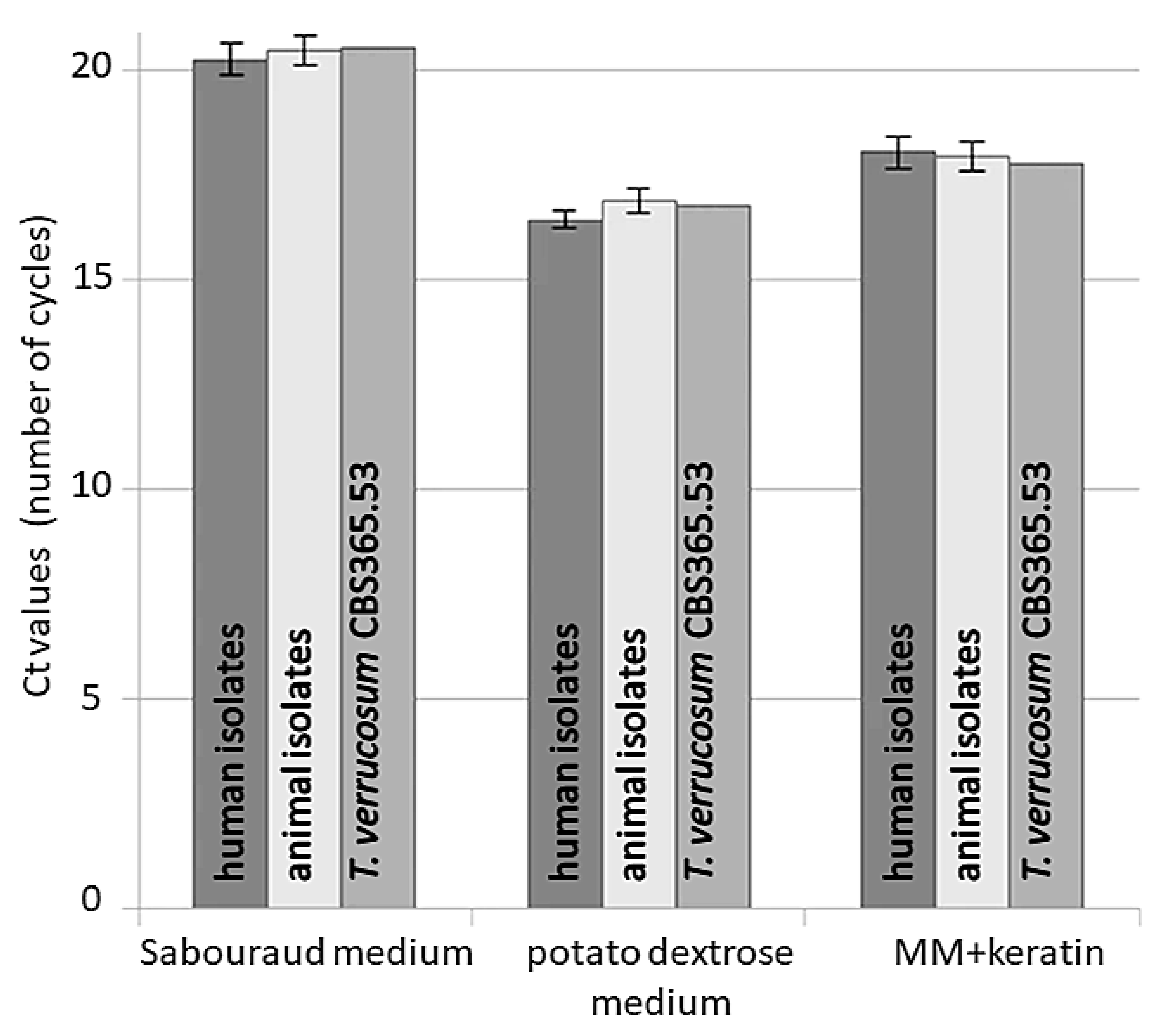
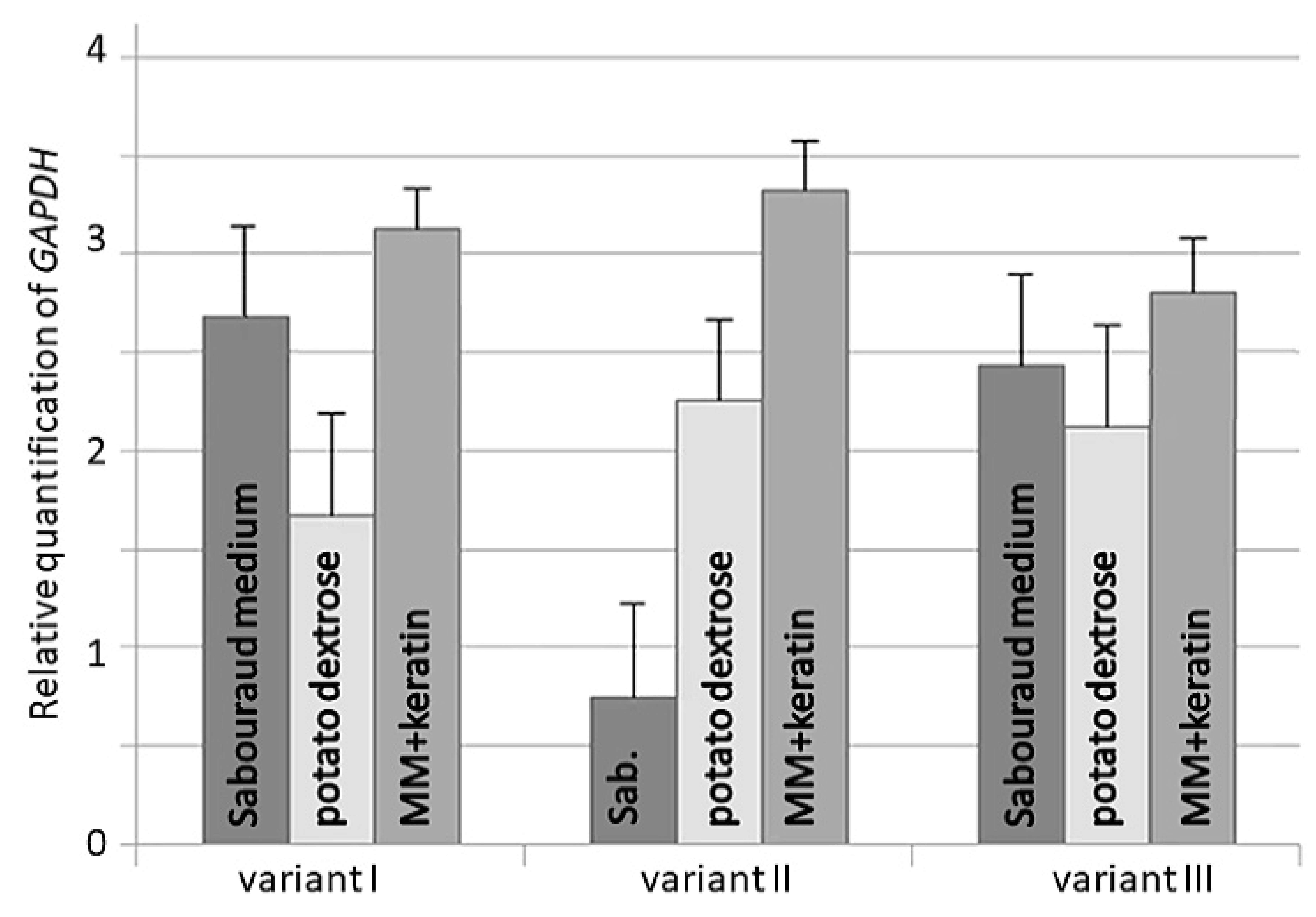
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gnat, S.; Nowakiewicz, A.; Zięba, P. Taxonomy of Dermatophytes—The Classification Systems May Change But the Identification Problems Remain the Same. Postępy Mikrobiol. Adv. Microbiol. 2019, 58, 49–58. [Google Scholar] [CrossRef] [Green Version]
- de Hoog, G.S.; Dukik, K.; Monod, M.; Packeu, A.; Stubbe, D.; Hendrickx, M.; Kupsch, C.; Stielow, J.B.; Freeke, J.; Göker, M.; et al. Toward a Novel Multilocus Phylogenetic Taxonomy for the Dermatophytes. Mycopathologia 2017, 182, 5–31. [Google Scholar] [CrossRef] [Green Version]
- Havlickova, B.; Czaika, V.A.; Friedrich, M. Epidemiological Trends in Skin Mycoses Worldwide. Mycoses 2008, 51, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Del Boz, J.; Crespo, V.; Rivas-Ruiz, F.; De Troya, M. A 30-Year Survey of Paediatric Tinea Capitis in Southern Spain. J. Eur. Acad. Dermatol. Venereol. 2011, 25, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Bitew, A. Dermatophytosis: Prevalence of Dermatophytes and Non-Dermatophyte Fungi from Patients Attending Arsho Advanced Medical Laboratory, Addis Ababa, Ethiopia. Dermatol. Res. Pract. 2018, 2018, 8164757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozkutuk, A.; Ergon, C.; Yulug, N. Species Distribution and Antifungal Susceptibilities of Dermatophytes during a One Year Period at a University Hospital in Turkey. Mycoses 2007, 50, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Bishnoi, A.; Vinay, K.; Dogra, S. Emergence of Recalcitrant Dermatophytosis in India. Lancet Infect. Dis. 2018, 18, 250–251. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Masih, A.; Khurana, A.; Singh, P.K.; Gupta, M.; Hagen, F.; Meis, J.F.; Chowdhary, A. High Terbinafine Resistance in Trichophyton interdigitale Isolates in Delhi, India Harbouring Mutations in the Squalene Epoxidase Gene. Mycoses 2018, 61, 477–484. [Google Scholar] [CrossRef]
- Łagowski, D.; Gnat, S.; Nowakiewicz, A.; Osińska, M. Comparison of in Vitro Activities of 11 Antifungal Agents against Trichophyton verrucosum Isolates Associated with a Variety Hosts and Geographical Origin. Mycoses 2020, 63, 294–301. [Google Scholar] [CrossRef]
