Anaplasma phagocytophilum and Anaplasma ovis–Emerging Pathogens in the German Sheep Population
Abstract
:1. Introduction
2. Results
2.1. Clinical and Clinicopathological Findings
2.2. Fecal Parasitological Examinations
2.3. Anaplasma phagocytophilum and Anaplasma ovis Laboratory Investigations
2.3.1. Antibody Detection
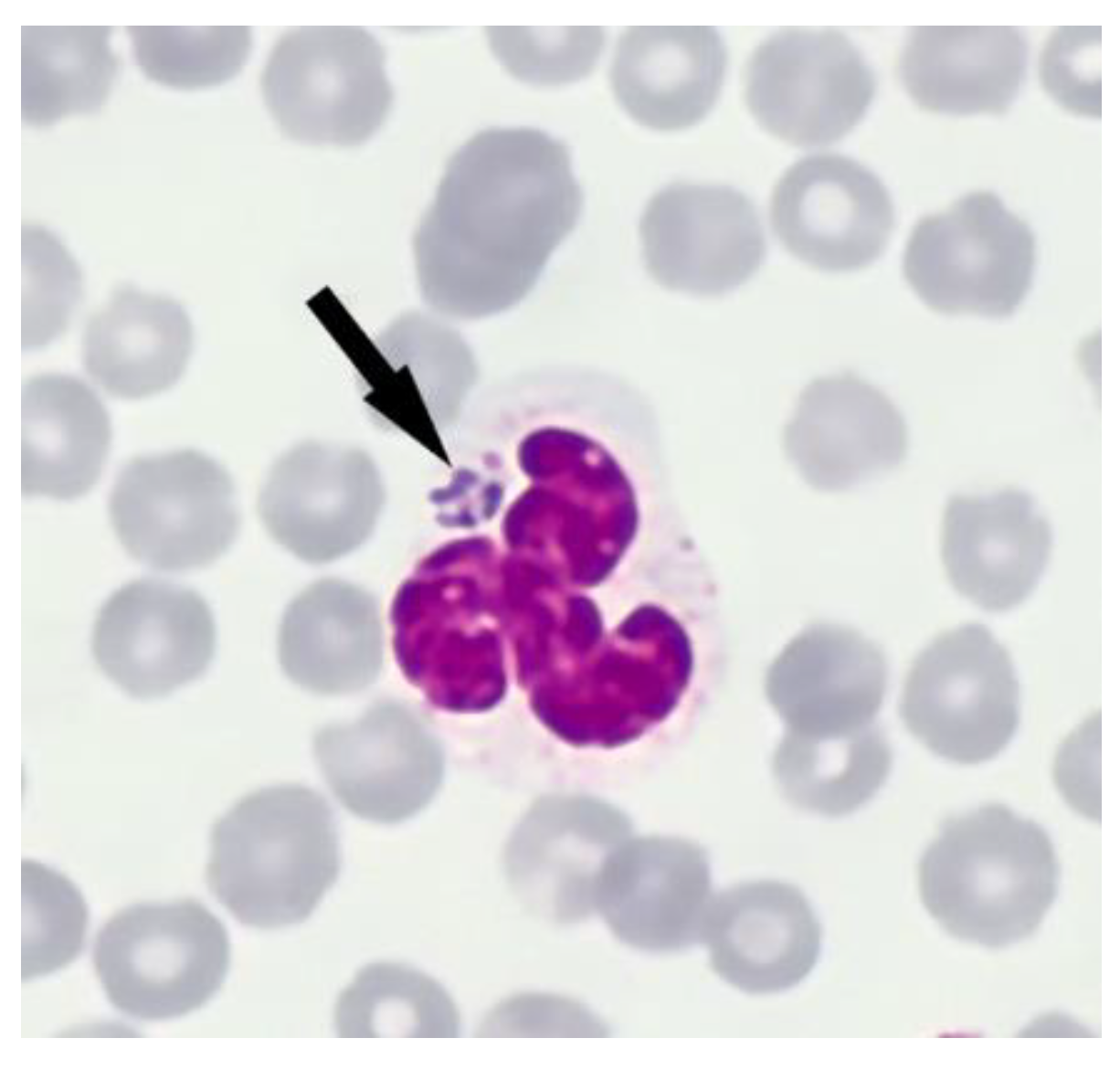
2.3.2. Detection by Microscopical Evaluation of Blood Smears
2.3.3. DNA Detection in Blood and in Ticks
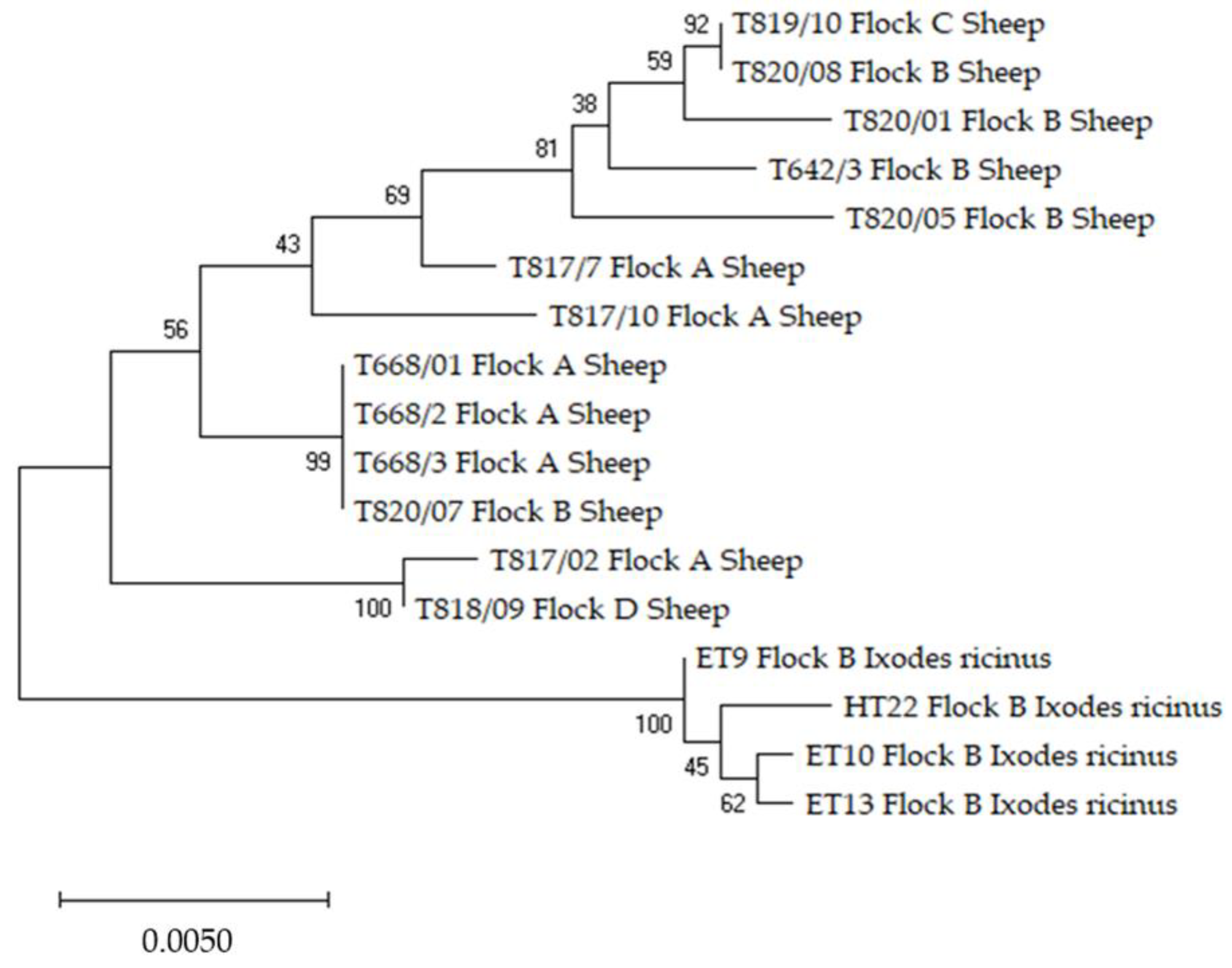
Anaplasma phagocytophilum and Anaplasma ovis Sequencing
2.4. Statistical Outcomes
3. Discussion
4. Materials and Methods
4.1. Animals’ History, Clinical Examination and Sampling
4.2. Clinicopathological and Fecal Parasitological Examinations
4.2.1. Complete Blood Cell Count and Microscopical Examinations of Blood Smears
4.2.2. Biochemical Profile
4.2.3. Fecal Parasitological Examination
4.3. Ticks Collection, Identification and Storage
4.4. Anaplasma phagocytophilum and Anaplasma ovis Laboratory Investigations
4.4.1. Antibody Detection
4.4.2. Microscopical Detection in Blood Smears
4.4.3. DNA Detection in Blood and Ticks
DNA Extraction
Real Time PCR
Nested PCR of Real Time Anaplasma ovis Postive Blood Samples
Sequence Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kauffmann, M.; Rehbein, S.; Hamel, D.; Lutz, W.; Heddergott, M.; Pfister, K.; Silaghi, C. Anaplasma phagocytophilum and Babesia spp. in roe deer (Capreolus capreolus), fallow deer (Dama dama) and mouflon (Ovis musimon) in Germany. Mol. Cell. Probes 2017, 31, 46–54. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, J.; Ruiz-Fons, F.; Naranjo, V.; Torina, A.; Rodríguez, O.; Gortázar, C. Evidence of Anaplasma infections in European roe deer (Capreolus capreolus) from southern Spain. Res. Vet. Sci. 2008, 84, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Scharf, W.; Schauer, S.; Freyburger, F.; Petrovec, M.; Schaarschmidt-Kiener, D.; Liebisch, G.; Runge, M.; Ganter, M.; Kehl, A.; Dumler, J.S.; et al. Distinct host species correlate with Anaplasma phagocytophilum ankA gene clusters. J. Clin. Microbiol. 2011, 49, 790–796. [Google Scholar] [CrossRef] [Green Version]
- Víchová, B.; Majláthová, V.; Nováková, M.; Stanko, M.; Hviščová, I.; Pangrácová, L.; Chrudimský, T.; Čurlík, J.; Peťko, B. Anaplasma infections in ticks and reservoir host from Slovakia. Infect. Genet. Evol. 2014, 22, 265–272. [Google Scholar] [CrossRef]
- Stuen, S. Haemoparasites in small ruminants in European countries: Challenges and clinical relevance. Small Rumin. Res. 2016, 142, 22–27. [Google Scholar] [CrossRef]
- Matei, I.A.; Estrada-Peña, A.; Cutler, S.J.; Vayssier-Taussat, M.; Varela-Castro, L.; Potkonjak, A.; Zeller, H.; Mihalca, A.D. A review on the eco-epidemiology and clinical management of human granulocytic anaplasmosis and its agent in Europe. Parasites Vectors 2019, 12, 599. [Google Scholar] [CrossRef]
- Stuen, S.; Granquist, E.; Silaghi, C. Anaplasma phagocytophilum—A widespread multi-host pathogen with highly adaptive strategies. Front. Cell. Infect. Microbiol. 2013, 3, 31. [Google Scholar] [CrossRef] [Green Version]
- Choi, K.S.; Garyu, J.; Park, J.; Dumler, J.S. Diminished adhesion of Anaplasma phagocytophilum-infected neutrophils to endothelial cells is associated with reduced expression of leukocyte surface selectin. Infect. Immun. 2003, 71, 4586–4594. [Google Scholar] [CrossRef] [Green Version]
- Stuen, S.; Van De Pol, I.; Bergström, K.; Schouls, L.M. Identification of Anaplasma phagocytophila (formerly Ehrlichia phagocytophila) variants in blood from sheep in Norway. J. Clin. Microbiol. 2002, 40, 3192–3197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almazán, C.; Fourniol, L.; Rouxel, C.; Alberdi, P.; Gandoin, C.; Lagrée, A.-C.; Boulouis, H.-J.; de la Fuente, J.; Bonnet, S.I. Experimental Ixodes ricinus-sheep cycle of Anaplasma phagocytophilum NV2Os propagated in tick cell cultures. Front. Vet. Sci. 2020, 7, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grøva, L.; Olesen, I.; Steinshamn, H.; Stuen, S. Prevalence of Anaplasma phagocytophilum infection and effect on lamb growth. Acta Vet. Scand. 2011, 53, 30. [Google Scholar] [CrossRef] [Green Version]
