Revisiting Persistent Salmonella Infection and the Carrier State: What Do We Know?
Abstract
:1. Introduction
2. What Is the Public Health and Economic Significance of These Diseases?
3. Are There Similarities and Trends in the Clinical Pictures in Acute Infections Produced by These Serovars?
3.1. S. Typhi, S. Gallinarum and S. Pullorum
3.2. S. Typhimurium
3.3. S. Dublin and S. Abortusovis
4. How Is Short- and Long-Term Carriage/Persistence Manifested?
4.1. S. Typhi
4.2. S. Gallinarum and S. Pullorum
4.3. S. Dublin
4.4. S. Abortusovis
4.5. S. Typhimurium
5. What Is/Are the Main Site(s) of Carriage and Dissemination?
6. Is There Anything Unique to These Serovars That Predisposes Them to Persistence?
7. Is There a Clear Host Genetic Element That Contributes to the Development of the Carrier State?
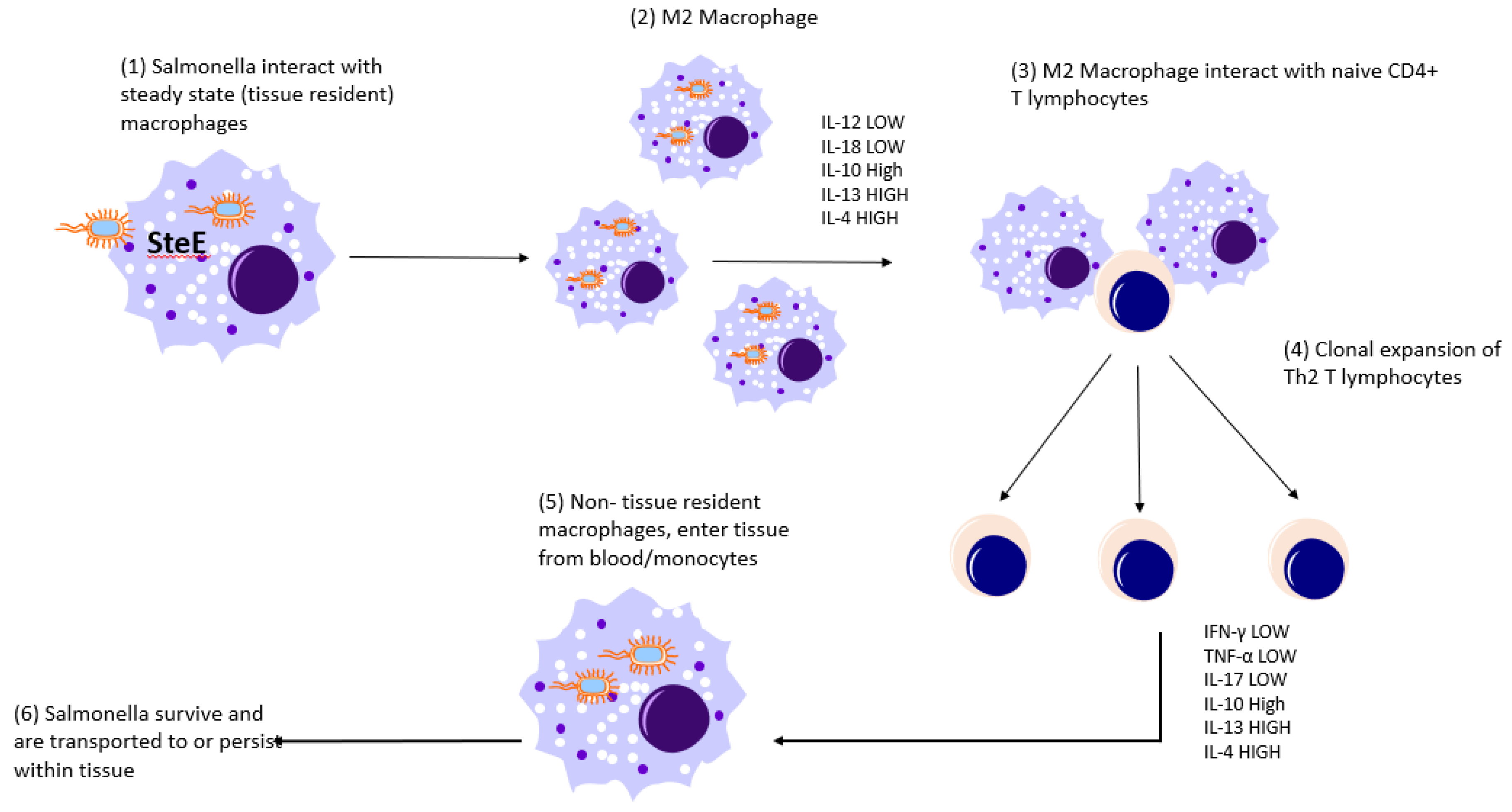
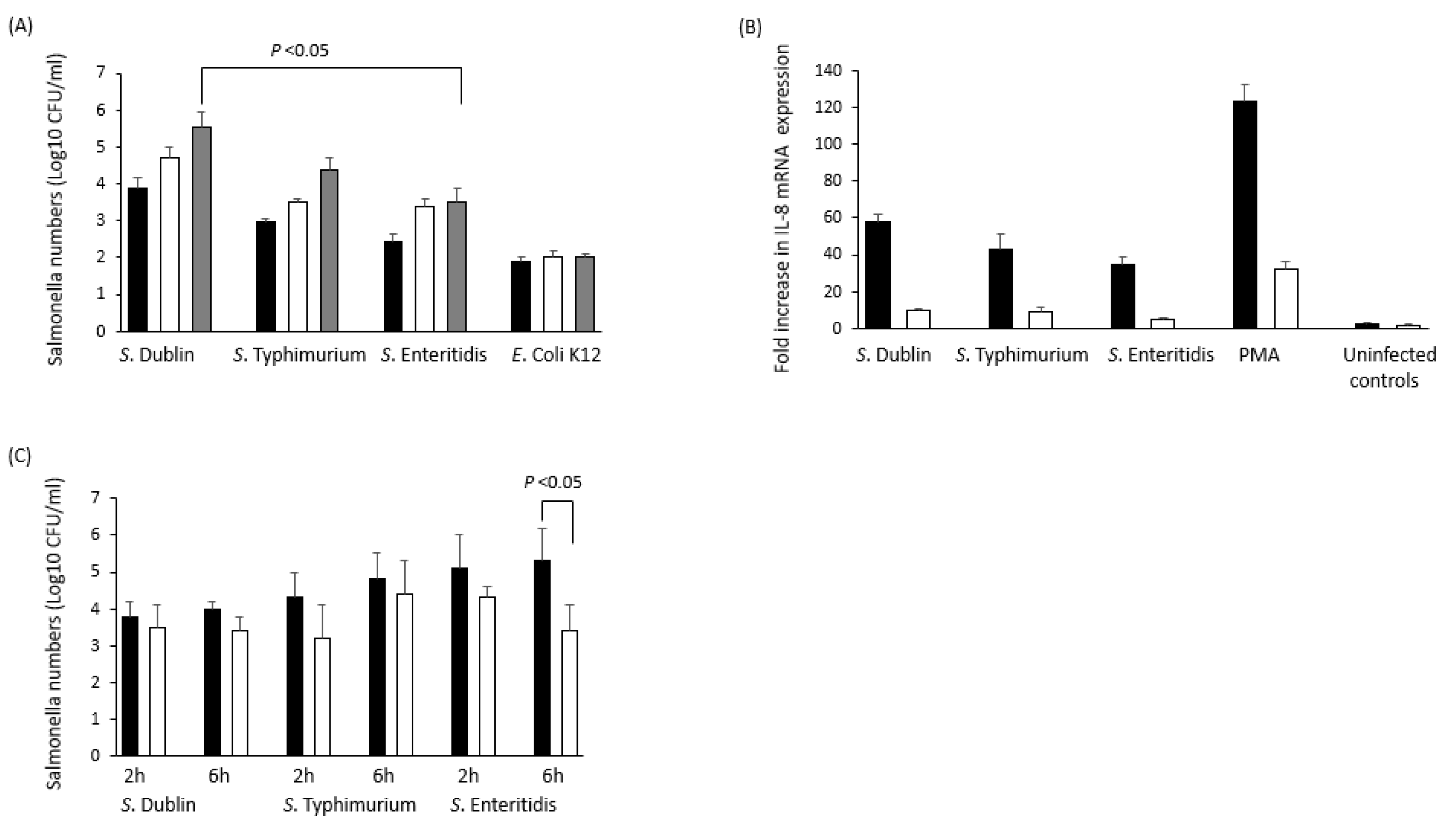
8. Is There Anything Characteristic to the Immune Response during Persistent Infections?
8.1. S. Typhimurium
8.2. S. Gallinarum and Pullorum
9. How Does This Information Apply to S. Typhi and the Remaining Typhoid Serovars?
9.1. S. Typhi
9.2. S. Dublin
10. Can Anything Be Done to Reduce the Impact of Persistent Infection by Remodulating the Immune Response?
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Coburn, B.; Grassl, G.A.; Finlay, B.B. Salmonella, the host and disease: A brief review. Immunol. Cell Biol. 2007, 85, 112–118. [Google Scholar] [CrossRef]
- Hornick, R.B.; Greisman, S.E.; Woodward, T.E.; DuPont, H.L.; Dawkins, A.T.; Snyder, M.J. Typhoid fever: Pathogenesis and immunologic control. N. Engl. J. Med. 1970, 283, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Rabsch, W.; Tschape, H.; Bäumler, A.J. Non-typhoidal salmonellosis: Emerging problems. Microbes Infect. 2001, 3, 237–247. [Google Scholar] [CrossRef]
- Monack, D.M.; Bouley, D.M.; Falkow, S. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNγ neutralization. J. Exp. Med. 2004, 1999, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Monack, D.M.; Mueller, A.; Falkow, S. Persistent bacterial infections: The interface of the pathogen and the host immune system. Nat. Rev. Microbiol. 2004, 2, 747–765. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.S.; Miles, A.A.; Topley, W.C.C. Topley and Wilson’s Principles of Bacteriology and Immunity, 5th ed.; Edward Arnold: London, UK, 1964. [Google Scholar]
- Uzzau, S.; Brown, D.J.; Wallis, T.; Rubino, S.; Leori, G.; Bernard, S.; Casadeus, J.; Platt, D.J.; Olsen, J.E. Host-adapted serotypes of Salmonella enterics. Epidemiol. Infect. 2000, 125, 229–255. [Google Scholar] [CrossRef]
- Barrow, P.A.; Methner, U. (Eds.) Salmonella in Domestic Animals, 2nd ed.; CABI Press: Wallingford, UK, 2013. [Google Scholar]
- Pang, T.; Levine, M.M.; Ivanoff, B.; Wain, J.; Finlay, B.B. Typhoid fever: Important issues still remain. Trends Microbiol. 1998, 6, 131–133. [Google Scholar] [CrossRef]
- Woc-Colburn, L.; Bobak, D.A. The expanding spectrum of disease due to Salmonella: An international perspective. Curr. Infect. Dis. Rep. 2009, 11, 120–124. [Google Scholar] [CrossRef]
- Miller, S.I.; Hohmann, E.L.; Pegues, D.A. Salmonella (including Salmonella Typhi). In Principles and Practice of Infectious Diseases; Mandell, G.L., Bennet, J.R., Dolin, R., Eds.; Livingstone: New York, NY, USA, 1994; pp. 2013–2033. [Google Scholar]
- Karkey, A.; Arjyal, A.; Anders, K.L.; Boni, M.F.; Dongol, S.; Koirala, S.; My, P.V.T.; Nga, T.V.T.; Clements, A.C.A.; Holt, K.E.; et al. The burden and characteristics of enteric fever at a healthcare facility in a densely populated area of kathmandu. PLoS ONE 2010, 5, e13988. [Google Scholar] [CrossRef] [Green Version]
- Abera, B.; Yitayew, G.; Amare, H. Salmonella serotype Typhi, Shigella, and intestinal parasites among food handlers at Bahir Dar University, Ethiopia. J. Infect. Dev. Ctries. 2016, 10, 121–126. [Google Scholar] [CrossRef] [Green Version]
- Lovane, L.; Martínez, M.J.; Massora, S.; Mandomando, I.; Ussene, E.; Jordao, D.; Castillo, P.; Ismail, M.R.; Lorenzoni, C.; Carrilho, C.; et al. Carriage prevalence of Salmonella enterica serotype Typhi in gallbladders of adult autopsy cases from Mozambique. J. Infect. Dev. Ctries. 2016, 10, 410–412. [Google Scholar] [CrossRef] [Green Version]
- Murdoch, D.A.; Banatvaia, N.; Bone, A.; Shoismatulloev, B.I.; Ward, L.R.; Threlfall, E.J. Epidemic ciprofloxacin-resistant Salmonella typhi in Tajikistan. Lancet 1998, 31, 339–351. [Google Scholar]
- Kariuki, S.; Revathi, G.; Kiiru, J.; Mengo, D.M.; Mwituria, J.; Muyodi, J.; Munyalo, A.; Teo, Y.Y.; Holt, K.E.; Kingsley, R.A.; et al. Typhoid in Kenya is associated with a dominant multidrug-resistant Salmonella enterica serovar Typhi haplotype that is also widespread in Southeast Asia. J. Clin. Microbiol. 2010, 48, 2171–2176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrasekaran, B.; Balakrishnan, S. Screening, phylogenetic analysis and antibiotic sensitivity pattern of Salmonella enterica serovar Typhi isolates from typhoid asymptomatic carriers. Asian Pac. J. Trop. Med. 2011, 4, 769–772. [Google Scholar] [PubMed] [Green Version]
- House, J.K.; Smith, B.P.; Kamiya, D. Serological distinction of bovine Salmonella carriers from vaccinated and acutely infected cows. J. Vet. Diagn. Investig. 2001, 13, 483–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, L.R.; Schukken, Y.H.; Gröhn, Y.T.; Ersbøll, A.K. Salmonella Dublin infection in dairy cattle: Risk factors for becoming a carrier. Prev. Vet. Med. 2004, 65, 47–62. [Google Scholar] [CrossRef]
- Vaessen, M.A.; Veling, J.; Frankena, K.; Graat, E.A.; Klunder, T. Risk factors for Salmonella dublin infection on dairy farms. Vet. Q. 1998, 20, 97–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrow, P.A.; Neto, O.C. Pullorum disease and fowl typhoid—New thoughts on old diseases: A review. Avian Pathol. 2011, 40, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Shivaprasad, H.L.; Methner, U.; Barrow, P.A. Salmonella infections in domestic fowl. In Salmonella in Domestic Animals, 2nd ed.; Barrow, P.A., Methner, U., Eds.; CABI Press: Wallingford, UK, 2013. [Google Scholar]
- Jack, K.J. Salmonella Abortusovis: An atypical Salmonella. Vet. Rec. 1968, 82, 1168–1174. [Google Scholar]
- Uzzau, S. Salmonella infections in sheep. In Salmonella in Domestic Animals; Barrow, P., Methner, U., Eds.; CABI Press: Wallingford, UK, 2013. [Google Scholar]
- Pardon, P.; Sanchis, R.; Marly, J.; Lantier, F.; Pepin, M.; Popoff, M.Y. Salmonellose ovine due to Salmonella Abortusovis. Ann. Rech. Vet. 1988, 19, 221–235. [Google Scholar]
