The Prevalence and Serological Association of Hepatitis D Virus Genotypes in Taiwan
Abstract
:1. Introduction
2. Results
2.1. Baseline Characteristics of HBV/HDV Superinfected Patients
2.2. Factors Associated with HDV Genotypes in HBV/HDV Superinfection Patients
2.3. Comparison of the Distribution of Different Biomarkers in HDV/HBV Patients with HDV-IV and Non-HDV-IV Genotype
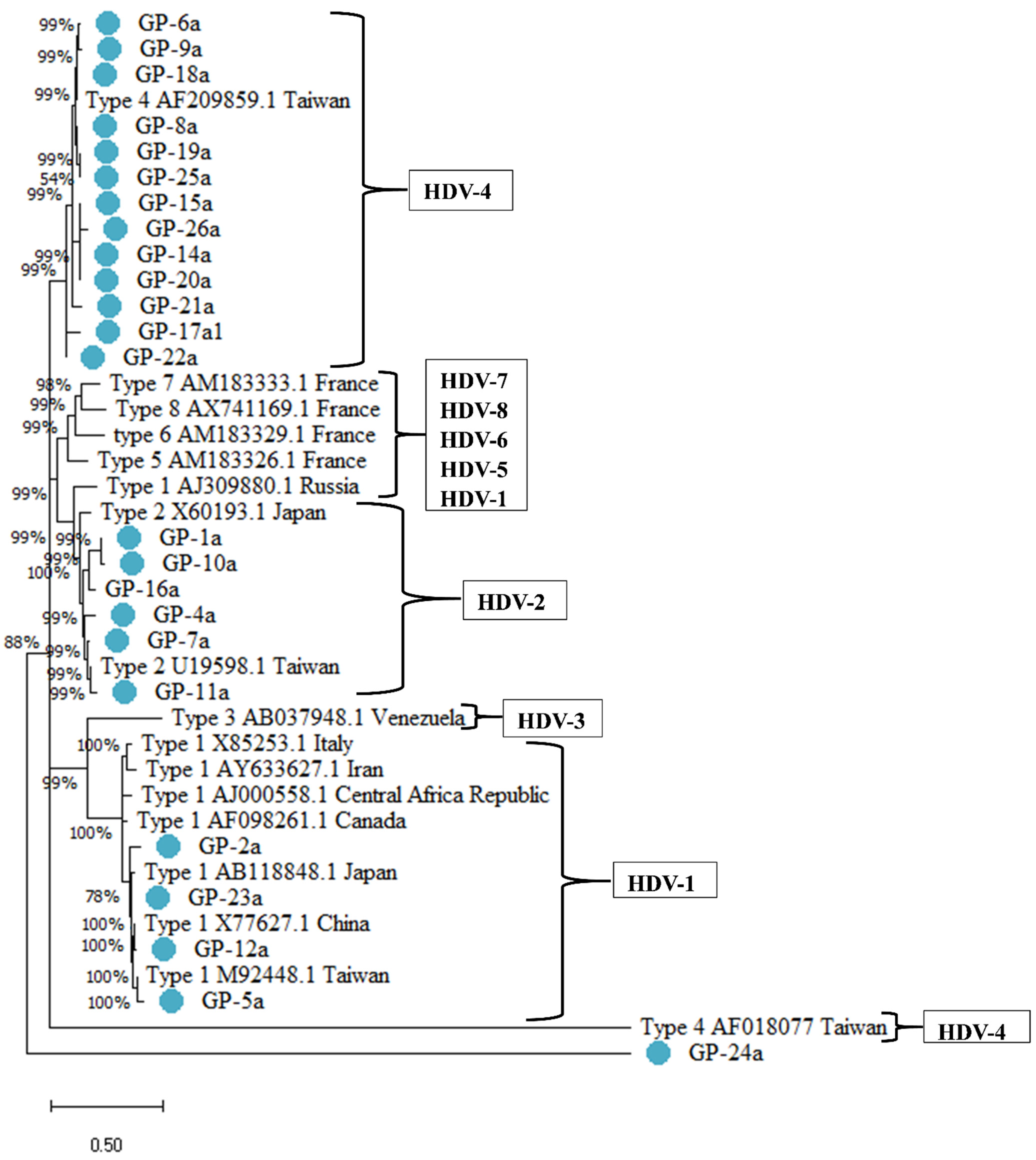
2.4. Distribution of HDV Genotypes among Baseline Factors by Group
3. Discussion
4. Materials and Methods
4.1. Study Subjects
4.2. Laboratory Analyses of HBV, HCV, HDV
4.3. HDV Genotyping
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Rizzetto, M.; Canese, M.G.; Arico, S.; Crivelli, O.; Trepo, C.; Bonino, F. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis b virus in liver and in serum of hbsag carriers. Gut 1977, 18, 997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfaiate, D.; Dény, P.; Durantel, D. Hepatitis delta virus: From biological and medical aspects to current and investigational therapeutic options. Antivir. Res. 2015, 122, 112–129. [Google Scholar] [CrossRef] [PubMed]
- Botelho-Souza, L.F.; Vasconcelos, M.; Santos, A.D.; Salcedo, J.; Vieira, D.S. Hepatitis delta: Virological and clinical aspects. Virol. J. 2017, 14, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mentha, N.; Clément, S.; Negro, F.; Alfaiate, D. A review on hepatitis D: From virology to new therapies. J. Adv. Res. 2019, 17, 3–15. [Google Scholar] [CrossRef]
- Puigvehí, M.; Moctezuma-Velázquez, C.; Villanueva, A.; Llovet, J.M. The oncogenic role of hepatitis delta virus in hepatocellular carcinoma. JHEP Rep. 2019, 1, 120–130. [Google Scholar] [CrossRef] [Green Version]
- Raffaella, R.; Arnolfo, P.; Isabel, P.E.; Floriana, F.; Riccardo, P.; Enrico, G.; Ullah, K.N.; Di, C.L.; Rocco, S.; Gerardo, B. Hepatitis delta virus and hepatocellular carcinoma: An update. Epidemiol. Infect. 2018, 146, 1612–1618. [Google Scholar]
- Jang, T.-Y.; Wei, Y.-J.; Liu, T.-W. Role of hepatitis D virus infection in development of hepatocellular carcinoma among chronic hepatitis B patients treated with nucleotide/nucleoside analogues. Sci. Rep. 2021, 11, 8184. [Google Scholar] [CrossRef]
- Yang, J.F.; Lin, C.I.; Huang, J.F.; Dai, C.Y.; Chang, W.Y. Viral hepatitis infections in southern Taiwan: A multicenter community-based study. Kaohsiung J. Med. Sci. 2010, 26, 461–469. [Google Scholar] [CrossRef] [Green Version]