- Gnat, S.; Łagowski, D.; Nowakiewicz, A.; Dyląg, M. Tinea Corporis Caused by Trichophyton equinum Transmitted from Asymptomatic Dogs to Two Siblings. Braz. J. Microbiol. 2020, 51, 1433–1438. [Google Scholar] [CrossRef]
- Gnat, S.; Łagowski, D.; Nowakiewicz, A.; Trościańczyk, A.; Zięba, P. Infection of Trichophyton verrucosum in Cattle Breeders, Poland: A 40-Year Retrospective Study on the Genomic Variability of Strains. Mycoses 2018, 61, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Courtellemont, L.; Chevrier, S.; Degeilh, B.; Belaz, S.; Gangneux, J.P.; Robert-Gangneux, F. Epidemiology of Trichophyton verrucosum Infection in Rennes University Hospital, France: A 12-Year Retrospective Study. Med. Mycol. 2017, 55, 720–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Łagowski, D.; Gnat, S.; Nowakiewicz, A.; Osińska, M.; Trościańczyk, A.; Zięba, P. In Search of the Source of Dermatophytosis: Epidemiological Analysis of Trichophyton verrucosum Infection in Llamas and the Breeder (Case Report). Zoonoses Public Health 2019, 66, 982–989. [Google Scholar] [CrossRef]
- Bassiri Jahromi, S.; Khaksar, A.A. Aetiological Agents of Tinea Capitis in Tehran (Iran). Mycoses 2006, 49, 65–67. [Google Scholar] [CrossRef] [PubMed]
- Gnat, S.; Łagowski, D.; Nowakiewicz, A.; Dyląg, M. Molekularne Metody Diagnostyki Dermatomykoz—Przegląd Dostępnych Technik Oraz Ocena Ich Zalet i Wad w Implementacji Do Rutynowego Stosowania. Adv. Microbiol. 2019, 58, 483–494. [Google Scholar] [CrossRef] [Green Version]
- Verrier, J.; Krähenbühl, L.; Bontems, O.; Fratti, M.; Salamin, K.; Monod, M. Dermatophyte Identification in Skin and Hair Samples Using a Simple and Reliable Nested Polymerase Chain Reaction Assay. Br. J. Dermatol. 2013, 168, 295–301. [Google Scholar] [CrossRef]
- Verrier, J.; Monod, M. Diagnosis of Dermatophytosis Using Molecular Biology. Mycopathologia 2017, 182, 193–202. [Google Scholar] [CrossRef]
- Rudramurthy, S.; Shaw, D. Overview and Update on the Laboratory Diagnosis of Dermatophytosis. Clin. Dermatol. Rev. 2017, 1, 3. [Google Scholar] [CrossRef]
- Kawai, M. Diagnosis of dermatophytoses: Conventional methods and recent molecular biological methods. Nihon Ishinkin Gakkai Zasshi 2003, 44, 261–264. [Google Scholar] [CrossRef]
