Relationship between Saliva and Sublingual Immunotherapy
Abstract
:1. Introduction
2. Biomarkers for Predicting the Efficacy of SLIT
3. Biomarkers for Monitoring the Efficacy of SLIT
4. Effects of Salivary pH on Immunity and SLIT
5. Salivary Microbiome and Allergic Diseases
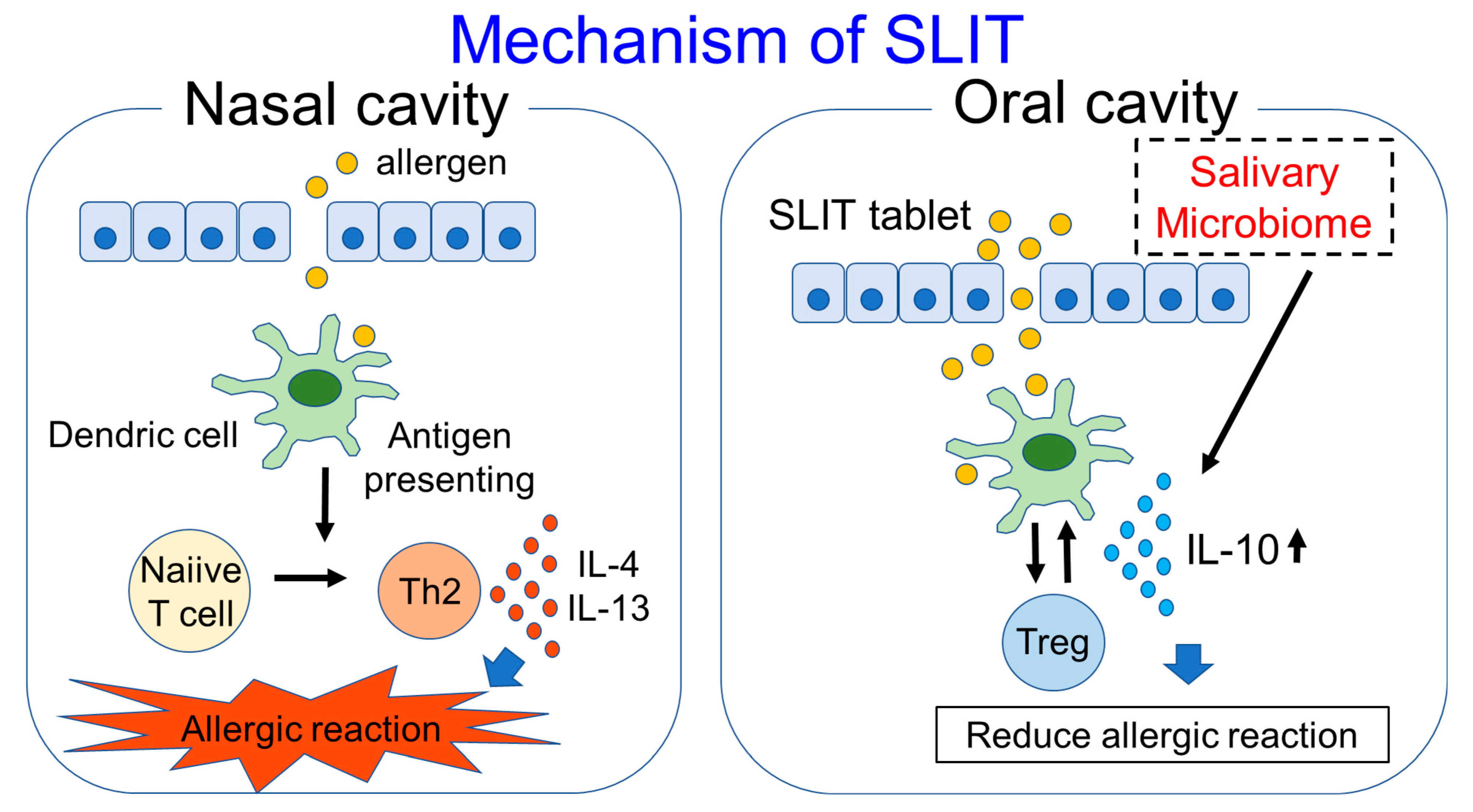
6. Salivary Microbiome and SLIT
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Roberts, G.; Pfaar, O.; Akdis, C.A.; Ansotegui, I.J.; Durham, S.R.; Halken, S.; Larenas-Linnemann, D.; Pawankar, R.; Pitsios, C.; Sheikh, A.; et al. EAACI Guidelines on Allergen Immunotherapy: Allergic rhinoconjunctivitis. Allergy 2018, 73, 765–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhami, S.; Kakourou, A.; Asamoah, F.; Agache, I.; Lau, S.; Jutel, M.; Muraro, A.; Roberts, G.; Akdis, C.A.; Bonini, M.; et al. Allergen immunotherapy for allergic asthma: A systematic review and meta-analysis. Allergy 2017, 72, 1825–1848. [Google Scholar] [CrossRef] [PubMed]
- Kucuksezer, U.C.; Ozdemir, C.; Cevhertas, L.; Ogulur, I.; Akdis, M.; Akdis, C.A. Mechanisms of allergen-specific immu-notherapy and allergen tolerance. Allergol. Int. 2020, 69, 549560. [Google Scholar] [CrossRef] [PubMed]
- Dhami, S.; Nurmatov, U.; Arasi, S.; Khan, T.; Asaria, M.; Zaman, H.; Agarwal, A.; Netuveli, G.; Roberts, G.; Pfaar, O.; et al. Allergen immunotherapy for allergic rhinoconjunctivitis: A systematic review and meta-analysis. Allergy 2017, 72, 1597–1631. [Google Scholar] [CrossRef] [Green Version]
- Lundmark, A.; Hu, Y.O.O.; Huss, M.; Johannsen, G.; Andersson, A.F.; Yucel-Lindberg, T. Identification of Salivary Mi-crobiota and Its Association with Host Inflammatory Mediators in Periodontitis. Front. Cell. Infect. Microbiol. 2019, 9, 216. [Google Scholar]
- Qi, Y.; Zang, S.-Q.; Wei, J.; Yu, H.-C.; Yang, Z.; Wu, H.-M.; Kang, Y.; Tao, H.; Yang, M.-F.; Jin, L.; et al. High-throughput sequencing provides insights into oral microbiota dysbiosis in association with inflammatory bowel disease. Genomics 2021, 113, 664–676. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Verma, M.; Panda, M. Role of Oral Microbiome Signatures in Diagnosis and Prognosis of Oral Cancer. Technol. Cancer Res. Treat. 2019, 18. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Zeng, Q.; Luo, R. Predictors for Short-Term Efficacy of Allergen-Specific Sublingual Immunotherapy in Children with Allergic Rhinitis. Mediat. Inflamm. 2020, 2020, 1847061. [Google Scholar] [CrossRef] [Green Version]
- Fujimura, T.; Yonekura, S.; Horiguchi, S.; Taniguchi, Y.; Saito, A.; Yasueda, H.; Inamine, A.; Nakayama, T.; Takemori, T.; Taniguchi, M.; et al. Increase of regulatory T cells and the ratio of specific IgE to total IgE are can-didates for response monitoring or prognostic biomarkers in 2-year sublingual immunotherapy (SLIT) for Japanese cedar pollinosis. Clin. Immunol. 2011, 139, 65–74. [Google Scholar] [CrossRef]