- Ni, Y.-H.; Chang, M.-H.; Jan, C.-F.; Hsu, H.Y.; Chen, D.S. Continuing decrease in hepatitis B virus infection 30 years after initiation of infant vaccination program in Taiwan. Clin. Gastroenterol. Hepatol. 2016, 14, 1324–1330. [Google Scholar] [CrossRef]
- Stockdale, A.J.; Kreuels, B.; Henrion, M.Y.R.; Giorgi, E.; Geretti, A.M. The global prevalence of hepatitis D virus infection: Systematic review and meta-analysis. J. Hepatol. 2020, 73, 523–532. [Google Scholar] [CrossRef]
- Miao, Z.; Zhang, S.; Ou, X. Estimating the global prevalence, disease progression, and clinical outcome of hepatitis delta virus infection. J. Infect. Dis. 2020, 221, 1677–1687. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.C.; Chen, T.K.; Han, H.F.; Lin, Y.C.; Liu, C.J. Investigating the prevalence and clinical effects of hepatitis delta viral infection in Taiwan. J. Microbiol. Immunol. Infect. 2021, 4, S1684-1182(21)00066-9. [Google Scholar]
- Negro, F. Hepatitis D virus coinfection and superinfection. Cold Spring Harb. Perspect. Med. 2014, 4, a021550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Höner zu Siederdissen, C.; Cornberg, M. Management of HBV and HBV/HDV-Associated Liver Cirrhosis. Visc. Med. 2016, 32, 86–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, T.Y.; Wei, Y.J.; Hsu, C.T.; Hsu, P.Y.; Yu, M.L. Serial serologic changes of hepatitis D virus in chronic hepatitis B patients receiving nucleos(t)ides analogues therapy. J. Gastroenterol. Hepatol. 2020, 35, 1886–1892. [Google Scholar] [CrossRef] [PubMed]
- Ghamari, S.; Alavian, S.M.; Rizzetto, M.; Olivero, A.; Jazayeri, S.M. Prevalence of Hepatitis Delta Virus (HDV) Infection in Chronic Hepatitis B Patients with Unusual Clinical Pictures. Hepat. Mon. 2013, 13, e6731. [Google Scholar] [CrossRef] [Green Version]
- Sy, B.T.; Ratsch, B.A.; Toan, N.L. High Prevalence and Significance of Hepatitis D Virus Infection among Treatment-Naïve HBsAg-Positive Patients in Northern Vietnam. PLoS ONE 2013, 8, e78094. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Sy, B.T.; Trung, N.T.; Hoan, N.X.; Bock, C.T. Prevalence and genotype distribution of hepatitis delta virus among chronic hepatitis B carriers in Central Vietnam. PLoS ONE 2017, 12, e0175304. [Google Scholar]