- Bouchara, J.P.; Mignon, B.; Chaturvedi, V. Dermatophytes and Dermatophytoses: A Thematic Overview of State of the Art, and the Directions for Future Research and Developments. Mycopathologia 2017, 182, 1–4. [Google Scholar] [CrossRef]
- Graser, Y.; Monod, M.; Bouchara, J.-P.; Dukik, K.; Nenoff, P.; Kargl, A.; Kupsch, C.; Zhan, P.; Packeu, A.; Chaturvedi, V.; et al. New Insights in Dermatophyte Research. Med. Mycol. 2018, 56, S2–S9. [Google Scholar] [CrossRef] [Green Version]
- Monod, M.; Feuermann, M.; Salamin, K.; Fratti, M.; Makino, M.; Alshahni, M.M.; Makimura, K.; Yamada, T. Trichophyton rubrum Azole Resistance Mediated by a New ABC Transporter, TruMDR3. Antimicrob. Agents Chemother. 2019, 63, e00863-19. [Google Scholar] [CrossRef] [Green Version]
- Kobylak, N.; Bykowska, B.; Nowicki, R.; Brillowska-Dabrowska, A. Real-Time PCR Approach in Dermatophyte Detection and Trichophyton rubrum Identification. Acta Biochim. Pol. 2015, 62, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Ohst, T.; Kupsch, C.; Graser, Y.; Gräser, Y. Detection of Common Dermatophytes in Clinical Specimens Using a Simple Quantitative Real-Time TaqMan Polymerase Chain Reaction Assay. Br. J. Dermatol. 2016, 174, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Alexander, C.L.; Shankland, G.S.; Carman, W.; Williams, C. Introduction of a Dermatophyte Polymerase Chain Reaction Assay to the Diagnostic Mycology Service in Scotland. Br. J. Dermatol. 2011, 164, 966–972. [Google Scholar] [CrossRef] [PubMed]
- Ciesielska, A.; Staczek, P. Selection and Validation of Reference Genes for QRT-PCR Analysis of Gene Expression in Microsporum canis Growing under Different Adhesion-Inducing Conditions. Sci. Rep. 2018, 8, 1197. [Google Scholar] [CrossRef] [PubMed]
- Cove, D.J. The Induction and Repression of Nitrate Reductase in the Fungus Aspergillus nidulans. Biochim. Biophys. Acta 1966, 113, 51–56. [Google Scholar] [CrossRef]
- Gnat, S.; Łagowski, D.; Nowakiewicz, A.; Zięba, P. The Host Range of Dermatophytes, It Is at All Possible? Phenotypic Evaluation of the Keratinolytic Activity of Trichophyton verrucosum Clinical Isolates. Mycoses 2019, 62, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. Quantification strategies in real-time PCR. In A-Z of Quantitative PCR; Bustin, S.A., Ed.; International University Line: La Jolla, CA, USA, 2004; Volume 5, pp. 87–112. ISBN 9780963681782. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Gnat, S.; Łagowski, D.; Nowakiewicz, A. Major Challenges and Perspectives in the Diagnostics and Treatment of Dermatophyte Infections. J. Appl. Microbiol. 2020, 129, 212–232. [Google Scholar] [CrossRef] [Green Version]