- Yonekura, S.; Okamoto, Y.; Sakurai, D.; Okubo, K.; Gotoh, M.; Kaneko, S.; Konno, A. An analysis of factors related to the effect of sublingual immunotherapy on Japanese cedar pollen induced allergic rhinitis. Allergol. Int. 2018, 67, 201–208. [Google Scholar] [CrossRef]
- Hoshino, M.; Akitsu, K.; Kubota, K.; Ohtawa, J. Association between biomarkers and house dust mite sublingual immu-notherapy in allergic asthma. Clin. Exp. Allergy 2020, 50, 1035–1043. [Google Scholar]
- Hoshino, M.; Akitsu, K.; Kubota, K.; Ohtawa, J. Serum Periostin as a Biomarker for Predicting Clinical Response to House Dust Mite Sublingual Immunotherapy in Allergic Rhinitis. J. Allergy Clin. Immunol. Pr. 2021, 9, 1864–1870. [Google Scholar] [CrossRef]
- Xie, S.; Jiang, S.; Zhang, H.; Wang, F.; Liu, Y.; She, Y.; Jing, Q.; Gao, K.; Fan, R.; Xie, S.; et al. Prediction of sublingual immunotherapy efficacy in allergic rhinitis by serum metabolomics analysis. Int. Immunopharmacol. 2021, 90, 107211. [Google Scholar] [CrossRef]
- Shamji, M.H.; Durham, S.R. Mechanisms of allergen immunotherapy for inhaled allergens and predictive biomarkers. J. Allergy Clin. Immunol. 2017, 140, 1485–1498. [Google Scholar] [CrossRef] [Green Version]
- Miranda, D.O.; Silva, D.A.; Fernandes, J.F.; Queirós, M.G.; Chiba, H.F.; Ynoue, L.H.; Resende, R.O.; Pena, J.D.; Sung, S.S.; Segundo, G.R.; et al. Serum and salivary IgE, IgA, and IgG4 antibodies to Dermatophagoides pteronyssinus and its major allergens, Der p1 and Der p2, in allergic and nonallergic children. Clin. Dev. Immunol. 2011, 2011, 302739. [Google Scholar] [CrossRef] [Green Version]
- Pereira, E.A.; Silva, D.A.; Cunha-Júnior, J.P.; Almeida, K.C.; Alves, R.; Sung, S.J.; Taketomi, E.A. IgE, IgG1, and IgG4 antibody responses to Blomia tropicalis in atopic patients. Allergy 2005, 60, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Oka, A.; Ninomiya, T.; Fujiwara, T.; Takao, S.; Sato, Y.; Gion, Y.; Minoura, A.; Haruna, S.-I.; Yoshida, N.; Sakuma, Y.; et al. Serum IgG4 as a biomarker reflecting pathophysiology and post-operative recurrence in chronic rhinosinusitis. Allergol. Int. 2020, 69, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Ladjemi, M.Z.; Gras, D.; Dupasquier, S.; Detry, B.; Lecocq, M.; Garulli, C.; Fregimilicka, C.; Bouzin, C.; Gohy, S.; Chanez, P.; et al. Bronchial Epithelial IgA Secretion Is Impaired in Asthma. Role of IL-4/IL-13. Am. J. Respir. Crit. Care Med. 2018, 197, 1396–1409. [Google Scholar] [CrossRef] [PubMed]
- Lúðvíksson, B.R.; Arason, G.J.; Thorarensen, O.; Árdal, B.; Valdimarsson, H. Allergic diseases and asthma in relation to serum immunoglobulins and salivary immunoglobulin A in pre-school children: A follow-up community-based study. Clin. Exp. Allergy 2005, 35, 64–69. [Google Scholar] [CrossRef]
- Smeekens, J.M.; Kulis, M.D. Evolution of Immune Responses in Food Immunotherapy. Immunol. Allergy Clin. North Am. 2020, 40, 87–95. [Google Scholar] [CrossRef]