- Tsatsralt-Od, B.; Takahashi, M.; Nishizawa, T.; Endo, K.; Inoue, J.; Okamoto, H. High prevalence of dual or triple infection of hepatitis B, C, and delta viruses among patients with chronic liver disease in Mongolia. J. Med. Virol. 2005, 77, 491–499. [Google Scholar] [CrossRef]
- Mumtaz, K.; Hamid, S.S.; Adil, S. Epidemiology and clinical pattern of hepatitis delta virus infection in Pakistan. J. Gastroenterol. Hepatol. 2005, 20, 1503–1507. [Google Scholar] [CrossRef]
- Lu, S.N.; Chen, T.M.; Lee, C.M.; Wang, J.H.; Tung, H.D.; Wu, J.C. Molecular epidemiological and clinical aspects of hepatitis D virus in a unique triple hepatitis viruses (B, C, D) endemic community in Taiwan. J. Med. Virol. 2003, 70, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Bartholomeusz, A.; Locarnini, S. Hepatitis B virus mutations associated with antiviral therapy. J. Med. Virol. 2006, 78, S52–S55. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-H.; Lee, S.S.-J.; Yu, M.-L. Changing hepatitis D virus epidemiology in a hepatitis B virus endemic area with a national vaccination program. Hepatology 2015, 61, 1870–1879. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-S.; Lai, M.-Y.; Sung, J.-L. δ Agent Infection in Patients with Chronic Liver Diseases and Hepatocellular Carcinoma-An Infrequent Finding in Taiwan. Hepatology 1984, 4, 502–503. [Google Scholar] [CrossRef]
- Huo, T.I.; Wu, J.C.; Lin, R.Y. Decreasing hepatitis D virus infection in Taiwan: An analysis of contributory factors. J. Gastroenterol. Hepatol. 1997, 12, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-C.; Lin, C.-L.; Hsu, C.-W. Decreasing seroprevalence of anti-hepatitis D virus antibodies in the antiviral era with inverse association with hepatitis B virus DNA, Taiwan, 2006 to 2019. J. Med. Virol. 2020, 92, 124–127. [Google Scholar] [CrossRef]
- Coghill, S.; McNamara, J.; Woods, M.; Hajkowicz, K. Epidemiology and clinical outcomes of hepatitis delta (D) virus infection in Queensland, Australia. Int. J. Infect. Dis. 2018, 74, 123–127. [Google Scholar] [CrossRef] [Green Version]
- Lima, D.S.; Murad, A.J.; Barreira, M.A. Liver transplantation in hepatitis delta: South America experience. Arq. Gastroenterol. 2018, 55, 14–17. [Google Scholar] [CrossRef] [Green Version]
- Tham, C.Y.L.; Kah, J.; Tan, A.T. Hepatitis Delta Virus Acts as an Immunogenic Adjuvant in Hepatitis B Virus-Infected Hepatocytes. Cell Rep. Med. 2020, 1, 100060. [Google Scholar] [CrossRef]