- Rezaei-Matehkolaei, A.; Makimura, K.; De Hoog, G.S.; Shidfar, M.R.; Satoh, K.; Najafzadeh, M.J.; Mirhendi, H. Discrimination of Trichophyton tonsurans and Trichophyton equinum by PCR-RFLP and by β-Tubulin and Translation Elongation Factor 1-α Sequencing. Med. Mycol. 2012, 50, 760–764. [Google Scholar] [CrossRef] [Green Version]
- Sherman, S.; Goshen, M.; Treigerman, O.; Ben-Zion, K.; Carp, M.J.; Maisler, N.; Ehrenreich, I.B.; Kimchi, A.; Lifshitz, S.; Smollan, G.; et al. Evaluation of Multiplex Real-Time PCR for Identifying Dermatophytes in Clinical Samples—A Multicentre Study. Mycoses 2018, 61, 119–126. [Google Scholar] [CrossRef]
- Emam, S.M.; Abd El-salam, O.H. Real-Time PCR: A Rapid and Sensitive Method for Diagnosis of Dermatophyte Induced Onychomycosis, a Comparative Study. Alex. J. Med. 2016, 52, 83–90. [Google Scholar] [CrossRef]
- Kupsch, C.; Ohst, T.; Pankewitz, F.; Nenoff, P.; Uhrlaß, S.; Winter, I.; Gräser, Y. The Agony of Choice in Dermatophyte Diagnostics—Performance of Different Molecular Tests and Culture in the Detection of Trichophyton rubrum and Trichophyton interdigitale. Clin. Microbiol. Infect. 2016, 22, 735.e11–735.e17. [Google Scholar] [CrossRef] [Green Version]
- Jacobson, L.S.; Giacinti, J.A.; Robertson, J. Medical Conditions and Outcomes in 371 Hoarded Cats from 14 Sources: A Retrospective Study (2011–2014). J. Feline Med. Surg. 2020, 22, 484–491. [Google Scholar] [CrossRef]
- Jacob, T.R.; Peres, N.T.A.; Persinoti, G.F.; Silva, L.G.; Mazucato, M.; Rossi, A.; Martinez-Rossi, N.M. Rpb2 Is a Reliable Reference Gene for Quantitative Gene Expression Analysis in the Dermatophyte Trichophyton rubrum. Med. Mycol. 2012, 50, 368–377. [Google Scholar] [CrossRef] [Green Version]