- Stuen, S.; Bergström, K.; Palmér, E. Reduced weight gain due to subclinical Anaplasma phagocytophilum (Formerly Ehrlichia phagocytophila) infection. Exp. Appl. Acarol. 2002, 28, 209–215. [Google Scholar] [CrossRef]
- Stuen, S.; Scharf, W.; Schauer, S.; Freyburger, F.; Bergström, K.; von Loewenich, F.D. Experimental infection in lambs with a red deer (Cervus elaphus) isolate of Anaplasma phagocytophilum. J. Wildl. Dis. 2010, 46, 803–809. [Google Scholar] [CrossRef]
- Gokce, H.I.; Woldehiwet, Z. Differential haematological effects of tick-borne fever in sheep and goats. Zentralbl. Veterinarmed. B 1999, 46, 105–115. [Google Scholar] [CrossRef]
- Daniel, R.G.; Carson, A.; Evans, C.; Cookson, R.; Wessels, M. Pathological observations of tick-borne fever and intercurrent bacterial infections in lambs. Vet. Rec. Case Rep. 2016, 4, e000367. [Google Scholar] [CrossRef]
- Overås, J.; Lund, A.; Ulvund, M.J.; Waldeland, H. Tick-borne fever as a possible predisposing factor in septicaemic pasteurellosis in lambs. Vet. Rec. 1993, 133, 398. [Google Scholar] [CrossRef]
- Sargison, N.; Edwards, G. Tick infestations in sheep in the UK. Practice 2009, 31, 58–65. [Google Scholar] [CrossRef]
- Jiménez, C.; Benito, A.; Arnal, J.L.; Ortín, A.; Gómez, M.; López, A.; Villanueva-Saz, S.; Lacasta, D. Anaplasma ovis in sheep: Experimental infection, vertical transmission and colostral immunity. Small Rumin. Res. 2019, 178, 7–14. [Google Scholar] [CrossRef]
- Pereira, A.; Parreira, R.; Nunes, M.; Casadinho, A.; Vieira, M.L.; Campino, L.; Maia, C. Molecular detection of tick-borne bacteria and protozoa in cervids and wild boars from Portugal. Parasites Vectors 2016, 9, 251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornok, S.; Elek, V.; de la Fuente, J.; Naranjo, V.; Farkas, R.; Majoros, G.; Földvári, G. First serological and molecular evidence on the endemicity of Anaplasma ovis and A. marginale in Hungary. Vet. Microbiol. 2007, 122, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Chochlakis, D.; Ioannou, I.; Tselentis, Y.; Psaroulaki, A. Human anaplasmosis and Anaplasma ovis variant. Emerg. Infect. Dis. 2010, 16, 1031. [Google Scholar] [CrossRef]
- Renneker, S.; Abdo, J.; Salih, D.E.; Karagenç, T.; Bilgiç, H.; Torina, A.; Oliva, A.G.; Campos, J.; Kullmann, B.; Ahmed, J.; et al. Can Anaplasma ovis in small ruminants be neglected any longer? Transbound. Emerg. Dis. 2013, 60 (Suppl. S2), 105–112. [Google Scholar] [CrossRef] [PubMed]
- Derdáková, M.; Štefančíková, A.; Špitalská, E.; Tarageľová, V.; Košťálová, T.; Hrkľová, G.; Kybicová, K.; Schánilec, P.; Majláthová, V.; Várady, M.; et al. Emergence and genetic variability of Anaplasma species in small ruminants and ticks from Central Europe. Vet. Microbiol. 2011, 153, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Friedhoff, K.T. Tick-borne diseases of sheep and goats caused by Babesia, Theileria or Anaplasma spp. Parassitologia 1997, 39, 99–109. [Google Scholar]
- Hornok, S.; de la Fuente, J.; Biró, N.; Fernández de Mera, I.G.; Meli, M.L.; Elek, V.; Gönczi, E.; Meili, T.; Tánczos, B.; Farkas, R. First molecular evidence of Anaplasma ovis and Rickettsia spp. in keds (Diptera: Hippoboscidae) of sheep and wild ruminants. Vector Borne Zoonotic Dis. 2011, 11, 1319–1321. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.X.; Wang, Y.; Li, Y.; Han, S.Y.; Wang, B.; Yuan, G.H.; Zhang, P.Y.; Yang, Z.W.; Wang, S.L.; Chen, J.Y.; et al. Vector-borne pathogens with veterinary and public health significance in Melophagus ovinus (Sheep Ked) from the Qinghai-Tibet Plateau. Pathogens 2021, 10, 249. [Google Scholar] [CrossRef]
- Lacasta, D.; Ferrer, L.M.; Sanz, S.; Labanda, R.; González, J.M.; Benito, A.; Ruiz, H.; Rodríguez-Largo, A.; Ramos, J.J. Anaplasmosis outbreak in lambs: First report causing carcass condemnation. Animals 2020, 10, 1851. [Google Scholar] [CrossRef]
- Torina, A.; Galindo, R.C.; Vicente, J.; Di Marco, V.; Russo, M.; Aronica, V.; Fiasconaro, M.; Scimeca, S.; Alongi, A.; Caracappa, S.; et al. Characterization of Anaplasma phagocytophilum and A. ovis infection in a naturally infected sheep flock with poor health condition. Trop. Anim. Health Prod. 2010, 42, 1327–1331. [Google Scholar] [CrossRef] [Green Version]