- Kulis, M.; Saba, K.; Kim, E.H.; Bird, J.A.; Kamilaris, N.; Vickery, B.P.; Staats, H.; Burks, A.W. Increased peanut-specific IgA in saliva correlates with food challenge outcomes following peanut sublingual immunotherapy. J. Allergy Clin. Immunol. 2012, 129, 1159–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huoman, J.; Papapavlou, G.; Pap, A.; Alm, J.; Nilsson, L.J.; Jenmalm, M.C. Sublingual immunotherapy alters salivary IgA and systemic immune mediators in timothy allergic children. Pediatr. Allergy Immunol. 2019, 30, 522–530. [Google Scholar] [CrossRef]
- James, L.K.; Till, S.J. Potential Mechanisms for IgG4 Inhibition of Immediate Hypersensitivity Reactions. Curr. Allergy Asthma Rep. 2016, 16, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, A.; James, L.; Bahnson, H.T.; Shamji, M.H.; Couto-Francisco, N.C.; Islam, S.; Houghton, S.; Clark, A.T.; Stephens, A.; Turcanu, V.; et al. IgG4 inhibits peanut-induced basophil and mast cell activation in peanut-tolerant children sensitized to peanut major allergens. J. Allergy Clin. Immunol. 2015, 135, 1249–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Neut Kolfschoten, M.; Schuurman, J.; Losen, M.; Bleeker, W.K.; Martínez-Martínez, P.; Vermeulen, E.; den Bleker, T.H.; Wiegman, L.; Vink, T.; Aarden, L.A.; et al. Anti-inflammatory activity of human IgG4 antibodies by dynamic Fab arm exchange. Science 2007, 317, 1554–1557. [Google Scholar] [CrossRef] [Green Version]
- Lycke, N.; Eriksen, L.; Holmgren, J. Protection against cholera toxin after oral immunisation is thymus dependent and associated with intestinal production of neutralising IgA antitoxin. Scand. J. Immunol. 1987, 25, 413–419. [Google Scholar] [CrossRef]
- Kadaoui, K.A.; Corthésy, B. Secretory IgA Mediates Bacterial Translocation to Dendritic Cells in Mouse Peyer’s Patches with Restriction to Mucosal Compartment. J. Immunol. 2007, 179, 7751–7757. [Google Scholar] [CrossRef] [Green Version]
- Varadhachary, A.; Chatterjee, D.; Garza, J.; Garr, R.P.; Foley, C.; Letkeman, A.F.; Dean, J.; Haug, D. Salivary anti-SARS-CoV-2 IgA as an accessible biomarker of mucosal immunity against COVID-19. medRxivPreprint 2020, arXiv:2020.08.07.20170258. [Google Scholar]
- Kotani, Y.; Kunisawa, J.; Suzuki, Y.; Sato, I.; Saito, T.; Toba, M.; Kohda, N.; Kiyono, H. Role of Lactobacillus pentosus Strain b240 and the Toll-Like Receptor 2 Axis in Peyer’s Patch Dendritic Cell-Mediated Immunoglobulin a Enhancement. PLoS ONE 2014, 9, e91857. [Google Scholar] [CrossRef]
- Kawashima, T.; Ikari, N.; Kouchi, T.; Kowatari, Y.; Kubota, Y.; Shimojo, N.; Tsuji, N.M. The molecular mechanism for activating IgA production by Pediococcus acidilactici K15 and the clinical impact in a randomized trial. Sci. Rep. 2018, 8, 5065. [Google Scholar] [CrossRef]
- Nakajima, S.; Gillespie, D.N.; Gleich, G.J. Differences between IgA and IgE as secretory proteins. Clin. Exp. Immunol. 1975, 21, 306–317. [Google Scholar]