- Nailis, H.; Coenye, T.; Van Nieuwerburgh, F.; Deforce, D.; Nelis, H.J. Development and Evaluation of Different Normalization Strategies for Gene Expression Studies in Candida albicans Biofilms by Real-Time PCR. BMC Mol. Biol. 2006, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Bohle, K.; Jungebloud, A.; Göcke, Y.; Dalpiaz, A.; Cordes, C.; Horn, H.; Hempel, D.C.C. Selection of Reference Genes for Normalisation of Specific Gene Quantification Data of Aspergillus niger. J. Biotechnol. 2007, 132, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Vera, M.; Pani, B.; Griffiths, L.A.; Muchardt, C.; Abbott, C.M.; Singer, R.H.; Nudler, E. The Translation Elongation Factor EEF1A1 Couples Transcription to Translation during Heat Shock Response. eLife 2014, 3, e03164. [Google Scholar] [CrossRef]
- Findeisen, P.; Mühlhausen, S.; Dempewolf, S.; Hertzog, J.; Zietlow, A.; Carlomagno, T.; Kollmar, M. Six Subgroups and Extensive Recent Duplications Characterize the Evolution of the Eukaryotic Tubulin Protein Family. Genome Biol. Evol. 2014, 6, 2274–2288. [Google Scholar] [CrossRef] [Green Version]
- Lim, A.; Bolin, S. Evaluation of Reference Genes for Differential Gene Expression Study of Bovine Tuberculosis. Adv. Zool. Bot. 2017, 5, 17–24. [Google Scholar] [CrossRef]
- Kreth, S.; Heyn, J.; Grau, S.; Kretzschmar, H.A.; Egensperger, R.; Kreth, F.W. Identification of Valid Endogenous Control Genes for Determining Gene Expression in Human Glioma. Neuro. Oncol. 2010, 12, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Cappelli, K.; Felicetti, M.; Capomaccio, S.; Spinsanti, G.; Silvestrelli, M.; Supplizi, A.V. Exercise Induced Stress in Horses: Selection of the Most Stable Reference Genes for Quantitative RT-PCR Normalization. BMC Mol. Biol. 2008, 9, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledderose, C.; Heyn, J.; Limbeck, E.; Kreth, S. Selection of Reliable Reference Genes for Quantitative Real-Time PCR in Human T Cells and Neutrophils. BMC Res. Notes 2011, 4, 427. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.Q.; Skinner, J.; Bennett, J.E. Evaluation of Reference Genes for Real-Time Quantitative PCR Studies in Candida glabrata Following Azole Treatment. BMC Mol. Biol. 2012, 13, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Wu, X.; Guo, X.; Bao, P.; Ding, X.; Chu, M.; Liang, C.; Yan, P. Identification of Optimal Reference Genes for Examination of Gene Expression in Different Tissues of Fetal Yaks. Czech J. Anim. Sci. 2017, 62, 426–434. [Google Scholar] [CrossRef] [Green Version]