- Yasini, S.P.; Khaki, Z.; Rahbari, S.; Kazemi, B.; Salar Amoli, J.; Gharabaghi, A.; Jalali, S.M. Hematologic and clinical aspects of experimental ovine anaplasmosis caused by Anaplasma ovis in Iran. Iran. J. Parasitol. 2012, 7, 91–98. [Google Scholar]
- Battilani, M.; De Arcangeli, S.; Balboni, A.; Dondi, F. Genetic diversity and molecular epidemiology of Anaplasma. Infect. Genet. Evol. 2017, 49, 195–211. [Google Scholar] [CrossRef]
- Jaarsma, R.I.; Sprong, H.; Takumi, K.; Kazimirova, M.; Silaghi, C.; Mysterud, A.; Rudolf, I.; Beck, R.; Földvári, G.; Tomassone, L.; et al. Anaplasma phagocytophilum evolves in geographical and biotic niches of vertebrates and ticks. Parasites Vectors 2019, 12, 328. [Google Scholar] [CrossRef]
- Lagrée, A.C.; Rouxel, C.; Kevin, M.; Dugat, T.; Girault, G.; Durand, B.; Pfeffer, M.; Silaghi, C.; Nieder, M.; Boulouis, H.J.; et al. Co-circulation of different A. phagocytophilum variants within cattle herds and possible reservoir role for cattle. Parasites Vectors 2018, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Langenwalder, D.B.; Silaghi, C.; Nieder, M.; Pfeffer, M.; von Loewenich, F.D. Co-infection, reinfection and superinfection with Anaplasma phagocytophilum strains in a cattle herd based on ankA gene and multilocus sequence typing. Parasites Vectors 2020, 13, 157. [Google Scholar] [CrossRef]
- Silaghi, C.; Liebisch, G.; Pfister, K. Genetic variants of Anaplasma phagocytophilum from 14 equine granulocytic anaplasmosis cases. Parasites Vectors 2011, 4, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silaghi, C.; Nieder, M.; Sauter-Louis, C.; Knubben-Schweizer, G.; Pfister, K.; Pfeffer, M. Epidemiology, genetic variants and clinical course of natural infections with Anaplasma phagocytophilum in a dairy cattle herd. Parasites Vectors 2018, 11, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silaghi, C.; Hamel, D.; Thiel, C.; Pfister, K.; Passos, L.M.; Rehbein, S. Genetic variants of Anaplasma phagocytophilum in wild caprine and cervid ungulates from the Alps in Tyrol, Austria. Vector Borne Zoonotic Dis. 2011, 11, 355–362. [Google Scholar] [CrossRef]
- Ladbury, G.A.F.; Stuen, S.; Thomas, R.; Bown, K.J.; Woldehiwet, Z.; Granquist, E.G.; Bergström, K.; Birtles, R.J. Dynamic transmission of numerous Anaplasma phagocytophilum genotypes among lambs in an infected sheep flock in an area of anaplasmosis endemicity. J. Clin. Microbiol. 2008, 46, 1686–1691. [Google Scholar] [CrossRef] [Green Version]
- Drehmann, M.; Springer, A.; Lindau, A.; Fachet, K.; Mai, S.; Thoma, D.; Schneider, C.R.; Chitimia-Dobler, L.; Bröker, M.; Dobler, G.; et al. The spatial distribution of Dermacentor ticks (Ixodidae) in Germany—Evidence of a continuing spread of Dermacentor reticulatus. Front. Vet. Sci. 2020, 7, 578220. [Google Scholar] [CrossRef]
- Overzier, E.; Pfister, K.; Herb, I.; Mahling, M.; Böck, G., Jr.; Silaghi, C. Detection of tick-borne pathogens in roe deer (Capreolus capreolus), in questing ticks (Ixodes ricinus), and in ticks infesting roe deer in southern Germany. Ticks Tick Borne Dis. 2013, 4, 320–328. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huo, B.; Wu, T.; Song, C.; Shen, X. Studies of selenium deficiency in the Wumeng semi-fine wool sheep. Biol. Trace Element Res. 2019, 194, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Alam, R.T.M.; Hassanen, E.A.A.; El-Mandrawy, S.A.M. Heamonchus contortus infection in sheep and goats: Alterations in haematological, biochemical, immunological, trace element and oxidative stress markers. J. Appl. Anim. Res. 2020, 48, 357–364. [Google Scholar] [CrossRef]
- Chapman, H. The effects of natural and artificially acquired infections of coccidia in lambs. Res. Vet. Sci. 1974, 16, 1–6. [Google Scholar] [CrossRef]
- Woldehiwet, Z. The natural history of Anaplasma phagocytophilum. Vet. Parasitol. 2010, 167, 108–122. [Google Scholar] [CrossRef] [PubMed]
- Chirek, A.; Silaghi, C.; Pfister, K.; Kohn, B. Granulocytic anaplasmosis in 63 dogs: Clinical signs, laboratory results, therapy and course of disease. J. Small Anim. Pract. 2018, 59, 112–120. [Google Scholar] [CrossRef] [Green Version]