- Scheuer, P.J. Classification of chronic viral hepatitis: A need for reassessment. J. Hepatol. 1991, 13, 372–374. [Google Scholar] [CrossRef]
- Castéra, L.; Vergniol, J.; Foucher, J.; Bail, B.L.; Chanteloup, E.; Haaser, M.; Darriet, M.; Couzigou, P.; Lédinghen, V.D. Prospective comparison of transient elastography, Fibrotest, APRI, and liver biopsy for the assessment of fibrosis in chronic hepatitis C. Gastroenterology 2005, 128, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.; Ebner, L.; Heverhagen, J.T.; Christe, A. State-of-the-art imaging of liver fibrosis and cirrhosis: A comprehensive review of current applications and future perspectives. Eur. J. Radiol. Open 2015, 2, 90–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.-Y.; Huang, J.-F.; Liu, S.-F.; Yu, M.-L.; Tsai, C.-H.; Yang, J.-F.; Lin, I.-L.; Dai, C.-Y.; Lin, Z.-Y.; Chen, S.-C. Performance characteristics of two real-time PCR assays for quantification of hepatitis B virus DNA. Scand. J. Infect. Dis. 2009, 41, 614–618. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | Total Cohort (n = 24) | Non-Cirrhosis (n = 13) | Cirrhosis (n = 11) | p Value |
|---|---|---|---|---|
| Gender (Male/Female) | 18/6 | 1.31 (0.480) | 1.09 (0.302) | 0.194 |
| Age (years, mean [SD]) | 52.42 (11.86) | 52.92 (13.23) | 51.82 (10.63) | 0.826 |
| AST (IU/L, mean [SD]) | 226.29 (316.23) | 271.38 (356.62) | 173.00 (267.64) | 0.449 |
| ALT (IU/L, mean [SD]) | 211.13 (321.96) | 289.69 (417.81) | 118.27 (106.87) | 0.176 |
| Bilirubin (mg/dL, mean [SD]) ‡ | 3.28 (4.64) | 4.61 (6.21) | 1.96 (1.63) | 0.198 |
| INR (mean, [SD]) | 1.21 (0.40) | 1.10 (0.20) | 1.34 (0.53) | 0.183 |
| WBC (cells/mL, mean [SD]) † | 6347 (2825.64) | 7841 (3205.97) | 5003 (1610.25) | * 0.024 |
| PLT(x103/μL, mean [SD]) | 157.38 (104.92) | 211.77 (113.01) | 93.09 (40.04) | * 0.003 |
| Hb (g/dL, mean [SD]) † | 12.81 (2.06) | 12.69 (1.96) | 12.92 (2.58) | 0.815 |
| HBV DNA (Log10 IU/L, mean [SD]) ‡ | 3.81 (2.25) | 3.58 (2.31) | 4.08 (2.27) | 0.617 |
| HBsAg (IU/mL, mean [SD]) †† | 1376.7 (1,616.95) | 750.80 (1133.97) | 2002.60 (1901.96) | 0.242 |
| HBeAg (positive/negative) | (2/22) | 0.92 (0.28) | 0.91 (0.30) | 0.907 |
| Anti-HCV (positive/negative) | (5/19) | 0.69 (0.48) | 0.91 (0.30) | 0.194 |
| HDV RNA (Log10 copies/mL, mean [SD]) | 2.15 (1.67) | 2.22 (1.93) | 2.06 (1.41) | 0.816 |