- Sojka, W.J.; Wray, C.; Schreeve, J.E.; Bell, J.C. The incidence of Salmonella infection in sheep in England and Wales, 1975 to 1981. Br. Vet. J. 1983, 139, 386–392. [Google Scholar] [CrossRef]
- Smith, H.W. Observations on experimental fowl typhoid. J. Comp. Pathol. 1955, 65, 37–54. [Google Scholar] [CrossRef]
- House, D.; Bishop, A.; Parry, C.; Dougan, G.; Wain, J. Typhoid fever: Pathogenesis and disease. Curr. Opin. Infect. Dis. 2001, 14, 573–578. [Google Scholar] [CrossRef]
- Jepson, M.A.; Clark, M.A. The role of M cells in Salmonella infection. Microbes Infect. 2001, 3, 1183–1190. [Google Scholar] [CrossRef]
- Barrow, P.A.; Lovell, M.A.; Stocker, B.A.D. Protection against experimental fowl typhoid by parenteral administration of live SL5928, an aroA-serC (aromatic dependent) mutant of a wild-type Salmonella Gallinarum strain made lysogenic for P22 sie. Avian Pathol. 2000, 29, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Wain, J.; Diep, T.S.; Ho, V.A.; Walsh, A.M.; Nguyen, T.T.; Parry, C.M.; White, N.J. Quantitation of bacteria in blood of typhoid fever patients and relationship between counts and clinical features, transmissibility, and antibiotic resistance. J. Clin. Microbiol. 1998, 36, 1683–1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vladoianu, I.R.; Chang, H.R.; Pechere, J.C. Expression of host resistance to Salmonella typhi and Salmonella typhimurium: Bacterial survival within macrophages of murine and human origin. Microb. Pathog. 1990, 8, 83–90. [Google Scholar] [CrossRef]
- van Basten, J.P.; Stockenbrugger, R. Typhoid perforation: A review of the literature since 1960. Trop. Geogr. Med. 1994, 46, 336–339. [Google Scholar] [PubMed]
- Azad, A.K.; Islam, R.; Salam, M.A.; Alam, A.N.; Islam, M.; Butler, T. Comparison of clinical features and pathologic findings in fatal cases of typhoid fever during the initial and later stages of the disease. Am. J. Trop. Med. Hyg. 1997, 56, 490–493. [Google Scholar] [CrossRef]
- Mukawi, T.J. Histopathological study of typhoid perforation of the small intestines. Southeast. Asian J. Trop. Med. Public Health 1978, 9, 252–255. [Google Scholar] [PubMed]
- Edelman, R.; Levine, M.M. Summary of an international workshop on typhoid fever. Rev. Infect. Dis. 1986, 8, 329–349. [Google Scholar] [CrossRef] [PubMed]
- Sansonetti, P. Phagocytosis of bacterial pathogens: Implications in the host response. Semin. Immunol. 2001, 13, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Torres, A.; Jones-Carson, J.; Bäumler, A.J.; Falkow, S.; Valdivia, R.; Brown, W.; Berggren, M.L.R.; Parks, W.T.; Fang, F.C. Extraintestinal dissemination of Salmonella via CD18-expressing phagocytes. Nature 1999, 401, 804–808. [Google Scholar] [CrossRef] [PubMed]
- Richter-Dahlfors, A.; Buchan, A.M.J.; Finlay, B.B. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med. 1997, 186, 569–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drecktrah, D.; Knodler, L.A.; Ireland, R.; Steele-Mortimer, O. The mechanism of Salmonella entry determines the vacuolar environment and intracellular gene expression. Traffic 2006, 7, 39–51. [Google Scholar] [CrossRef]
- Mackaness, G.B. Cellular resistance to infection. J. Exp. Med. 1962, 116, 381–406. [Google Scholar] [CrossRef]
- Vazquez-Torres, A.; Xu, Y.; Jones-Carson, J.; Holden, D.W.; Lucia, S.M.; Dinauer, M.C.; Mastroeni, P.; Fang, F.C. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 2000, 287, 1655–1658. [Google Scholar] [CrossRef]
- Helaine, S.; Thompson, J.A.; Watson, K.G.; Liu, M.; Boyle, C.; Holden, D.W. Dynamics of intracellular bacterial replication at the single cell level. Proc. Natl. Acad. Sci. USA 2010, 107, 3746–3751. [Google Scholar] [CrossRef] [Green Version]
- Fink, S.L.; Cookson, B.T. Pyroptosis and host cell death responses during Salmonella infection. Cell Microbiol. 2007, 9, 2562–2570. [Google Scholar] [CrossRef]
- Royle, M.C.; Totemeyer, S.; Alldridge, L.C.; Maskell, D.J.; Bryant, C.E. Stimulation of toll-like receptor 4 by lipopolysaccharide during cellular invasion by live Salmonella Typhimurium is a critical but not exclusive event leading to macrophage responses. J. Immunol. 2003, 170, 5445–5454. [Google Scholar] [CrossRef] [Green Version]
- Vazquez-Torres, A.; Jones-Carson, J.; Mastroeni, P.; Ischiropoulos, H.; Fang, F.C. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J. Exp. Med. 2000, 192, 227–236. [Google Scholar] [CrossRef] [Green Version]
- Mastroeni, P. Mechanisms of immunity to Salmonella infection. In Salmonella Infections: Clinical, Immunological, and Molecular Aspects; Mastroeni, P., Maskell, D., Eds.; Cambridge University Press: Cambridge, MA, USA, 2006; pp. 207–254. [Google Scholar]
- Dougan, G.; John, V.; Palmer, S.; Mastroeni, P. Immunity to salmonellosis. Immunol. Rev. 2011, 240, 196–210. [Google Scholar] [CrossRef] [PubMed]
- Hess, J.; Ladel, C.; Miko, D.; Kaufmann, S.H.E. Salmonella typhimurium aroA infection in gene-targeted immunodeficient mice: Major role of CD4 TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J. Immunol. 1996, 156, 3321–3326. [Google Scholar] [PubMed]
- Kupz, A.; Scott, T.A.; Belz, G.T.; Andrews, D.M.; Greyer, M.; Lew, A.M.; Brooks, A.G.; Smyth, M.J.; Curtiss, R.; Bedoui, S.; et al. Contribution of Thy1 NK cells to protective IFN-production during Salmonella typhimurium infections. Proc. Natl. Acad. Sci. USA 2013, 110, 2252–2257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Ragione, R.; Metcalfe, H.J.; Villarreal-Ramos, B.; Werling, D. Salmonella Infections in Cattle. In Salmonella in Domestic Animals, 2nd ed.; Barrow, P.A., Meth-ner, U., Eds.; CABI Press: Wallingford, UK, 2013. [Google Scholar]
- Nielsen, L.R. Review of pathogenesis and diagnostic methods of immediate relevance for epidemiology and control of Salmonella Dublin in cattle. Vet. Microbiol. 2013, 162, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pullinger, G.D.; Paulin, S.M.; Charleston, B.; Watson, P.R.; Bowen, A.J.; Dziva, F.; Morgan, E.; Villarreal-Ramos, B.; Wallis, T.S.; Stevens, M.P. Systemic translocation of Salmonella enterica serovar Dublin in cattle occurs predominantly via efferent lymphatics in a cell-free niche and requires type III secretion system 1 (T3SS-1) but not T3SS-2. Infect. Immun. 2007, 75, 5191–5199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinton, M. The diagnosis of Salmonella abortion in cattle with particular reference to Salmonella dublin. A review. J. Hyg. Camb. 1977, 79, 25–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osborne, A.D.; Pearson, H.; Linton, A.H.; Shimeld, C. Epidemiology of Salmonella infection in calves: The source of calfhood infection by Salmonella Dublin. Vet. Rec. 1977, 101, 513–516. [Google Scholar] [PubMed]