- Franzén, P.; Aspan, A.; Egenvall, A.; Gunnarsson, A.; Aberg, L.; Pringle, J. Acute clinical, hematologic, serologic, and polymerase chain reaction findings in horses experimentally infected with a European strain of Anaplasma phagocytophilum. J. Vet. Intern. Med. 2005, 19, 232–239. [Google Scholar] [CrossRef]
- Foster, W.N.M.; Cameron, A.E. Thrombocytopenia in sheep associated with experimental tick-borne fever infection. J. Comp. Pathol. 1968, 78, 251–254. [Google Scholar] [CrossRef]
- Rankins, D.L.; Pugh, D.G. Feeding and nutrition. In Sheep and Goat Medicine, 2nd ed.; Pugh, D.G., Baird, A.N., Eds.; Elsevier Saunders: Maryland Heights, USA, MO, 2012; pp. 18–49. [Google Scholar]
- Polizopoulou, Z. Haematological tests in sheep health management. Small Rumin. Res. 2010, 92, 88–91. [Google Scholar] [CrossRef]
- Katsogiannou, E.G.; Athanasiou, L.V.; Christodoulopoulos, G.; Polizopoulou, Z.S. Diagnostic approach of anemia in ruminants. J. Hell. Vet. Med Soc. 2018, 69, 1033–1046. [Google Scholar] [CrossRef] [Green Version]
- Plummer, P.J.; Plummer, C.L.; Still, K.M. Diseases of the respiratory system. In Sheep and Goat Medicine, 2nd ed.; Pugh, D.G., Baird, A.N., Eds.; Elsevier Saunders: Maryland Heights, MO, USA, 2012; pp. 126–149. [Google Scholar]
- Siska, W.D.; Tuttle, R.E.; Messick, J.B.; Bisby, T.M.; Toth, B.; Kritchevsky, J.E. Clinicopathologic characterization of six cases of equine granulocytic anaplasmosis in a nonendemic area (2008–2011). J. Equine Vet. Sci. 2013, 33, 653–657. [Google Scholar] [CrossRef]
- Chow, C.K.; Chen, C.J. Dietary selenium and age-related susceptibility of rat erythrocytes to oxidative damage. J. Nutr. 1980, 110, 2460–2466. [Google Scholar] [CrossRef] [Green Version]
- Nagababu, E.; Chrest, F.J.; Rifkind, J.M. Hydrogen-peroxide-induced heme degradation in red blood cells: The protective roles of catalase and glutathione peroxidase. Biochim. et Biophys. Acta (BBA) Gen. Subj. 2003, 1620, 211–217. [Google Scholar] [CrossRef]
- Kümper, H. Hypericum poisoning in sheep. Tierarztl. Prax. 1989, 17, 257–261. [Google Scholar]
- Kako, M.D.; al-Sultan, I.I.; Saleem, A.N. Studies of sheep experimentally poisoned with Hypericum perforatum. Vet. Hum. Toxicol. 1993, 35, 298–300. [Google Scholar]
- Navarre, C.B.; Baird, A.; Pugh, D. Diseases of the gastrointestinal system. In Sheep and Goat Medicine, 2nd ed.; Pugh, D.G., Baird, A.N., Eds.; Elsevier Saunders: Maryland Heights, MO, USA, 2012; pp. 71–105. [Google Scholar]
- Mason, K.L.; Gonzalez, M.V.; Chung, C.; Mousel, M.R.; White, S.N.; Taylor, J.B.; Scoles, G.A. Validation of an improved Anaplasma antibody competitive ELISA for detection of Anaplasma ovis antibody in domestic sheep. J. Vet. Diagn. Investig. 2017, 29, 763–766. [Google Scholar] [CrossRef] [Green Version]
- Shabana, I.I.; Alhadlag, N.M.; Zaraket, H. Diagnostic tools of caprine and ovine anaplasmosis: A direct comparative study. BMC Vet. Res. 2018, 14, 165. [Google Scholar] [CrossRef]
- Kocan, K.M.; Busby, A.T.; Allison, R.W.; Breshears, M.A.; Coburn, L.; Galindo, R.C.; Ayllón, N.; Blouin, E.F.; de la Fuente, J. Sheep experimentally infected with a human isolate of Anaplasma phagocytophilum serve as a host for infection of Ixodes scapularis ticks. Ticks Tick Borne Dis. 2012, 3, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Dreher, U.M.; de la Fuente, J.; Hofmann-Lehmann, R.; Meli, M.L.; Pusterla, N.; Kocan, K.M.; Woldehiwet, Z.; Braun, U.; Regula, G.; Staerk, K.D.C.; et al. Serologic cross-reactivity between Anaplasma marginale and Anaplasma phagocytophilum. Clin. Diagn. Lab. Immunol. 2005, 12, 1177–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorman, J.K.; Hoar, B.R.; Nieto, N.C.; Foley, J.E. Evaluation of Anaplasma phagocytophilum infection in experimentally inoculated sheep and determination of Anaplasma spp. seroprevalence in 8 free-ranging sheep flocks in California and Oregon. Am. J. Vet. Res. 2012, 73, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Stuen, S.; Bergström, K.; Petrovec, M.; Van de Pol, I.; Schouls, L.M. Differences in clinical manifestations and hematological and serological responses after experimental infection with genetic variants of Anaplasma phagocytophilum in sheep. Clin. Diagn. Lab. Immunol. 2003, 10, 692–695. [Google Scholar] [CrossRef] [Green Version]