| Anti-HDV (mean [SD]) | 14.54 (7.51) | 13.69 (8.56) | 15.55 (6.30) | 0.556 |
| Variables | Non-HDV-IV (n = 10) | HDV-IV (n = 14) | p Value | Logistic Regression Analysis OR 95% CI p Value |
|---|---|---|---|---|
| Age (years, mean [SD]) | 54.10 (13.96) | 51.21 (10.50) | 0.568 | |
| AST (IU/L, mean [SD]) | 390.30 (404.35) | 109.14 (168.13) | 0.062 | |
| ALT (IU/L, mean [SD]) | 334.50 (403.11) | 123.00 (225.44) | 0.114 | |
| Bilirubin (mg/dL, mean [SD]) ‡ | 4.90 (6.23) | 1.93 (2.21) | 0.181 | |
| INR (mean, [SD]) | 1.31 (0.553) | 1.14 (0.239) | 0.32 | |
| WBC (cells/mL, mean [SD]) † | 7593.75 (3,509.24) | 5440.91 (1898.62) | 0.102 | |
| PLT(x103 /μL, mean [SD]) | 170.90 (137.51) | 147.71 (78.39) | 0.605 | |
| Hb (g/dL, mean [SD]) † | 12.90 (2.20) | 12.75 (2.07) | 0.877 | |
| HBV DNA Log10 (IU/L, mean [SD]) ‡ | 2.35 (1.87) | 1.27 (2.06) | 0.298 | |
| HBsAg (IU/mL, mean [SD]) †† | 2467.64 (1259.46) | 909.19 (1594.61) | 0.175 | |
| HDV RNA (Log10 copies/mL, mean [SD]) | 3.30 (1.82) | 1.34 (0.99) | * 0.009 | 0.37 0.164–0.83 ** 0.017 |
| Anti-HDV (mean [SD]) | 13.96 (9.24) | 14.96 (6.34) | 0.756 |
| Variables | HDV-I | HDV-II | HDV-IV | p Value |
|---|---|---|---|---|
| Age (years, mean [SD])) | 53.5 (9.88) | 54.50 (17.09) | 51.21 (10.50) | 0.846 |
| AST (IU/L, mean [SD]) | 443.00 (435.97) | 355.17 (420.18) | 119.92 (180.33) | 0.086 |
| ALT (IU/L, mean [SD]) | 487.75 (545.40) | 232.33 (287.59) | 133.83 (243.21) | 0.132 |
| Bilirubin (mg/dL, mean [SD]) ‡ | 5.74 (7.47) | 4.34 (5.96) | 1.93 (2.21) | 0.308 |
| Creatinine (mg/dL, mean [SD]) | 2.69 (3.88) | 3.91 (3.65) | 0.96 (0.50) | * 0.047 |
| INR (mean [SD]) | 1.50 (0.84) | 1.19 (0.28) | 1.14 (0.24) | 0.304 |
| WBC (cells/mL, mean [SD]) † | 8460.00 (4919.16) | 7074.00 (2924.26) | 5440.91 (1898.62) | 0.216 |
| PLT(x10 3 /μL, mean [SD]) | 196.00 (79.99) | 154.17 (171.34) | 147.71 (78.39) | 0.734 |
| Hb (g/dL, mean [SD]) † | 14.03 (1.39) | 12.22 (2.44) | 12.75 (2.07) | 0.505 |
| HBV DNA (Log10 IU/L, mean [SD]) ‡ | 3.72 (2.60) | 4.50 (2.48) | 3.49 (2.16) | 0.686 |
| HBsAg (IU/mL, mean [SD]) †† | 251 (0.00) | 1581 (1506) | 711.73 (1340) | 0.338 |
| HDV RNA (Log10 copies/mL, mean [SD]) | 3.19 (2.22) | 3.34 (1.72) | 1.34 (0.99) | * 0.012 |
| Anti-HDV (mean [SD]) | 12.88 (9.38) | 14.69 (9.96) | 14.96 (6.34) | 0.895 |
| Non-HDV-IV | HDV-IV | Total | p Value | ||