- Costa, R.A.; Casaux, M.L.; Caffarena, R.D.; Macıas-Rioseco, M.; Schild, C.O.; Fraga, M.; Riet-Correa, F.; Giannitti, F. Urocystitis and Ureteritis in Holstein Calves with septicaemia caused by Salmonella enterica serotype Dublin. J. Comp. Pathol. 2018, 164, 32–36. [Google Scholar] [CrossRef] [PubMed]
- House, J.K.; Smith, B.P.; Dilling, G.W.; Roden, L.D. Enzyme linked immunosorbent assay for serologic detection of Salmonella dublin carriers on a large dairy. Am. J. Vet. Res. 1993, 54, 1391–1399. [Google Scholar]
- Nielsen, L.R.; Borne, B.; Schaik, G. Salmonella Dublin infection in young dairy calves: Transmission parameters estimated from field data and an SIR-model. Prev. Vet. Med. 2007, 79, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Pardon, P.; Sanchis, R.; Marly, J.; Lantier, F.; Guilloteau, L.; Buzoni-Gatel, D.; Oswald, P.; Pepin, M.; Kaeffer, B.; Berthon, P.; et al. Experimental ovine salmonellosis (Salmonella abortusovis): Pathogenesis and vaccination. Res. Microbiol. 1990, 141, 945–953. [Google Scholar] [CrossRef]
- Sanchlis, R.; Pardon, P.; Abadie, G. Abortion and serological reaction of ewes after conjunctival instillation of Salmonella enterica subsp. enterica ser. abortusovis. Ann. Rech. Vet. 1991, 22, 59–64. [Google Scholar]
- Nikbaht, G.H.; Raffatellu, M.; Uzzau, S.; Tadjbakhsh, H.; Rubino, S. IS200 fingerprinting of Salmonella enterica serotype Abortusovis strains isolated in Iran. Epidemiol. Infect. 2002, 128, 333–336. [Google Scholar] [CrossRef]
- Young, D.; Hussell, T.; Dougan, G. Chronc bacterial infections: Living with unwanted guests. Nat. Immmunol. 2002, 3, 1026–1032. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.W.; Hamblen, A.D.; Smith, H.M. Typhoid carriers—A study of their disease producing potentialities over a series of years as indicated by a study of cases. Am. J. Public Health Nations Health 1936, 26, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Vogelsang, T.M.; Boe, J. Temporary and chronic carriers of Salmonella typhi and Salmonella paratyphi B. J. Hyg. 1948, 46, 252–261. [Google Scholar] [CrossRef] [Green Version]
- Shpargel, J.S.; Berardi, R.S.; Lenz, D. Salmonella typhi carrier state 52 years after illness with typhoid fever: A case study. J. Infect. Control 1985, 13, 122–123. [Google Scholar] [CrossRef]
- Parry, C.M.; Hien, T.T.; Dougan, G.; White, N.J.; Farrar, J.J. Typhoid fever. N. Engl. J. Med. 2002, 347, 1770–1782. [Google Scholar] [CrossRef] [Green Version]
- Levine, M.M.; Black, R.E.; Lanata, C. Precise estimation of the numbers of chronic carriers of Salmonella typhi in Santiago, Chile, an endemic area. J. Infect. Dis. 1982, 146, 724–726. [Google Scholar] [CrossRef]
- Mohan, U.; Mohan, V.; Raj, K. A study of carrier state of S. typhi, intestinal parasites & personal hygiene amongst food handlers in Amritsar city. Indian J. Comm. Med. 2006, 31, 60–61. [Google Scholar]
- Merselis, J.G., Jr.; Kaye, D.; Connolly, C.S.; Hook, E.W. Quantitative bacteriology of the typhoid carrier state. Am. J. Trop. Med. Hyg. 1964, 13, 425–429. [Google Scholar] [CrossRef]
- Nath, G.; Mauryal, P.; Gulati, A.K.; Singh, T.B.; Srivastava, R.; Kumar, K.; Tripathi, S.K. Comparison of Vi serology and nested PCR in diagnosis of chronic typhoid carriers in two different study populations in typhoid endemic area of India. Southeast. Asian J. Trop. Med. Public Health 2010, 41, 636–640. [Google Scholar] [PubMed]
- Thompson, L.C.; Dunstan, S.J.; Dolecek, C.; Perkin, T.; House, D.; Dougan, G.; Hue, N.T.; La, T.T.P.; Du, D.C.; Phuong, L.T.; et al. Transcriptional response in the peripheral blood of patients infected with Salmonella enterica serovar Typhi. Proc. Natl. Acad. Sci. USA 2009, 52, 22433–22438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berchieri, A., Jr.; Murphy, C.K.; Marston, K.; Barrow, P.A. Observations on the persistence and vertical transmission of Salmonella enterica serovars Pullorum and Gallinarum in chickens; effect of bacterial and host genetic background. Avian Pathol. 2001, 30, 229–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wigley, P.; Hulme, S.D.; Powers, C.; Beal, R.K.; Berchieri, A., Jr.; Smith, A.; Barrow, P. Infection of the reproductive tract and eggs by Salmonella enterica serovar Pullorum in the chicken is associated with suppression of cellular immunity at sexual maturity. Infect. Immun. 2005, 73, 2986–2990. [Google Scholar] [CrossRef] [Green Version]
- Wigley, P.; Berchieri, A., Jr.; Page, K.L.; Smith, A.L.; Barrow, P.A. Salmonella enterica serovar Pullorum persists in splenic macrophages and in the reproductive tract during persistent, disease-free carriage in chickens. Infect. Immun. 2001, 69, 7873–7879. [Google Scholar] [CrossRef] [Green Version]
- Field, H.I. A survey of bovine salmonellosis in Mid and West Wales. Vet. J. 1948, 104, 323–339. [Google Scholar]
- Gibson, E.A. Studies on the Epidemiology of Salmonella Infection in Cattle. Ph.D. Thesis, University of London, London, UK, 1958. [Google Scholar]
- Richardson, A. Transmission of Salmonella Dublin to Calves from Adult Carrier Cows. Vet. Rec. 1973, 92, 112–115. [Google Scholar] [CrossRef]
- Sojka, W.J.; Thomson, P.D.; Hudson, E.B. Excretion of Salmonella dublin by adult bovine carriers. Br. Vet. J. 1974, 130, 482–488. [Google Scholar] [CrossRef]
- Sojka, W.J.; Field, H.I. Salmonellosis in England and Wales 1958–1967. Vet. Bull. 1970, 40, 515–531. [Google Scholar]
- Wray, C.; Sojka, W.J. Salmonella dublin infection of calves: Use of small doses to simulate natural infection on the farm. J. Hyg. 1981, 87, 501–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wray, C.; Wadsworth, Q.C.; Richards, D.W.; Morgan, J.H. A three-year study of Salmonella dublin infection in a closed dairy herd. Vet. Rec. 1989, 124, 532–537. [Google Scholar] [CrossRef]
- Belloy, L.; Decrausaz, L.; Boujon, P.; Hächler, H.; Waldvogel, A.S. Diagnosis by culture and PCR of Salmonella Abortusovis infection under clinical conditions in aborting sheep in Switzerland. Vet. Microbiol. 2009, 138, 373–377. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, D.; Maskell, D.J.; Liew, F.Y.; Easmon, C.F.; Dougan, G. Characterization of aromatic-and purine-dependent Salmonella typhimurium: Attenuation, persistence, and ability to induce protective immunity in BALB/c mice. Infect. Immun. 1988, 56, 419–423. [Google Scholar] [CrossRef] [Green Version]
- Sukupolvi, S.; Edelstein, A.; Rhen, M.; Normack, S.; Pfeifer, J.D. Development of a murine model of chronic Salmonella Infection. Infect. Immun. 1997, 65, 838–842. [Google Scholar] [CrossRef] [Green Version]