- Ebo, D.G.; Bridts, C.H.; Mertens, C.H.; Hagendorens, M.M.; Stevens, W.J.; De Clerck, L.S. Analyzing histamine release by flow cytometry (HistaFlow): A novel instrument to study the degranulation patterns of basophils. J. Immunol. Methods 2012, 375, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Nullens, S.; Sabato, V.; Faber, M.; Leysen, J.; Bridts, C.H.; De Clerck, L.S.; Falcone, F.H.; Maurer, M.; Ebo, D.G. Basophilic histamine content and release during venom immunotherapy: Insights by flow cytometry. Cytom. Part B Clin. Cytom. 2013, 84, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Foglio-Bonda, P.L.; Migliario, M.; Rocchetti, V.; Pattarino, F.; Foglio-Bonda, A. Daily and annually variation of unstimu-lated whole saliva flow rate and pH and their relation with body profile in healthy young adults. Eur. Rev. Med. Pharmacol. Sci. 2013, 17, 2538–2545. [Google Scholar] [PubMed]
- Takahashi, N. Oral Microbiome Metabolism: From “Who Are They?” to “What Are They Doing?” . J. Dent. Res. 2015, 94, 1628–1637. [Google Scholar]
- Abranches, J.; Zeng, L.; Kajfasz, J.K.; Palmer, S.R.; Chakraborty, B.; Wen, Z.T.; Richards, V.P.; Brady, L.J.; Lemos, J. Biology of Oral Streptococci. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, K.; Deng, M.; Exterkate, R.A.M.; Liu, C.; Zhou, X.; Cheng, L.; Cate, J.M.T. Effect of arginine on the growth and biofilm formation of oral bacteria. Arch. Oral Biol. 2017, 82, 256–262. [Google Scholar] [CrossRef]
- Nascimento, M.M. Potential Uses of Arginine in Dentistry. Adv. Dent. Res. 2018, 29, 98–103. [Google Scholar] [CrossRef] [Green Version]
- Bikandi, J.; Moragues, M.D.; Quindós, G.; Polonelli, L.; Pontón, J. Influence of environmental pH on the reactivity of Candida albicans with salivary IgA. J. Dent. Res. 2000, 79, 1439–1442. [Google Scholar] [CrossRef]
- Motamayel, F.A.; Amjad, S.V.; Goodarzi, M.T.; Poorolajal, J. Evaluation of Salivary Uric Acid and pH in Human Immunodeficiency Virus Infected Patients: A Historical Cohort Study. Infect. Disord. Drug Targets 2018, 18, 35–40. [Google Scholar] [CrossRef]
- Koppelman, S.J.; Smits, M.; Tomassen, M.; de Jong, G.A.H.; Baumert, J.; Taylor, S.L.; Witkamp, R.; Veldman, R.J.; Pieters, R.; Wichers, H. Release of Major Peanut Allergens from Their Matrix under Various pH and Simulated Saliva Condi-tions-Ara h2 and Ara h6 Are Readily Bio-Accessible. Nutrients 2018, 10, 1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sathe, S.K.; Venkatachalam, M.; Sharma, G.M.; Kshirsagar, H.H.; Teuber, S.S.; Roux, K.H. Solubilization and elec-trophoretic characterization of select edible nut seed proteins. J. Agric. Food Chem. 2009, 57, 7846–7856. [Google Scholar] [CrossRef] [PubMed]