|---|---|---|---|---|---|
| Variables | Category | N (%) | N (%) | N (%) | |
| Gender | Male | 8 (44.4) | 10 (55.6) | 18 (100.0) | 0.269 |
| Female | 2 (33.3) | 4 (66.7) | 6 (100.0) | ||
| Total | 10 (41.7) | 14 (58.3) | 24 (100.0) | ||
| LC group | Non-LC | 5 (38.5) | 8 (61.5) | 13 (100.0) | 0.729 |
| LC | 5 (45.5) | 6 (54.5) | 11 (100.0) | ||
| Total | 10 (41.7) | 14 (58.3) | 24 (100.0) | ||
| ALT group | ALT < 400 IU/L | 7 (35.0) | 13 (65.0) | 20 (100.0) | 0.139 |
| ALT ≥ 400 IU/L | 3 (75.0) | 1 (25.0) | 4 (100.0) | ||
| Total | 10 (41.7) | 14 (58.3) | 24 (100.0) | ||
| Bilirubin group | BIL < 2 mL/dL | 6 (42.9) | 8 (57.1) | 14 (100.0) | 0.746 |
| BIL ≥ 2 mL/dL | 4 (50.0) | 4 (50.0) | 8 (100.0) | ||
| Total | 10 (45.5) | 12 (54.5) | 22 (100.0) | ||
| HBV DNA group | HBV DNA < 2000 IU/mL | 4 (50.0) | 4 (50.0) | 8 (100.0) | 0.746 |
| HBV DNA ≥ 2000 IU/mL | 6 (42.9) | 8 (57.1) | 14 (100.0) | ||
| Total | 10 (45.5) | 12 (54.5) | 22 (100.0) | ||
| HBsAg group | HBsAg < 250 IU/mL | 5 (41.7) | 7 (58.3) | 12 (100.0) | 0.737 |
| HBsAg ≥ 250 IU/mL | 3 (50.0) | 3 (50.0) | 6 (100.0) | ||
| Total | 8 (44.4) | 10 (55.6) | 18 (100.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joseph, K.; Shabangu, C.S.; Jang, T.-Y.; Huang, C.-F.; Dai, C.-Y.; Huang, J.-F.; Chuang, W.-L.; Yu, M.-L.; Wang, S.-C. The Prevalence and Serological Association of Hepatitis D Virus Genotypes in Taiwan. Pathogens 2021, 10, 1227. https://doi.org/10.3390/pathogens10101227
Joseph K, Shabangu CS, Jang T-Y, Huang C-F, Dai C-Y, Huang J-F, Chuang W-L, Yu M-L, Wang S-C. The Prevalence and Serological Association of Hepatitis D Virus Genotypes in Taiwan. Pathogens. 2021; 10(10):1227. https://doi.org/10.3390/pathogens10101227
Chicago/Turabian StyleJoseph, Keva, Ciniso Sylvester Shabangu, Tyng-Yuan Jang, Chung-Feng Huang, Chia-Yen Dai, Jee-Fu Huang, Wan-Long Chuang, Ming-Lung Yu, and Shu-Chi Wang. 2021. "The Prevalence and Serological Association of Hepatitis D Virus Genotypes in Taiwan" Pathogens 10, no. 10: 1227. https://doi.org/10.3390/pathogens10101227
APA StyleJoseph, K., Shabangu, C. S., Jang, T.-Y., Huang, C.-F., Dai, C.-Y., Huang, J.-F., Chuang, W.-L., Yu, M.-L., & Wang, S.-C. (2021). The Prevalence and Serological Association of Hepatitis D Virus Genotypes in Taiwan. Pathogens, 10(10), 1227. https://doi.org/10.3390/pathogens10101227