- Caron, J.; Loredo-Osti, J.C.; Laroche, L.; Skamene, E.K.; Morgan, K.; Malo, D. Identification of genetic loci controlling bacterial clearance in experimental Salmonella enteritidis infection: An unexpected role of Nramp1 (Slc11a1) in the persistence of infection in mice. Genes Immun. 2002, 3, 196–204. [Google Scholar] [CrossRef] [Green Version]
- Marzel, A.; Desai, P.T.; Goren, A.; Schorr, Y.I.; Nissan, I.; Porwollik, S.; Valinsky, L.; McClelland, M.; Rahav, G.; Gal-Mor, O. Persistent infections by nontyphoidal Salmonella in humans: Epidemiology and genetics. Clin. Infect. Dis. 2016, 62, 879–886. [Google Scholar] [CrossRef] [Green Version]
- Obgol’ts, A.A.; Kononov, A.V.; Konev, V.P. Pathogenetic and immunological characteristics of different forms of the Salmonella typhi carrier state. Zh. Mikrobiol. Epidemiol. Immunobiol. 1987, 4, 71–76. [Google Scholar]
- Karaki, K.; Matsubara, Y. Surgical treatment of chronic biliary typhoid and paratyphoid carriers. Nippon Shokakibyo Gakkai Zasshi 1984, 81, 2978–2985. [Google Scholar]
- Nath, G.; Singh, Y.K.; Kumar, K.; Gulati, A.K.; Shukla, V.K.; Khanna, A.K.; Tripathi, S.K.; Jain, A.K.; Kumar, M.; Singh, T.B. Association of carcinoma of the gallbladder with typhoid carriage in a typhoid endemic area using nested PCR. J. Infect. Dev. Ctries. 2008, 2, 302–307. [Google Scholar] [CrossRef] [Green Version]
- Caygill, C.P.; Hill, M.J.; Braddick, M.; Sharp, J.C. Cancer mortality in chronic typhoid and paratyphoid carriers. Lancet 1994, 343, 83–84. [Google Scholar] [CrossRef]
- Dutta, U.; Garg, P.K.; Kumar, R.; Tandon, R.K. Typhoid carriers among patients with gallstones are at increased risk for carcinoma of the gallbladder. Am. J. Gastroenterol. 2000, 95, 784–787. [Google Scholar] [CrossRef]
- Nagaraja, V.; Eslick, G.D. Systematic review with meta-analysis: The relationship between chronic Salmonella typhi carrier status and gall-bladder cancer. Aliment. Pharmacol. Therapeut. 2014, 39, 745–750. [Google Scholar] [CrossRef]
- Nath, G.; Singh, H.; Shukla, V.K. Chronic typhoid carriage and carcinoma of the gall bladder. J. Cancer Prev. 1997, 6, 557–559. [Google Scholar] [CrossRef] [PubMed]
- Welton, J.C.; Marr, J.S.; Friedman, S.M. Association between hepatobiliary cancer and typhoid carrier state. Lancet 1979, 14, 791–794. [Google Scholar] [CrossRef]
- Blanchette, F.; Day, R.; Dong, W.; Laprise, M.H.; Dubois, C.M. TGFbeta1 regulates gene expression of its own converting enzyme furin. J. Clin. Investig. 1997, 99, 1974–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galdiero, M.; Marcatili, A.; Cipollaro de l’Ero, G.; Nuzzo, I.; Bentivoglio, C.; Romano Carratelli, C. Effect of transforming growth factor beta on experimental Salmonella Typhimurium infection in mice. Infect. Immun. 1999, 67, 1432–1438. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Escobedo, G.; Marshall, J.M.; Gunn, J.S. Chronic and acute infection of the gall bladder by Salmonella Typhi: Understanding the carrier state. Nat. Rev. Microbiol. 2011, 9, 9–14. [Google Scholar] [CrossRef] [Green Version]
- Dongol, S.; Thompson, C.N.; Clare, S.; Nga, T.V.T.; Duy, P.T.; Karkey, A.; Arjyal, A.; Koirala, S.; Khatri, N.S.; Maskey, P.; et al. The microbiological and clinical characteristics of invasive Salmonella in gallbladders from cholecystectomy patients in Kathmandu, Nepal. PLoS ONE 2012, 7, e47342. [Google Scholar] [CrossRef] [Green Version]
- González, J.F.; Halley Alberts, H.; Joel Lee, J.; Doolittle, L.; Gunn, J.S. Bioflm Formation Protects Salmonella from the antibiotic ciprofoxacin in vitro and in vivo in the mouse model of chronic carriage. Sci. Rep. 2018, 8, 222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, R.W.; Gibson, D.L.; Kay, W.W.; Gunn, J.S. Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect. Immun. 2008, 76, 5341–5349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, R.W.; Rosales-Reyes, R.; de la Luz, M.; de la Ramírez-Aguilarb, M.L.; Chapa-Azuelac, C.O.; Alpuche Arandad, C.; Gunn, J.S. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc. Natl. Acad. Sci. USA 2010, 107, 4353–4358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, R.W.; Reeve, K.E.; Gunn, J.S. Flagellated but not hyper-fimbriated Salmonella enterica serovar Typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J. Bacteriol. 2010, 192, 2981–2990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunn, J.S.; Marshall, J.M.; Baker, S.; Dongol, S.; Charles, R.C.; Ryan, E.T. Salmonella chronic carriage: Epidemiology, diagnosis and gallbladder persistence. Trends Microbiol. 2014, 22, 648–655. [Google Scholar] [CrossRef] [Green Version]
- Barrow, P.A. Further observations on the effect of feeding diets containing avoparcin on the excretion of salmonellas by experimentally infected chickens. Epidemiol. Infect. 1989, 102, 239–252. [Google Scholar] [CrossRef] [Green Version]
- Vogelsang, T.M. The campaign against typhoid and paratyphoid B in western Norway. Results of cholecystectomy. J. Hyg. 1964, 62, 443–449. [Google Scholar] [CrossRef] [Green Version]
- Gaines, S.; Sprinz, H.; Tully, J.G.; Tigertt, W.D. Studies on infection and immunity in experimental typhoid fever. VII. The distribution of Salmonella typhi in chimpanzee tissue following oral challenge, and the relationship between the numbers of bacilli and morphologic lesions. J. Infect. Dis. 1968, 118, 293–306. [Google Scholar] [CrossRef]
- Ristori, C.; Rodríguez, H.; Vicent, P.; Ferreccio, C.; García, J.; Lobos, H.; D’Ottone, K. Persistence of the Salmonella typhi-paratyphi carrier state after gallbladder removal. Bull. Pan. Am. Health Organ. 1982, 16, 361–366. [Google Scholar]
- Ristori, C.; Rodríguez, H.; Vicent, P.; Lobos, L.; D’Ottone, K.; García, J.; Pinto, M.E.; Nercelles, P.; Cisneros, L. Investigation of the Salmonella typhi-paratyphi carrier state in cases of surgical intervention for gallbladder disease. Bull. Pan. Am. Health Organ. 1982, 16, 161–171. [Google Scholar]
- Lawson, G.H.; McPherson, E.A.; Laing, A.H.; Wooding, P. The epidemiology of Salmonella dublin infection in a dairy herd. I. Excretion and persistence of the organism. J. Hyg. 1974, 72, 311–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menendez, A.; Arena, E.T.; Guttman, J.A.; Thorson, L.; Vallance, B.A.; Vogl, W.; Finlay, B.B. Salmonella infection of gallbladder epithelial cells drives local inflammation and injury in a model of acute typhoid fever. J. Infect. Dis. 2009, 200, 1703–1713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tischler, A.D.; McKinney, J.D. Contrasting persistence strategies in Salmonella and Mycobacterium. Curr. Opin. Microbiol. 2010, 13, 93–99. [Google Scholar] [CrossRef] [Green Version]