- Walczyk, N.E.; Smith, P.M.C.; Tovey, E.R.; Roberts, T.H. Peanut protein extraction conditions strongly influence yield of allergens Ara h1and 2 and sensitivity of immunoassays. Food Chem. 2017, 221, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, Y.; Takeshita, T. The oral microbiome and human health. J. Oral Sci. 2017, 59, 201–206. [Google Scholar] [CrossRef] [Green Version]
- De Luca, F.; Shoenfeld, Y. The microbiome in autoimmune diseases. Clin. Exp. Immunol. 2019, 195, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Said, H.S.; Suda, W.; Nakagome, S.; Chinen, H.; Oshima, K.; Kim, S.; Kimura, R.; Iraha, A.; Ishida, H.; Fujita, J.; et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its associ-ation with oral immunological biomarkers. DNA Res. 2014, 21, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Bellando-Randone, S.; Russo, E.; Venerito, V.; Matucci-Cerinic, M.; Iannone, F.; Tangaro, S.; Amedei, A. Exploring the Oral Microbiome in Rheumatic Diseases, State of Art and Future Prospective in Personalized Medicine with an AI Approach. J. Pers. Med. 2021, 11, 625. [Google Scholar] [CrossRef]
- Dzidic, M.; Abrahamsson, T.R.; Artacho, A.; Collado, M.C.; Mira, A.; Jenmalm, M.C. Oral microbiota maturation during the first 7 years of life in relation to allergy development. Allergy 2018, 73, 2000–2011. [Google Scholar] [CrossRef] [Green Version]
- Arweiler, N.B.; Rahmel, V.; Alhamwe, B.A.; Alhamdan, F.; Zemlin, M.; Boutin, S.; Dalpke, A.; Renz, H. Dental Biofilm and Saliva Microbiome and Its Interplay with Pediatric Allergies. Microorganisms 2021, 9, 1330. [Google Scholar] [CrossRef]
- Carrasco Pro, S.; Lindestam Arlehamn, C.S.; Dhanda, S.K.; Carpenter, C.; Lindvall, M.; Faruqi, A.A.; Santee, C.A.; Renz, H.; Renz, H.; Sette, A. Microbiota epitope similarity either dampens or enhances the immunogenicity of disease-associated antigenic epitopes. PLoS ONE 2018, 13, e0196551. [Google Scholar] [CrossRef] [Green Version]
- Macia, L.; Nanan, R.; Hosseini-Beheshti, E.; Grau, G.E. Host- and Microbiota-Derived Extracellular Vesicles, Immune Function, and Disease Development. Int. J. Mol. Sci. 2019, 21, 107. [Google Scholar] [CrossRef] [Green Version]
- Liaskos, M.; Ferrero, R.L. Immune modulation by bacterial outer membrane vesicles. Nat. Rev. Immunol. 2015, 15, 375–387. [Google Scholar] [CrossRef]
- Pérez-Cruz, C.; Delgado, L.; López-Iglesias, C.; Mercade, E. Outer-Inner Membrane Vesicles Naturally Secreted by Gram-Negative Pathogenic Bacteria. PLoS ONE 2015, 10, e0116896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cecil, J.D.; O’Brien-Simpson, N.M.; Lenzo, J.C.; Holden, J.A.; Singleton, W.; Gonzalez, A.P.; Mansell, A.; Reynolds, E.C. Outer Membrane Vesicles Prime and Activate Macrophage Inflammasomes and Cytokine Secretion In Vitro and In Vivo. Front. Immunol. 2017, 8, 1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haruna, T.; Kariya, S.; Fujiwara, T.; Yuta, A.; Higaki, T.; Zhao, P.; Ogawa, Y.; Kanai, K.; Hirata, Y.; Oka, A.; et al. Role of whole saliva in the efficacy of sublingual immunotherapy in seasonal allergic rhinitis. Allergol. Int. 2019, 68, 82–89. [Google Scholar] [CrossRef]