- Kozera, B.; Rapacz, M. Reference Genes in Real-Time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Chandra, S.; Sharma, M. Difference in Keratinase Activity of Dermatophytes at Different Environmental Conditions Is an Attribute of Adaptation to Parasitism. Mycoses 2012, 55, 410–415. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Chi, W.; Shi, Y.; Chen, S.; Lin, D.; Jin, Y. Metalloprotease Genes of Trichophyton mentagrophytes Are Important for Pathogenicity. Med. Mycol. 2014, 52, 36–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peres, N.T.; Sanches, P.R.; Falco, J.P.; Silveira, H.C.; Paião, F.G.; Maranhão, F.C.; Gras, D.E.; Segato, F.; Cazzaniga, R.A.; Mazucato, M.; et al. Transcriptional Profiling Reveals the Expression of Novel Genes in Response to Various Stimuli in the Human Dermatophyte Trichophyton rubrum. BMC Microbiol. 2010, 10, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Jonge, H.J.M.; Fehrmann, R.S.N.; de Bont, E.S.J.M.; Hofstra, R.M.W.; Gerbens, F.; Kamps, W.A.; de Vries, E.G.E.; van der Zee, A.G.J.; te Meerman, G.J.; ter Elst, A. Evidence Based Selection of Housekeeping Genes. PLoS ONE 2007, 2, e898. [Google Scholar] [CrossRef] [Green Version]
| Gene | Name | Primers (5′-3′) | Length of Product (bp) | Tm (°C) | Ct Range |
|---|---|---|---|---|---|
| TUBB | β-tubulin | AAGAGTTCCCAGACCGTT TGTTGTACAAGGCCTCAT | 161 | 59.5 | 15.9–18.4 |
| ACTB | β-actin | TCCTGAGGCTCTCTTCC GTAGTACCGCCGGACAT | 144 | 60.0 | 16.3–18.8 |
| ADPRF | ADP ribosylation factor | TTCTCATGGTCGGTCTC CGTTGAATCCGATGGTG | 101 | 58.5 | 16.1–19.8 |
| RPL2 | ribosomal protein L2 | GATCTATATTCACGGCTC ATGATGTTCTTCACGACA | 111 | 60.5 | 16.4–19.3 |
| SDHA | succinate dehydrogenase complex flavoprotein subunit A | TCTAGGAAACATGCACAT TTCGATAACACTCTGAGT | 130 | 60.5 | 16.0–18.9 |
| EEF1A1 | elongation factor 1-α | CCTAAGTTCGTCAAGTCT CTTCTCGACAGCCTTGAT | 161 | 60.0 | 16.2–19.1 |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase | AACGGCTTCGGTCGTATTG TATTCGGCGTATTTGGTCTCA | 110 | 59.5 | 15.8–18.7 |
| Gene | Condition | geNorm | NormFinder | BestKeeper | RefFinder | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| M-Value | SV-Value | r-Index | CV (%Ct) | Ranking Value | ||||||
| TUBB | Sabouraud | 0.593 | 0.561 | 0.256 | 0.231 | 0.832 | 0.78 | 8.18 | 6.732 | 6.329 |
| MM+keratin | 0.489 | 0.203 | 0.721 | 6.021 | ||||||