- Holt, K.E.; Parkhill, J.; Mazzoni, C.J.; Roumagnac, P.; Weill, F.X.; Goodhead, I.; Rance, R.; Baker, S.; Maskell, D.J.; Wain, J.; et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 2008, 40, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Johnston, R.N.; Liu, G.-R.; Liu, S.-L. Genomic comparison between Salmonella Gallinarum and Pullorum: Differential pseudogene formation under common host restriction. PLoS ONE 2013, 8, e59427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langridge, G.C.; Fookes, M.; Connor, T.R.; Feltwell, T.; Feasey, N.; Parsons, B.N.; Seth-Smith, H.M.B.; Barquist, L.; Stedman, A.; Humphrey, T.J.; et al. Patterns of genome evolution that have accompanied host adaptation in Salmonella. Proc. Natl. Acad. Sci. USA 2015, 112, 863–868. [Google Scholar] [CrossRef] [Green Version]
- Thomson, N.R.; Clayton, D.J.; Windhorst, D.; Davidson, S.; Vernikos, G.; Churcher, C.; Quail, M.; Stevens, M.J.; Jones, M.A.; Lord, A.; et al. Comparative genome analysis of Salmonella enteritidis PT4 and Salmonella gallinarum 287/91 provides insights into host adaptation in zoonotic bacterial pathogens. Genome Res. 2008, 18, 1624–1637. [Google Scholar] [CrossRef] [Green Version]
- Matthews, T.D.; Schmieder, R.; Silva, G.G.Z.; Busch, J.; Cassman, N.; Dutilh, B.E.; Green, D.; Matlock, B.; Heffernan, B.; Olsen, G.J.; et al. Genomic comparison of the closely-related Salmonella enterica serovars Enteritidis, Dublin and Gallinarum. PLoS ONE 2015, 10, e0126883. [Google Scholar] [CrossRef]
- Deligios, M.; Bacciu, D.; Deriu, E.; Corti, G.; Bordoni, R.; De Bellis, G.; Leori, G.S.; Rubino, S.; Uzzau, S. Draft genome sequence of the host-restricted Salmonella enterica serovar Abortusovis strain SS44. Genome Announc. 2014, 2, e00261-14. [Google Scholar] [CrossRef] [Green Version]
- Matthews, T.D.; Rabsch, W.; Maloy, S. Chromosomal rearrangements in Salmonella enterica serovar Typhi strains isolated from asymptomatic human carriers. mBio 2011, 2, e00060-11. [Google Scholar] [CrossRef] [Green Version]
- Chiou, C.-S.; Wei, H.-L.; Mu, J.-L.; Liao, Y.-S.; Liang, S.-Y.; Liao, C.-H.; Tsao, C.-S.; Wang, S.-C. Salmonella enterica serovar Typhi variants in long-term carriers. J. Clin. Microbiol. 2013, 51, 669–672. [Google Scholar] [CrossRef] [Green Version]
- Duy, T.P.; Thieu, N.T.V.; Nguyen, T.; Nguyen, T.; Thanh, H.N.D.; Dongol, S.; Karkey, A.; Carey, M.; Basnyat, B.; Dougan, G.; et al. Gallbladder carriage generates genetic variation and genome degradation in Salmonella Typhi. PLoS Pathog. 2020, 16, e1008998. [Google Scholar] [CrossRef]
- Roumagnac, P.; Weill, F.X.; Dolecek, C.; Baker, S.; Brisse, S.; Chinh, N.T.; Le, T.A.H.; Acosta, C.J.; Farrar, J.; Dougan, G.; et al. Evolutionary history of Salmonella Typhi. Science 2006, 314, 1301–1304. [Google Scholar] [CrossRef] [Green Version]
- Batista, D.F.A.; Neto, O.C.F.; de Almeida, A.M.; Maboni, G.; de Carvalho, T.F.; Barrow, P.A.; Junior, A.B. Evaluation of pathogenicity of Salmonella Gallinarum strains harbouring deletions in genes whose orthologues are conserved pseudogenes in S. pullorum. PLoS ONE 2018, 13, e0200585. [Google Scholar]
- Parkhill, J.; Dougan, G.; James, K.D.; Thomson, N.R.; Pickard, D.; Wain, J.; Churcher, C.; Mungall, K.L.; Bentley, S.D.; Holden, M.T.G.; et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 2001, 413, 848–852. [Google Scholar] [CrossRef]
- Baddam, R.; Kumar, N.; Shaik, S.; Lankapalli, A.K.; Ahmed, N. Genome dynamics and evolution of Salmonella Typhi strains from the typhoid-endemic zones. Sci. Rep. 2014, 4, 7457. [Google Scholar] [CrossRef]
- Lawley, T.D.; Chan, K.; Thompson, L.J.; Kim, C.C.; Govoni, G.R.; Monack, D.M. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2006, 2, e11. [Google Scholar] [CrossRef] [Green Version]
- Jones, M.A.; Wigley, P.; Page, K.L.; Hulme, S.D.; Barrow, P.A. Salmonella enterica serovar Gallinarum requires the Salmonella pathogenicity island 2 type III secretion system but not the Salmonella pathogenicity island 1 type III secretion system for virulence in the chicken. Infect. Immun. 2001, 69, 5471–5476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodsky, I.E.; Ghori, N.; Falkow, S.; Monack, D. Mig-14 is an inner membrane-associated protein that promotes Salmonella typhimurium resistance to CRAMP, survival within activated macrophages and persistent infection. Mol. Microbiol. 2005, 55, 954–972. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Sashinami, H.; Takaya, A.; Toshifumi, T.; Matsui, H.; Kikuchi, Y.; Hanawa, T.; Kamiya, S.; Nakane, A. Disruption of the genes for ClpXP protease in Salmonella enterica serovar Typhimurium results in persistent infection in mice, and development of persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect. Immun. 2001, 69, 3164–3174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knudsen, G.M.; Olsen, J.E.; Aabo, S.; Barrow, P.; Rychlik, I.; Thomsen, L.E. ClpP deletion causes attenuation of Salmonella Typhimurium virulence through mis-regulation of RpoS and indirect control of CsrA and the SPI genes. Microbiology 2013, 159, 1497–1509. [Google Scholar] [CrossRef] [Green Version]
- Kudirkiene, E.; Sørensen, G.; Torpdahl, M.; Knegt, L.V.; Nielsen, L.R.; Rattenborg, E.; Ahmed, S.; Olsen, J.E. Epidemiology of Salmonella enterica serovar Dublin in cattle and humans in denmark, 1996 to 2016: A retrospective whole-genome-based study. Appl. Environ. Microbiol. 2020, 86, e01894-19. [Google Scholar] [CrossRef]
- Pinheiro Vilela, F.; dos Rodrigues, D.P.; Costa, R.G.; Casas, M.R.T.; Falcão, J.P.; Campioni, F. High similarity and high frequency of virulence genes among Salmonella Dublin strains isolated over a 33-year period in Brazil. Braz. J. Microbiol. 2020, 51, 497–509. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, Z.; Qiang, B.; Xu, Y.; Chen, X.; Li, Q.; Jiao, X. Loss and gain in the evolution of the Salmonella enterica serovar Gallinarum biovar Pullorum genome. mSphere 2019, 4, e00627–e00718. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Hu, Y.; Xu, Y.; Yin, K.; Li, Y.; Chen, Y.; Xia, J.; Xu, L.; Liu, Z.; Geng, S.; et al. Genetic analysis of Salmonella enterica serovar Gallinarum biovar Pullorum based on characterization and evolution of CRISPR sequence. Vet. Microbiol. 2017, 203, 81–87. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Xia, J.; Wang, X.; Yin, K.; Hu, Y.; Yin, C.; Liu, Z.; Jiao, X. Virulence of Salmonella enterica serovar Pullorum isolates compared using cell-based and chicken embryo infection models. Poult. Sci. 2019, 98, 1488–1493. [Google Scholar] [CrossRef] [PubMed]