- Thomas, R.; Birtles, R.; Radford, A.; Woldehiwet, Z. Recurrent bacteraemia in sheep infected persistently with Anaplasma phagocytophilum. J. Comp. Pathol. 2012, 147, 360–367. [Google Scholar] [CrossRef]
- Kiilerich, A.M.; Christensen, H.; Thamsborg, S.M. Anaplasma phagocytophilum in Danish sheep: Confirmation by DNA sequencing. Acta Vet. Scand. 2009, 51, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuen, S.; Pettersen, K.S.; Granquist, E.G.; Bergström, K.; Bown, K.; Birtles, R. Anaplasma phagocytophilum variants in sympatric red deer (Cervus elaphus) and sheep in southern Norway. Ticks Tick-borne Dis. 2013, 4, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Cabezas-Cruz, A.; Gallois, M.; Fontugne, M.; Allain, E.; Denoual, M.; Moutailler, S.; Devillers, E.; Zientara, S.; Memmi, M.; Chauvin, A. Epidemiology and genetic diversity of Anaplasma ovis in goats in Corsica, France. Parasites Vectors 2019, 12, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacasta, D.; Lorenzo, M.; González, J.M.; Ruiz de Arcaute, M.; Benito, A.; Baselga, C.; Milian, M.E.; Lorenzo, N.; Jiménez, C.; Villanueva-Saz, S.; et al. Epidemiological study related to the first outbreak of ovine anaplasmosis in Spain. Animals 2021, 11, 2036. [Google Scholar] [CrossRef]
- Silaghi, C.; Fröhlich, J.; Reindl, H.; Hamel, D.; Rehbein, S. Anaplasma phagocytophilum and Babesia species of sympatric roe deer (Capreolus capreolus), fallow deer (Dama dama), sika deer (Cervus nippon) and red deer (Cervus elaphus) in Germany. Pathogens 2020, 9, 968. [Google Scholar] [CrossRef]
- Tegtmeyer, P.; Ganter, M.; von Loewenich, F.D. Simultaneous infection of cattle with different Anaplasma phagocytophilum variants. Ticks Tick-borne Dis. 2019, 10, 1051–1056. [Google Scholar] [CrossRef]
- Overzier, E.; Pfister, K.; Thiel, C.; Herb, I.; Mahling, M.; Silaghi, C. Anaplasma phagocytophilum in questing Ixodes ricinus ticks: Comparison of prevalences and partial 16S rRNA gene variants in urban, pasture, and natural habitats. Appl. Environ. Microbiol. 2012, 79, 1730–1734. [Google Scholar] [CrossRef] [Green Version]
- Schorn, S.; Pfister, K.; Reulen, H.; Mahling, M.; Manitz, J.; Thiel, C.; Silaghi, C. Prevalence of Anaplasma phagocytophilum in Ixodes ricinus in Bavarian public parks, Germany. Ticks Tick-borne Dis. 2011, 2, 196–203. [Google Scholar] [CrossRef]
- Silaghi, C.; Kohn, B.; Chirek, A.; Thiel, C.; Nolte, I.; Liebisch, G.; Pfister, K. Relationship of molecular and clinical findings on Anaplasma phagocytophilum involved in natural infections of dogs. J. Clin. Microbiol. 2011, 49, 4413–4414. [Google Scholar] [CrossRef] [Green Version]
- Adamska, M. The role of different species of wild ungulates and Ixodes ricinus ticks in the circulation of genetic variants of Anaplasma phagocytophilum in a forest biotope in north-western Poland. Ticks Tick-borne Dis. 2020, 11, 101465. [Google Scholar] [CrossRef] [PubMed]
- Jahfari, S.; Coipan, E.C.; Fonville, M.; van Leeuwen, A.D.; Hengeveld, P.; Heylen, D.; Heyman, P.; van Maanen, C.; Butler, C.M.; Földvári, G.; et al. Circulation of four Anaplasma phagocytophilum ecotypes in Europe. Parasites Vectors 2014, 7, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrovec, M.; Sumner, J.W.; Nicholson, W.L.; Childs, J.E.; Strle, F.; Barlicč, J.; Lotricč-Furlan, S.; Županc, T.A. Identity of ehrlichial DNA sequences derived from Ixodes ricinus ticks with those obtained from patients with human granulocytic ehrlichiosis in Slovenia. J. Clin. Microbiol. 1999, 37, 209–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liz, J.S.; Sumner, J.W.; Pfister, K.; Brossard, M. PCR detection and serological evidence of granulocytic ehrlichial infection in roe deer (Capreolus capreolus) and chamois (Rupicapra rupicapra). J. Clin. Microbiol. 2002, 40, 892–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrovec, M.; Bidovec, A.; Sumner, J.W.; Nicholson, W.L.; Childs, J.E.; Avsic-Zupanc, T. Infection with Anaplasma phagocytophila in cervids from Slovenia: Evidence of two genotypic lineages. Wien. Klin. Wochenschr. 2002, 114, 641–647. [Google Scholar] [PubMed]