| potato dextrose | 0.603 | 0.234 | 0.787 | 6.234 | ||||||
| SDHA | Sabouraud | 0.497 | 0.455 | 0.098 | 0.092 | 0.886 | 0.887 | 8.57 | 2.120 | 2.083 |
| MM+keratin | 0.413 | 0.087 | 0.854 | 2.013 | ||||||
| potato dextrose | 0.455 | 0.091 | 0.921 | 2.115 | ||||||
| EEF1A1 | Sabouraud | 0.765 | 0.746 | 0.403 | 0.36 | 0.907 | 0.906 | 8.47 | 7.234 | 7.112 |
| MM+keratin | 0.712 | 0.299 | 0.890 | 6.983 | ||||||
| potato dextrose | 0.761 | 0.378 | 0.921 | 7.120 | ||||||
| ACTB | Sabouraud | 0.595 | 0.581 | 0.377 | 0.349 | 0.968 | 0.934 | 8.54 | 6.321 | 6.128 |
| MM+keratin | 0.546 | 0.304 | 0.879 | 5.954 | ||||||
| potato dextrose | 0.601 | 0.367 | 0.954 | 6.110 | ||||||
| ADPRF | Sabouraud | 0.598 | 0.57 | 0.254 | 0.217 | 0.921 | 0.906 | 8.65 | 7.012 | 6.714 |
| MM+keratin | 0.499 | 0.176 | 0.854 | 6.121 | ||||||
| potato dextrose | 0.612 | 0.221 | 0.943 | 7.010 | ||||||
| RPL2 | Sabouraud | 0.621 | 0.57 | 0.205 | 0.178 | 0.896 | 0.867 | 8.97 | 6.755 | 6.476 |
| MM+keratin | 0.564 | 0.146 | 0.812 | 5.999 | ||||||
| potato dextrose | 0.605 | 0.185 | 0.901 | 6.675 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gnat, S.; Łagowski, D.; Nowakiewicz, A.; Trościańczyk, A.; Dyląg, M. New Reference Genes for qRT-PCR Analysis as a Potential Target for Identification of Trichophyton verrucosum in Different Culture Conditions. Pathogens 2021, 10, 1361. https://doi.org/10.3390/pathogens10111361
Gnat S, Łagowski D, Nowakiewicz A, Trościańczyk A, Dyląg M. New Reference Genes for qRT-PCR Analysis as a Potential Target for Identification of Trichophyton verrucosum in Different Culture Conditions. Pathogens. 2021; 10(11):1361. https://doi.org/10.3390/pathogens10111361
Chicago/Turabian StyleGnat, Sebastian, Dominik Łagowski, Aneta Nowakiewicz, Aleksandra Trościańczyk, and Mariusz Dyląg. 2021. "New Reference Genes for qRT-PCR Analysis as a Potential Target for Identification of Trichophyton verrucosum in Different Culture Conditions" Pathogens 10, no. 11: 1361. https://doi.org/10.3390/pathogens10111361
APA StyleGnat, S., Łagowski, D., Nowakiewicz, A., Trościańczyk, A., & Dyląg, M. (2021). New Reference Genes for qRT-PCR Analysis as a Potential Target for Identification of Trichophyton verrucosum in Different Culture Conditions. Pathogens, 10(11), 1361. https://doi.org/10.3390/pathogens10111361