- Dunstan, S.J.; Hawn, T.R.; Hue, N.T.; Parry, C.P.; Ho, V.A.; Vinh, H.; Diep, T.S.; House, D.; Wain, J.; Aderem, A.; et al. Host susceptibility and clinical outcomes in toll-like receptor 5-deficient patients with typhoid fever in Vietnam. J. Infect. Dis. 2005, 191, 1068–1071. [Google Scholar] [CrossRef] [Green Version]
- Dunstan, S.J.; Hue, N.T.; Han, B.; Li, Z.; Tram, T.T.B.; Sim, K.S.; Parry, C.M.; Chinh, N.T.; Ha Vinh Lan, N.P.H.; Thieu, N.Y.V.; et al. Variation at HLA-DRB1 is associated with resistance to enteric fever. Nat. Genet. 2014, 46, 1333–1336. [Google Scholar] [CrossRef] [PubMed]
- Alvareza, M.I.; Glovera, L.C.; Luoa, P.; Wanga, L.; Theuschb, E.; Oehlersa, S.H.; Waltona, E.M.; Trame, T.T.B.; Kuang, Y.-L.; Rotterf, J.I.; et al. Human genetic variation in VAC14 regulates Salmonella invasion and typhoid fever through modulation of cholesterol. Proc. Natl. Acad. Sci. USA 2017, 114, E7746–E7755. [Google Scholar] [CrossRef] [Green Version]
- Ma, P.Y.; Tan, J.E.; Hee, E.Y.; Yong, D.W.X.; Heng, Y.S.; Low, W.X.; Wu, X.H.; Cletus, C.; Chellappan, D.K.; Aung, K.; et al. Human genetic variation influences enteric fever progression. Cells 2021, 10, 345. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.F.; Malo, D. Genetic regulation of host responses to Salmonella infection in mice. Genes Immun. 2002, 3, 381–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calenge, F.; Kaiser, P.; Vignal, A.; Beaumont, C. Genetic control of resistance to salmonellosis and to Salmonella carrier-state in fowl: A review. Genet. Select. Evol. 2010, 42, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Psifidi, A.; Russell, K.M.; Matika, O.; Sánchez-Molano, E.; Wigley, P.; Fulton, J.E.; Stevens, M.P.; Fife, M.S. The genomic architecture of fowl typhoid resistance in commercial layers. Front. Genet. 2018, 9, 519. [Google Scholar] [CrossRef] [PubMed]
- Loomis, W.P.; Johnson, M.L.; Brasfield, A.; Blanc, M.P.; Yi, J.; Miller, S.I.; Cookson, B.T.; Hajjar, A.M. Temporal and anatomical host resistance to chronic Salmonella infection is quantitatively dictated by Nramp1 and influenced by host genetic background. PLoS ONE 2014, 9, e111763. [Google Scholar] [CrossRef] [PubMed]
- Mastroeni, P.; Harrison, J.A.; Robinson, J.H.; Clare, S.; Khan, S.; Maskell, D.J.; Dougan, G.; Hormaeche, C.E. Interleukin-12 is required for control of the growth of attenuated aromatic-compound-dependent salmonellae in BALB/c mice: Role of gamma interferon and macrophage activation. Infect. Immun. 1998, 66, 4767–4776. [Google Scholar] [CrossRef] [Green Version]
- Kohno, K.; Kataoka, J.; Ohtsuki, T.; Suemoto, Y.; Okamoto, I.; Usui, M.; Ikeda, M.; Kurimoto, M. IFN-gamma-inducing factor (IGIF) is a costimulatory factor on the activation of Th1 but not Th2 cells and exerts its effect independently of IL-12. J. Immunol. 1997, 158, 1541–1550. [Google Scholar] [PubMed]
- Robinson, D.; Shibuya, K.; Mui, A.; Zonin, F.; Murphy, E.; Sana, T.; Hartley, S.B.; Menon, S.; Kastelein, R.; Bazan, F.; et al. IGIF does not drive Th1 development but synergizes with IL-12 for interferon-gamma production and activates IRAK and NFkappaB. Immunity 1997, 7, 571–581. [Google Scholar] [CrossRef] [Green Version]
- Beal, R.K.; Powers, C.; Wigley, P.; Barrow, P.A.; Smith, A.L. Temporal dynamics of the cellular, humoral and cytokine responses in chickens during primary and secondary infection with Salmonella enterica serovar Typhimurium. Avian Pathol. 2004, 33, 25–33. [Google Scholar] [CrossRef]
- Beal, R.K.; Wigley, P.; Powers, C.; Hulme, S.D.; Barrow, P.A.; Smith, A.L. Age at primary infection with Salmonella enterica serovar Typhimurium in the chicken influences persistence of infection and subsequent immunity to re-challenge. Vet. Immunol. Immunopathol. 2004, 100, 151–164. [Google Scholar] [CrossRef]
- Berndt, A.; Wilhelm, A.; Jugert, C.; Pieper, J.; Sachse, K.; Methner, U. Chicken cecum immune response to Salmonella enterica serovars of different levels of invasiveness. Infect. Immun. 2007, 75, 5993–6007. [Google Scholar] [CrossRef] [Green Version]
- Withanage, G.S.; Wigley, P.; Kaiser, P.; Mastroeni, P.; Brooks, H.; Powers, C.; Beal, R.; Barrow, P.; Maskell, D.; McConnell, I. Cytokine and chemokine responses associated with clearance of a primary Salmonella enterica serovar Typhimurium infection in the chicken and in protective immunity to rechallenge. Infect. Immun. 2005, 73, 5173–5182. [Google Scholar] [CrossRef] [Green Version]
- Hulme, S.D.; Barrow, P.A.; Foster, N. Inhibited Production of iNOS by Murine J774 Macrophages Occurs via a phoP-regulated differential expression of NFκB and AP-1. Interdisc. Persp. Infect. Dis. 2012, 14, 483170. [Google Scholar]
- Ruby, T.; McLaughlin, L.; Gopinath, S.; Monack, D. Salmonella’s long-term relationship with its host. FEMS Microbiol. Rev. 2012, 36, 600–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, R.W.; McLachlan, J.B.; Kurtz, J.R.; Jenkins, M.K. CD4+ T cell persistence and function after infection is maintained by low-level peptide: MHCII presentation1. J. Immunol. 2013, 190, 2828–2834. [Google Scholar] [CrossRef] [Green Version]
- Goggins, A.; Kurtz, J.R.; McLachlan, J.B. Control of persistent Salmonella infection relies on constant thymic output despite increased peripheral antigen-specific T cell immunity. Pathogens 2020, 9, 605. [Google Scholar] [CrossRef]
- Johansson, C.; Ingman, M.; Wick, M.J. Elevated neutrophil, macrophage and dendritic cell numbers characterize immune cell populations in mice chronically infected with Salmonella. Microbial. Pathog. 2006, 4, 49–58. [Google Scholar] [CrossRef]
- Eisele, N.A.; Ruby, T.; Jacobson, A.; Manzanillo, P.S.; Cox, J.S.; Lam, L.; Mukundan, L.; Chawla, A.; Monack, D.M. Salmonella require the fatty acid regulator PPARδ for the establishment of a metabolic environment essential for long term persistence. Cell Host Microbe 2013, 14, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Pham, T.H.M.; Brewer, S.M.; Thurston, T.; Massis, L.M.; Honeycutt, J.; Lugo, K.; Jacobson, A.R.; Vilches-Moure, J.G.; Hamblin, M.; Helaine, S.; et al. Salmonella-driven polarization of granuloma macrophages antagonizes TNF-mediated pathogen restriction during persistent infection. Cell Host Microbe 2020, 27, 54–67. [Google Scholar] [CrossRef]
- Jaslow, S.L.; Gibbs, K.D.; Fricke, W.F.; Wang, L.; Pittman, K.J.; Mammel, M.K.; Thaden, J.T.; Fowler, V.G., Jr.; Hammer, G.E.; Elfenbein, J.R.; et al. Salmonella activation of STAT3 signaling by SarA effector promotes intracellular replication and production of IL-10. Cell Rep. 2018, 23, 3525–3536. [Google Scholar] [CrossRef]