- Oka, A.; Kidoguchi, M.; Kariya, S.; Fujiwara, T.; Yuta, A.; Miyashita, H.; Higaki, T.; Ogawa, Y.; Kanai, K.; Makihara, S.; et al. Role of salivary microbiome in IL-10 production and efficacy of sublingual immunotherapy. Allergy 2021, 76, 2617–2620. [Google Scholar] [CrossRef] [PubMed]
- Klimek, L.; Bergmann, K.C.; Biedermann, T.; Bousquet, J.; Hellings, P.; Jung, K.; Merk, H.; Olze, H.; Schlenter, W.; Stock, P.; et al. Visual analogue scales (VAS): Measuring instruments for the documentation of symptoms and therapy monitoring in cases of allergic rhinitis in everyday health care. Allergo J. Int. 2017, 26, 16–24. [Google Scholar] [CrossRef] [Green Version]
- Hisbergues, M.; Magi, M.; Rigaux, P.; Steuve, J.; Garcia, L.; Goudercourt, D.; Pot, B.; Pestel, J.; Jacquet, A. In vivo and in vitro immunomodulation of Der p 1 allergen-specific response by Lactobacillus plantarum bacteria. Clin. Exp. Allergy 2007, 37, 1286–1295. [Google Scholar] [CrossRef] [PubMed]
- Van Overtvelt, L.; Moussu, H.; Horiot, S.; Samson, S.; Lombardi, V.; Mascarell, L.; van de Moer, A.; Bourdet-Sicard, R.; Moingeon, P. Lactic acid bacteria as adjuvants for sublingual allergy vaccines. Vaccine 2010, 28, 2986–2992. [Google Scholar] [CrossRef]
- Moussu, H.; Van Overtvelt, L.; Horiot, S.; Tourdot, S.; Airouche, S.; Zuercher, A.; Holvoet, S.; Prioult, G.; Nutten, S.; Mercenier, A.; et al. Bifidobacterium bifidum NCC453 promotes tolerance induction in murine models of sublingual immunotherapy. Int. Arch. Allergy Immunol. 2012, 158, 35–42. [Google Scholar] [CrossRef]
- Brisbin, J.T.; Gong, J.; Orouji, S.; Esufali, J.; Mallick, A.I.; Parvizi, P.; Shewen, P.E.; Sharif, S. Oral treatment of chickens with lactobacilli influences elicitation of immune responses. Clin. Vaccine Immunol. 2011, 18, 1447–1455. [Google Scholar] [CrossRef] [PubMed]
- Bender, M.E.; Read, T.D.; Edwards, T.S.; Hargita, M.; Cutler, A.J.; Wissel, E.F.; Wise, S.K. A Comparison of the Bacterial Nasal Microbiome in Allergic Rhinitis Patients Before and After Immunotherapy. Laryngoscope 2020, 130, E882–E888. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oka, A.; Okano, M. Relationship between Saliva and Sublingual Immunotherapy. Pathogens 2021, 10, 1358. https://doi.org/10.3390/pathogens10111358
Oka A, Okano M. Relationship between Saliva and Sublingual Immunotherapy. Pathogens. 2021; 10(11):1358. https://doi.org/10.3390/pathogens10111358
Chicago/Turabian StyleOka, Aiko, and Mitsuhiro Okano. 2021. "Relationship between Saliva and Sublingual Immunotherapy" Pathogens 10, no. 11: 1358. https://doi.org/10.3390/pathogens10111358
APA StyleOka, A., & Okano, M. (2021). Relationship between Saliva and Sublingual Immunotherapy. Pathogens, 10(11), 1358. https://doi.org/10.3390/pathogens10111358