- Rar, V.A.; Epikhina, T.I.; Livanova, N.N.; Panov, V.V.; Doroschenko, E.K.; Pukhovskaya, N.M.; Vysochina, N.P.; Ivanov, L.I. Genetic variability of Anaplasma phagocytophilum in Ixodes persulcatus ticks and small mammals in the Asian part of Russia. Vector Borne Zoonotic Dis. 2011, 11, 1013–1021. [Google Scholar] [CrossRef]
- de la Fuente, J.; Bussche, R.A.V.D.; Kocan, K.M. Molecular phylogeny and biogeography of North American isolates of Anaplasma marginale (Rickettsiaceae: Ehrlichieae). Vet. Parasitol. 2001, 97, 65–76. [Google Scholar] [CrossRef]
- Chartier, C.; Paraud, C. Coccidiosis due to Eimeria in sheep and goats, a review. Small Rumin. Res. 2012, 103, 84–92. [Google Scholar] [CrossRef]
- Sargison, N. Sheep Flock Health: A Planned Approach; Blackwell Publishing: Oxford, UK, 2008; pp. 143–302. [Google Scholar]
- Sargison, N.; Scott, P. The implementation and value of diagnostic procedures in sheep health management. Small Rumin. Res. 2010, 92, 2–9. [Google Scholar] [CrossRef]
- West, D.M.; Bruère, A.N.; Ridler, A.L. The Sheep—Health, Disease and Production, 4th ed.; Massey University Press: Auckland, New Zealand, 2018; p. 407. [Google Scholar]
- Bath, G.F.; van Wyk, J.A. The Five Point Check© for targeted selective treatment of internal parasites in small ruminants. Small Rumin. Res. 2009, 86, 6–13. [Google Scholar] [CrossRef]
- Piaton, E.; Fabre, M.; Goubin-Versini, I.; Bretz-Grenier, M.; Courtade-Saïdi, M.; Vincent, S.; Belleannée, G.; Thivolet, F.; Boutonnat, J.; Debaque, H.; et al. Guidelines for May-Grünwald-Giemsa staining in haematology and non-gynaecological cytopathology: Recommendations of the French Society of Clinical Cytology (SFCC) and of the French Association for Quality Assurance in Anatomic and Cytologic Pathology (AFAQAP). Cytopathology 2016, 27, 359–368. [Google Scholar] [CrossRef]
- Lepherd, M.; Canfield, P.; Hunt, G.; Bosward, K. Haematological, biochemical and selected acute phase protein reference intervals for weaned female Merino lambs. Aust. Vet. J. 2009, 87, 5–11. [Google Scholar] [CrossRef]
- Ganter, M. Referenzwerte. In Klinik der Schaf-und Ziegenkrankheiten; Bostedt, H., Ganter, M., Hiepe, T., Eds.; Georg Thieme Verlag KG: Stuttgart, Germany, 2019; pp. 688–697. [Google Scholar]
- World Health Organization. Haemoglobin Concentrations for the Diagnosis of Anaemia and Assessment of Severity; World Health Organization: Geneva, Switzerland, 2011; Available online: https://apps.who.int/iris/handle/10665/85839 (accessed on 24 September 2021).
- Humann-Ziehank, E.; Ganter, M.; Michalke, B. Selenium speciation in paired serum and cerebrospinal fluid samples of sheep. J. Trace Elements Med. Biol. 2016, 33, 14–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puls, R. Mineral Levels in Animal Health: Diagnostic Data, 2nd ed.; Sherpa International: Clearbrook, Canada, 1994. [Google Scholar]
- Eckert, J.; Braun, R.; Shirley, M.W.; Coudert, P. Guidelines on Techniques in Coccidiosis Research; COST 89/820; European Commission, DGXII: Brussels, Belgium, 1995; pp. 103–117. [Google Scholar]
- Estrada-Peña, A.; Bouattour, A.; Camicas, J.-L.; Walker, A.R. Ticks of Domestic Animals in the Mediterranean Region: A Guide to Identification of Species; University of Zaragoza: Zaragoza, Spain, 2004. [Google Scholar]
- Stuen, S.; Bergström, K. Serological investigation of granulocytic Ehrlichia infection in sheep in Norway. Acta Vet. Scand. 2001, 42, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Courtney, J.W.; Kostelnik, L.M.; Zeidner, N.S.; Massung, R.F. Multiplex real-time PCR for detection of Anaplasma phagocytophilum and Borrelia burgdorferi. J. Clin. Microbiol. 2004, 42, 3164–3168. [Google Scholar] [CrossRef] [Green Version]
- Michelet, L.; Delannoy, S.; Devillers, E.; Umhang, G.; Aspan, A.; Juremalm, M.; Chirico, J.; van der Wal, F.J.; Sprong, H.; Boye Pihl, T.P.; et al. High-throughput screening of tick-borne pathogens in Europe. Front. Cell. Infect. Microbiol. 2014, 4, 103. [Google Scholar] [CrossRef]