- Mattila, J.T.; Olabisi, O.O.; Kepka-Lenhart, D.; Marino, S.; Kim, J.H.; Eum, S.Y.; Via, L.E.; Barry, C.E., 3rd; Klein, E.; Kirschner, D.E.; et al. Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J. Immunol. 2013, 191, 773–784. [Google Scholar] [CrossRef] [Green Version]
- Munder, M.; Eichmann, K.; Moran, J.M.; Centeno, F.; Soler, G.; Modolell, M. Th1/Th2-regulated expression of arginase isoforms in murine macrophages and dendritic cells. J. Immunol. 1999, 163, 3771–3777. [Google Scholar] [PubMed]
- Mills, C.D.; Kincaid, K.; Alt, J.M.; Heilman, M.J.; Hill, A.M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2000, 164, 6166–6173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rath, M.; Muller, I.; Kropf, P.; Closs, E.I.; Munder, M. Metabolism via arginase or nitric oxide synthase: Two competing arginine pathways in macrophages. Front. Immunol. 2014, 5, 532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastroeni, P.; Grant, A.; Restif, O.; Maskell, D. A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat. Rev. Microbiol. 2009, 7, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, S.A.; Niedergang, F.; Corthesy-Theulaz, I.E.; Kraehenbuhl, J.P. A recombinant Salmonella typhimurium vaccine strain is taken up and survives within murine Peyer’s patch dendritic cells. Cell Microbiol. 2000, 2, 59–68. [Google Scholar] [CrossRef]
- Rescigno, M.; Urbano, M.; Valzasina, B.; Francolini, M.; Rotta, G.; Bonasio, R.; Granucci, F.; Kraehenbuhl, J.P.; Ricciardi-Castagnoli, P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2001, 2, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Cheminay, C.; Schoen, M.; Hensel, M.; Wandersee-Steinhäuser, A.; Ritter, U.; Körner, H.; Röllinghoff, M.; Hein, J. Migration of Salmonella typhimurium-harboring bone marrow--derived dendritic cells towards the chemokines CCL19 and CCL21. Microb. Pathog. 2002, 32, 207–218. [Google Scholar] [CrossRef] [Green Version]
- Halici, S.; Zenk, S.F.; Jantsch, J.; Hensel, M. Functional analysis of the Salmonella pathogenicity island 2-mediated inhibition of antigen presentation in dendritic cells. Infect. Immun. 2008, 76, 4924–4933. [Google Scholar] [CrossRef] [Green Version]
- Lapaque, N.; Hutchinson, J.L.; Jones, D.C.; Méresse, S.; Holden, D.W.; Trowsdale, J.; Kelly, A.P. Salmonella regulates polyubiquitination and surface expression of MHC class II antigens. Proc. Natl. Acad. Sci. USA 2009, 106, 14052–14057. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.; Herrero-Fresno, A.; Thofner, I.; Skov, S.; Olsen, J.E. Interaction differences of the avian host-specific Salmonella enterica serovar Gallinarum, the host-generalist S. Typhimurium, and the cattle host-adapted S. Dublin with chicken primary macrophage. Infect. Immun 2019, 87, e00552-19. [Google Scholar] [CrossRef]
- Bimczok, D.; Sowa, E.N.; Faber-Zuschratter, H.; Pabst, R.; Rothkötter, H.J. Site-specific expression of CD11b and SIRPalpha (CD172a) on dendritic cells: Implications for their migration patterns in the gut immune system. Eur. J. Immunol. 2005, 35, 1418–1427. [Google Scholar] [CrossRef]
- Gurka, S.; Hartung, E.; Becker, M.; Kroczek, R.A. Mouse conventional dendritic cells can be universally classified based on the mutually exclusive expression of XCR1 and SIRPα. Front. Immunol. 2015, 6, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, J.F.; Hahan, M.M.; Gunn, J.S. Chronic biofilm-based infections: Skewing of the immune response. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, J.F.; Kurtz, J.; Bauer, D.L.; Hitt, R.; Fitch, J.; Wetzel, A.; La Perle, K.; McLachlan, J.; Gunn, J.S. Establishment of chronic typhoid infection in a mouse carriage model involves a type 2 immune shift and T and B call recruitment to the gallbladder. mBio. 2019, 10, e02262-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Foster, N.; Jones, M.A.; Barrow, P.A. A model of persistent Salmonella infection: Salmonella Pullorum modulates the immune response of the chicken from a Th17 towards a Th2-type response. Infect. Immun. 2018, 86, e00307–e00318. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Jones, M.A.; Barrow, P.A.; Foster, N. Immune modulation and the development of fowl typhoid: A model of human disease? Pathogens 2020, 9, 843. [Google Scholar] [CrossRef]
- Chappell, L.; Kaiser, P.; Barrow, P.; Jones, M.A.; Johnston, C.; Wigley, P. The immunobiology of avian systemic salmonellosis. Vet. Immunol. Immunopathol. 2009, 128, 53–59. [Google Scholar] [CrossRef]
- Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003, 3, 23–35. [Google Scholar] [CrossRef]
- Martinez, F.O.; Sica, A.; Mantovani, A.; Locati, M. Macrophage activation and polarization. Front. Biosci. 2008, 13, 453–461. [Google Scholar] [CrossRef] [Green Version]
- Levine, M.M.; Tacket, C.; Sztein, M.B. Host-Salmonella interaction: Human trials. Microbes Infect. 2001, 3, 1271–1279. [Google Scholar] [CrossRef]
- Salerno-Gonçalves, R.; Fernandez-Viña, M.; Lewinsohn, D.M.; Sztein, M.B. Identification of a human HLA-E-Restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J. Immunol. 2004, 173, 5852–5862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sztein, M.B. Cell-mediated immunity and antibody responses elicited by attenuated Salmonella enterica serovar Typhi strains used as live oral vaccines in humans. Clin. Infect. Dis. 2007, 45, S15–S19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salerno-Goncalves, R.; Wyant, T.L.; Pasetti, M.F.; Fernandez-Vina, M.; Tacket, C.O.; Levine, M.M.; Sztein, M.B. Concomitant induction of CD4 and CD8 T cell responses in volunteers immunized with Salmonella enterica serovar typhi strain CVD 908-htrA. J. Immunol. 2003, 170, 2734–2741. [Google Scholar] [CrossRef] [Green Version]
- McArthur, M.A.; Sztein, M.B. Heterogeneity of multifunctional IL-17A producing S.Typhi-specific CD8 T cells in volunteers following Ty21a typhoid immunization. PLoS ONE 2012, 7, e38408. [Google Scholar] [CrossRef] [Green Version]
- Bhuiyan, S.; Sayeed, A.; Khanam, F.; Leung, D.T.; Rahman Bhuiyan, T.; Sheikh, A.; Salma, U.; LaRocque, R.C.; Harris, J.B.; Pacek, M.; et al. Cellular and cytokine responses to Salmonella enterica serotype Typhi proteins in patients with typhoid fever in Bangladesh. Am. J. Trop. Med. Hyg. 2014, 90, 1024–1030. [Google Scholar] [CrossRef] [Green Version]
- McArthur, M.A.; Fresnay, S.; Magde, L.S.; Darton, T.C.; Jones, C.; Waddington, C.S.; Blohmke, C.J.; Dougan, G.; Angus, B.; Levine, M.M.; et al. Activation of Salmonella Typhi-Specific regulatory T cells in typhoid disease in a wild-type S. typhi challenge model. PLoS Pathog. 2015, 11, e1004914. [Google Scholar] [CrossRef] [PubMed]
- Looney, R.J.; Steigbigel, R.T. Role of the Vi antigen of Salmonella typhi in resistance to host defense in vitro. J. Lab. Clin. Med. 1986, 108, 506–516. [Google Scholar] [PubMed]
- Raffatellu, M.; Chessa, D.; Wilson, R.P.; Dusold, R.; Rubino, S.; Baumler, A.J. The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-Like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect. Immun. 2005, 73, 3367–3374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansen, A.M.; Hall, L.J.; Clare, S.; Goulding, D.; Holt, K.E.; Grant, A.J.; Mastroeni, P.; Dougan, G.; Kingsley, R.A. A Salmonella Typhimurium-Typhi genomic chimera: A model to study Vi polysaccharide capsule function in vivo. PLoS Pathog. 2011, 7, e1002131. [Google Scholar] [CrossRef] [PubMed]
- Froh, M.; Thurman, R.G.; Wheeler, M.D. Molecular evidence for a glycine-gated chloride channel in macrophages and leukocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 283, G856–G863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falkow, S. Is persistent bacterial infection good for your health? Cell 2006, 124, 699–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwacha, M.G.; Meissler, J.J., Jr.; Eisenstein, T.K. Salmonella typhimurium infection in mice induces nitric oxide-mediated immunosuppression through a natural killer cell-dependent pathway. Infect. Immun. 1998, 66, 5862–5866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Velden, A.W.; Copass, M.K.; Starnbach, M.N. Salmonella inhibit T cell proliferation by a direct, contact-dependent immunosuppressive effect. Proc. Natl. Acad. Sci. USA 2005, 102, 17769–17774. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, A.; McSorley, S.J. Pivotal advance: Exposure to LPS suppresses CD4 T cell cytokine production in Salmonella-infected mice and exacerbates murine typhoid. J. Leukocite Biol. 2007, 81, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Lee, S.P. The role of bacteria in gallstone pathogenesis. Front. Biosci. 2001, 6, E93–E103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Singh, S.; Ahirwar, S.K.; Nath, G. Proteomics-based identification of plasma proteins and their association with the host–pathogen interaction in chronic typhoid carriers. Int. J. Infect. Dis. 2014, 19, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Dubois, C.M.; Blanchette, F.; Laprise, M.H.; Leduc, R.; Grondin, F.; Seidah, N.G. Evidence that furin is an authentic transforming growth factor-beta1-converting enzyme. Am. J. Pathol. 2001, 158, 305–316. [Google Scholar] [CrossRef]
- Pan, D.; Rostagno, M.H.; Ebner, P.D.; Eicher, S.D. Differential innate immune responses of bovine peripheral blood leukocytes to Salmonella enterica serovars Dublin, Typhimurium, and Enteritidis. Vet. Immunol. Immunopathol. 2015, 165, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, G.; Mathuri, N.K.; Job, C.K.; Nath, I.; Cohn, Z. Effect of multiple interferon y injections on the disposal of Mycobacterium leprae. Proc. Natl. Acad. Sci. USA 1989, 86, 8073–8077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nathan, C.F.; Kaplan, G.; Levis, W.R.; Nusrat, A.; Witmer, M.D.; Sherwin, S.A.; Job, C.K.; Horowitz, C.R.; Steinman, R.M.; Cohn, Z.A. Local and systemic effects of intradermal recombinant interferon-gamma in patients with lepromatous leprosy. N. Engl. J. Med. 1986, 315, 6–15. [Google Scholar] [CrossRef]
- Heinzel, F.P.; Schoenhaut, D.S.; Rerko, R.M.; Rosser, L.E.; Gately, M.K. Recombinant interleukin 12 cures mice infected with Leishmania major. J. Exp. Med. 1993, 177, 1505–1509. [Google Scholar] [CrossRef]
- Finkelman, F.D.; Madden, K.B.; Cheever, A.W.; Katona, I.M.; Morris, S.C.; Gately, M.K.; Hubbard, B.R.; Gause, W.C.; Urban, J.F. Effects of interleukin 12 on immune responses and host protection in mice infected with intestinal nematode parasites. J. Exp. Med. 1994, 179, 1563–1572. [Google Scholar] [CrossRef] [Green Version]
- Park, J.H.; Sung, H.W.; Yoon, B.I.; Kwon, H.M. Protection of chicken against very virulent IBDV provided by in ovo priming with DNA vaccine and boosting with killed vaccine and the adjuvant effects of plasmid-encoded chicken interleukin-2 and interferon-γ. J. Vet. Sci. 2009, 10, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Guo, P.; Thomas, J.D.; Bruce, M.P.; Hinton, T.M.; Bean, A.G.D.; Lowenthal, J.W. The chicken Th1 response: Potential therapeutic applications of ChIFN-γ. Dev. Comp. Immunol. 2013, 41, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Barrow, P. Bacterial Infections of Poultry—New Approaches to Vaccination and Other Methods of Infection Control; Global Alliance for Research on Avian Diseases: London, UK, 2015. [Google Scholar]
- Okamura, M.; Lillehoj, H.S.; Raybourne, R.B.; Babu, U.S.; Heckert, R.A.; Tani, H.; Sasai, K.; Baba, E.; Lillehoj, E.P. Differential responses of macrophages to Salmonella enterica serovars Enteritidis and Typhimurium. Vet. Immunol. Immunopathol. 2005, 107, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Narang, T.; Kaur, I.; Kumar, B.; Radotra, B.D.; Dogra, S. Comparative evaluation of immunotherapeutic efficacy of BCG and mw vaccines in patients of borderline lepromatous and lepromatous leprosy. Int. J. Lepr. Other Mycobact. Dis. 2005, 73, 105–114. [Google Scholar]
- Foster, N.; Berndt, A.; Lalmanach, A.C.; Methner, U.; Pasquali, P.; Rychlik, I.; Velge, P.; Zhou, X.; Barrow, P. Emergency and therapeutic vaccination—Is stimulating innate immunity an option? Res. Vet. Sci. 2012, 93, 7–12. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foster, N.; Tang, Y.; Berchieri, A.; Geng, S.; Jiao, X.; Barrow, P. Revisiting Persistent Salmonella Infection and the Carrier State: What Do We Know? Pathogens 2021, 10, 1299. https://doi.org/10.3390/pathogens10101299
Foster N, Tang Y, Berchieri A, Geng S, Jiao X, Barrow P. Revisiting Persistent Salmonella Infection and the Carrier State: What Do We Know? Pathogens. 2021; 10(10):1299. https://doi.org/10.3390/pathogens10101299
Chicago/Turabian StyleFoster, Neil, Ying Tang, Angelo Berchieri, Shizhong Geng, Xinan Jiao, and Paul Barrow. 2021. "Revisiting Persistent Salmonella Infection and the Carrier State: What Do We Know?" Pathogens 10, no. 10: 1299. https://doi.org/10.3390/pathogens10101299
APA StyleFoster, N., Tang, Y., Berchieri, A., Geng, S., Jiao, X., & Barrow, P. (2021). Revisiting Persistent Salmonella Infection and the Carrier State: What Do We Know? Pathogens, 10(10), 1299. https://doi.org/10.3390/pathogens10101299