- Yousefi, A. Phylogenetic analysis of Anaplasma marginale and Anaplasma ovis isolated from small ruminant based on MSP4 gene in western regions of Iran. Comp. Clin. Pathol. 2018, 27, 1161–1165. [Google Scholar] [CrossRef]
- Massung, R.F.; Slater, K.; Owens, J.H.; Nicholson, W.L.; Mather, T.N.; Solberg, V.B.; Olson, J.G. Nested PCR assay for detection of granulocytic ehrlichiae. J. Clin. Microbiol. 1998, 36, 1090–1095. [Google Scholar] [CrossRef] [Green Version]
- de la Fuente, J.; Massung, R.F.; Wong, S.J.; Chu, F.K.; Lutz, H.; Meli, M.; von Loewenich, F.D.; Grzeszczuk, A.; Torina, A.; Caracappa, S.; et al. Sequence analysis of the msp4 gene of Anaplasma phagocytophilum strains. J. Clin. Microbiol. 2005, 43, 1309–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberti, A.; Zobba, R.; Chessa, B.; Addis, M.F.; Sparagano, O.; Pinna Parpaglia, M.L.; Cubeddu, T.; Pintori, G.; Pittau, M. Equine and canine Anaplasma phagocytophilum strains isolated on the island of Sardinia (Italy) are phylogenetically related to pathogenic strains from the United States. Appl. Environ. Microbiol. 2005, 71, 6418–6422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Flock | Collection Date | Sample ID | cELISA | IFAT | A. phagocytophilum qPCR | A. ovis qPCR |
|---|---|---|---|---|---|---|
| A | May 2020 | T668.1 ewe | 81.7 | ≥1:640 | 23.5 | 33.3 |
| T668.2 lamb | 46.9 | 1:40 | 17.9 | - | ||
| T668.3 lamb | 52.1 | 1:320 | 21.5 | - | ||
| July 2020 | T817.1 lamb | 92.4 | ≥1:640 | - | 23.3 | |
| T817.2 lamb | 23.1 | ≥1:640 | 33.9 | - | ||
| T817.3 lamb | 64.8 | ≥1:640 | 33.1 | - | ||
| T817.4 lamb | 25.2 | 1:40 | - | - | ||
| T817.5 lamb | 84 | 1:320 | - | 29.3 | ||
| T817.6 lamb | 51.6 | 1:80 | - | - | ||
| T817.7 lamb | 86.6 | 1:160 | 25.4 | - | ||
| T817.8 lamb | 22.1 | ≥1:640 | - | - | ||
| T817.9 lamb | 44.8 | 1:320 | - | - | ||
| T817.10 lamb | 71.3 | ≥1:640 | 26.0 | - | ||
| T817.11 ewe | 94.1 | ≥1:640 | - | 20.6 | ||
| T817.12 ewe | 87.6 | ≥1:640 | - | - | ||
| T817.13 ewe | 92.6 | 1:160 | - | 21 | ||
| T817.14 lamb | 42 | 1:40 | - | - | ||
| T817.15 lamb | 31.5 | 1:160 | - | - | ||
| November 2020 | T1339 ram | n/a | n/a | - | 18 | |
| B | May 2020 | T642.1 lamb | −8.9 | 1:320 | 36.2 | - |
| T642.2 lamb | −2.9 | - | - | - | ||
| T642.3 lamb | −11.1 | 1:140 | 23.5 | - | ||
| T642.4 lamb | −14.5 | - | - | - | ||
| T642.5 lamb | −48.2 | - | - | |||
| T642.8 ewe | 54.6 | 1:160 | - | - | ||
| T642.9 lamb | −5.1 | - | - | - | ||
| T642.10 lamb | −4.4 | - | - | - | ||
| July 2020 | T820.1 lamb | 59.9 | ≥1:640 | 37.0 | - | |
| T820.2 lamb | −0.7 | - | - | - | ||
| T820.3 lamb | 48.5 | ≥1:640 | - | - | ||
| T820.4 lamb | 12.9 | - | - | - | ||
| T820.5 lamb | 81 | ≥1:640 | 29.1 | - | ||
| T820.6 lamb | 55.9 | ≥1:640 | - | - | ||
| T820.7 lamb | 60.2 | ≥1:640 | 20.8 | - | ||
| T820.8 lamb | 77.1 | ≥1:640 | 21.7 * | - | ||
| T820.9 lamb | 63.9 | ≥1:640 | - | - | ||
| T820.10 lamb | 44.9 | 1:320 | 32.7 | - | ||
| C | July 2020 | T819.1 lamb | 38.6 | ≥1:640 | - | - |
| T819.2 lamb | 26.7 | - | - | - | ||
| T819.3 lamb | 22.9 | - | - | - | ||
| T819.4 lamb | 12.3 | - | - | - | ||
| T819.5 lamb | 59.1 | - | - | - | ||
| T819.6 lamb | 38.1 | - | - | - | ||
| T819.7 lamb | 19.2 | - | - | - | ||
| T819.8 lamb | 21 | - | - | - | ||
| T819.9 lamb | 7.2 | - | - | - | ||
| T819.10 lamb | 29.5 | ≥1:640 | 16.6 * | - | ||
| D | July 2020 | T818.1 lamb | 21.2 | - | - | - |
| T818.2 lamb | 36.3 | 1:40 | - | - | ||
| T818.3 lamb | 38.8 | ≥1:640 | - | - | ||
| T818.4 lamb | 27.5 | ≥1:640 | - | - | ||
| T818.5 lamb | 23 | ≥1:640 | - | - | ||
| T818.6 lamb | 12.3 | - | - | - | ||
| T818.7 lamb | 12.7 | - | - | - | ||
| T818.8 lamb | 18.6 | - | - | - | ||
| T818.9 lamb | 49.7 | ≥1:640 | 28.1 | - | ||
| T818.10 lamb | 15.5 | - | - | - | ||
| E | July 2020 | T921.1 lamb | 9.2 | - | - | - |
| T921.2 lamb | −4.5 | - | 35.5 | - | ||
| T921.3 lamb | −33.2 | - | - | - | ||
| T921.4 lamb | −9.5 | - | - | - | ||
| T921.5 lamb | 19.1 | - | - | - | ||
| T921.6 lamb | 13.8 | - | - | - | ||
| T921.7 lamb | 14.7 | - | - | - | ||
| T921.8 lamb | 31.9 | - | - | - | ||
| T921.9 lamb | 12.4 | - | - | - | ||
| T921.10 lamb | −11.1 | - | - | - |
| Flock | Sample ID | Sample Date | Cq-Value | 16S rRNA | groEL | msp4 |
|---|---|---|---|---|---|---|
| A | T668.1 ewe | May 2020 | 23.5 | 16S-16 (S) | g-35 | New (shorter than other sequences (25 nt at beginning) |
| T668.2 lamb | 17.9 | 16S-16 (S) | g-35 | new | ||
| T668.3 lamb | 21.5 | 16S-16 (S) | g-35 | new | ||
| T817.2 lamb | July 2020 | 33.9 | 16S-2 (B) | g-2 (B) | m4-20 | |
| T817.3 lamb | 33.1 | n/a | n/a | n/a | ||
| T817.7 lamb | 25.4 | 16S-16 (S) | g-13 | m4-18 | ||
| T817.10 lamb | 26.0 | 16S-20 (W) | new | new | ||
| B | T642.1 lamb | May 2020 | 36.2 | n/a | n/a | n/a |
| T642.3 lamb | 23.5 | 16S-16 (S) | new | new | ||
| T820.1 lamb | July 2020 | 37.0 | 16S-20 (W) | Not clear, ambiguous pos. 473 (W) | m4-5 (I) | |
| T820.5 lamb | 29.1 | 16S-16 (S) | g-24 | new | ||
| T820.7 lamb | 20.8 | 16S-16 (S) | g-35 | New (shorter than other sequences (25 nt at beginning) | ||
| T820.8 lamb | 21.7 | 16S-20 (W) | new | m4-5 (I) | ||
| T820.10 lamb | 32.7 | 16S-20 (W) | New, ambiguous pos. 515 (K) | n/a | ||
| C | T819.10 lamb | July 2020 | 16.6 | 16S-20 (W) | g-24 | m4-5 (I) |
| D | T818.9 lamb | July 2020 | 28.1 | 16S-2 (B) | g-2 (B) | m4-20 |
| E | T921.2 lamb | July 2020 | 35.5 | 16S-21 (X) | n/a | new |
| Flock | Sample ID | Sample Date | Cq-value | 16S rRNA | groEL | msp4 |
|---|---|---|---|---|---|---|
| B | ET-9 | July 2020 | 28 | 16S-21 (X) | new | m4-13 (N) |
| ET-10 | 26.6 | 16S-21 (X) | g-7 (G) | m4-13 (N) | ||
| ET-13 | 27.3 | 16S-21 (X) | new | m4-13 (N) | ||
| HT-22 | 23.2 | 16S-22 (Y) | g-4 (D) | m4-13 (N) | ||
| HT-24 | 29 | 16S-22 (Y) | n/a | new |
| Target Gene | Reaction | Sequence (5′-3′) | Amplicon Size (bp) | Annealing | Reference |
|---|---|---|---|---|---|
| Anaplasma phagocytophilum | |||||
| msp2 | qPCR | ApMSP2f: TGGAAGGTAGTGTTGGTTATGGTATT ApMSP2r: TTGGTCTTGAAGCGCTCGTA ApMSP2p: TGGTGCCAGGGTTGAGCTTGAGATTG | 77 | 60 °C | [96] |
| 16S rRNA | Nested PCR | First PCR: | [99] | ||
| Ge3a: CACATGCAAGTCGAACGGATTATTC | 932 | 55 °C | |||
| Ge10r: TTCCGTTAAGAAGGATCTAATCTCC | |||||
| Nested PCR *: | |||||
| Ge9f: AACGGATTATTCTTTATAGCTTGCT | 546 | 55 °C | |||
| Ge2: GGCAGTATTAAAAGCAGCTCCAGG | |||||
| msp4 | Nested PCR | First PCR: | [100] | ||
| Msp4AP5: ATGAATTACAGAGAATTGCTTGTAGG | 849 | 54 °C | |||
| Msp4AP3: TTAATTGAAAGCAAATCTTGCTCC | |||||
| TATG | |||||
| Nested PCR *: | |||||
| Msp4f: CTATTGGYGGNGCYAGAGT | |||||
| Msp4r: GTTCATCGAAAATTCCGTGGTA | 362 | 54 °C | |||
| groEL | Nested PCR | First PCR: | [101] | ||
| EphplgroEL-F: ATGGTATGCAGTTTGATCGC | 624 | 55 °C | |||
| EphplgroEL-R: TCTACTCTGTCTTTGCGTTC | |||||
| Nested PCR *: | |||||
| EphplgroEL-F: ATGGTATGCAGTTTGATCGC | 573 | 55 °C | |||
| EphgroEL-R: TTGAGTACAGCAACACCACCGGAA | |||||
| Anaplasma ovis | |||||
| msp4 | qPCR | A_ov_msp4_F: TCATTCGACATGCGTGAGTCAA_ov_msp4_R: TTTGCTGGCGCACTCACATCA_ov_msp4_P: AGCAGAGAGACCTCGTATGTTAGAGGC | 92 | 60 °C | [97] |
| msp4 | Nested PCR | First PCR: | [98] | ||
| M-OM F: GGGAGCTCCTATGAATTACAGAGAATTGTTTAC | 870 | 60 °C | |||
| M-OM R: CCGGATCCTTAGCTGAACAGGAATCTTGC | |||||
| Nested PCR *: | |||||
| M-OV F: TGAAGGGAGCGGGGTCATGGG | 346 | 60 °C | |||
| M-OV R: GGTAATTGCAGCCAGGGACTCT | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauer, B.U.; Răileanu, C.; Tauchmann, O.; Fischer, S.; Ambros, C.; Silaghi, C.; Ganter, M. Anaplasma phagocytophilum and Anaplasma ovis–Emerging Pathogens in the German Sheep Population. Pathogens 2021, 10, 1298. https://doi.org/10.3390/pathogens10101298
Bauer BU, Răileanu C, Tauchmann O, Fischer S, Ambros C, Silaghi C, Ganter M. Anaplasma phagocytophilum and Anaplasma ovis–Emerging Pathogens in the German Sheep Population. Pathogens. 2021; 10(10):1298. https://doi.org/10.3390/pathogens10101298
Chicago/Turabian StyleBauer, Benjamin Ulrich, Cristian Răileanu, Oliver Tauchmann, Susanne Fischer, Christina Ambros, Cornelia Silaghi, and Martin Ganter. 2021. "Anaplasma phagocytophilum and Anaplasma ovis–Emerging Pathogens in the German Sheep Population" Pathogens 10, no. 10: 1298. https://doi.org/10.3390/pathogens10101298
APA StyleBauer, B. U., Răileanu, C., Tauchmann, O., Fischer, S., Ambros, C., Silaghi, C., & Ganter, M. (2021). Anaplasma phagocytophilum and Anaplasma ovis–Emerging Pathogens in the German Sheep Population. Pathogens, 10(10), 1298. https://doi.org/10.3390/pathogens10101298







