Dynamics and Malleability of Plant DNA Methylation During Abiotic Stresses
Abstract
1. Introduction
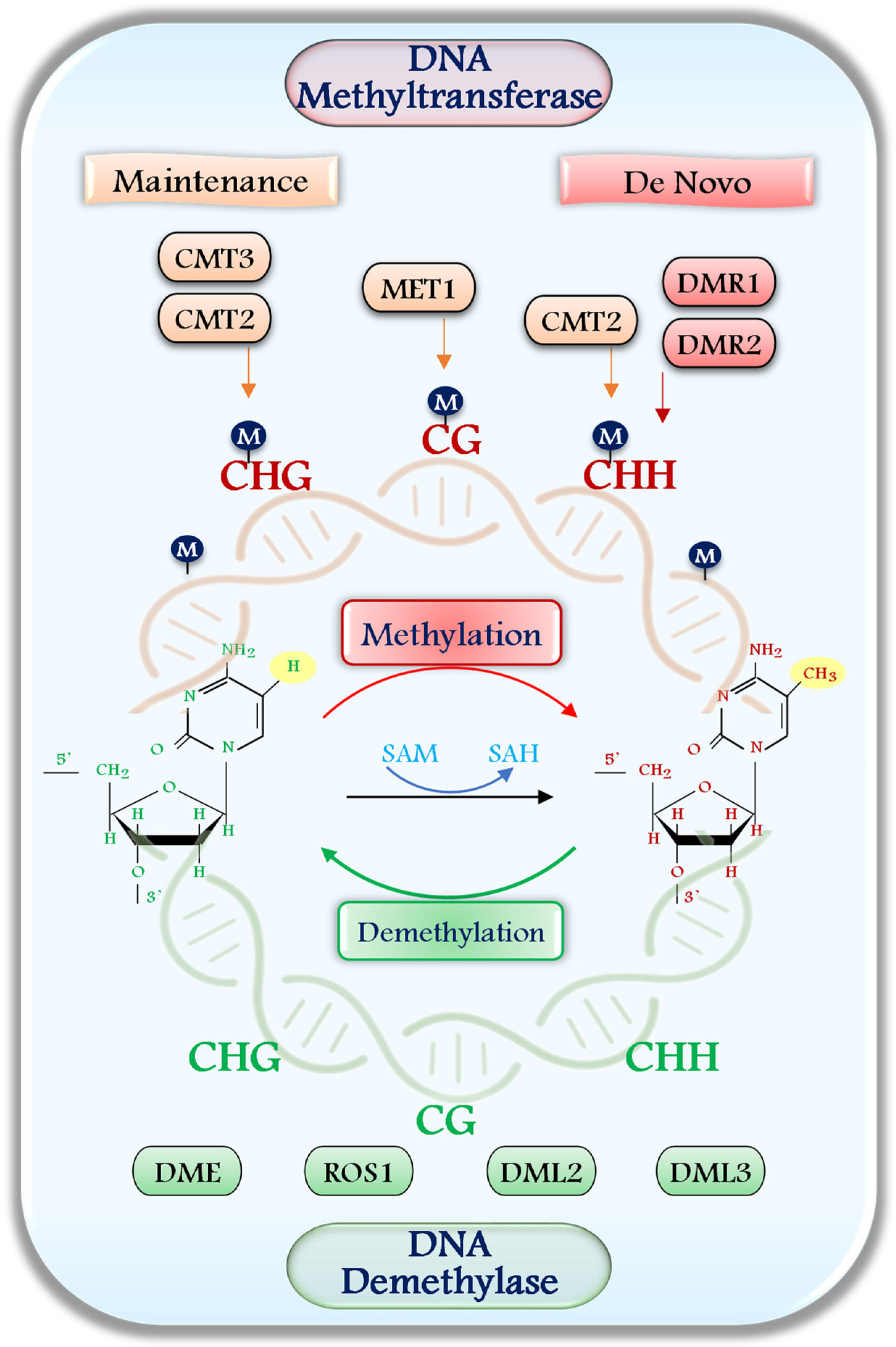
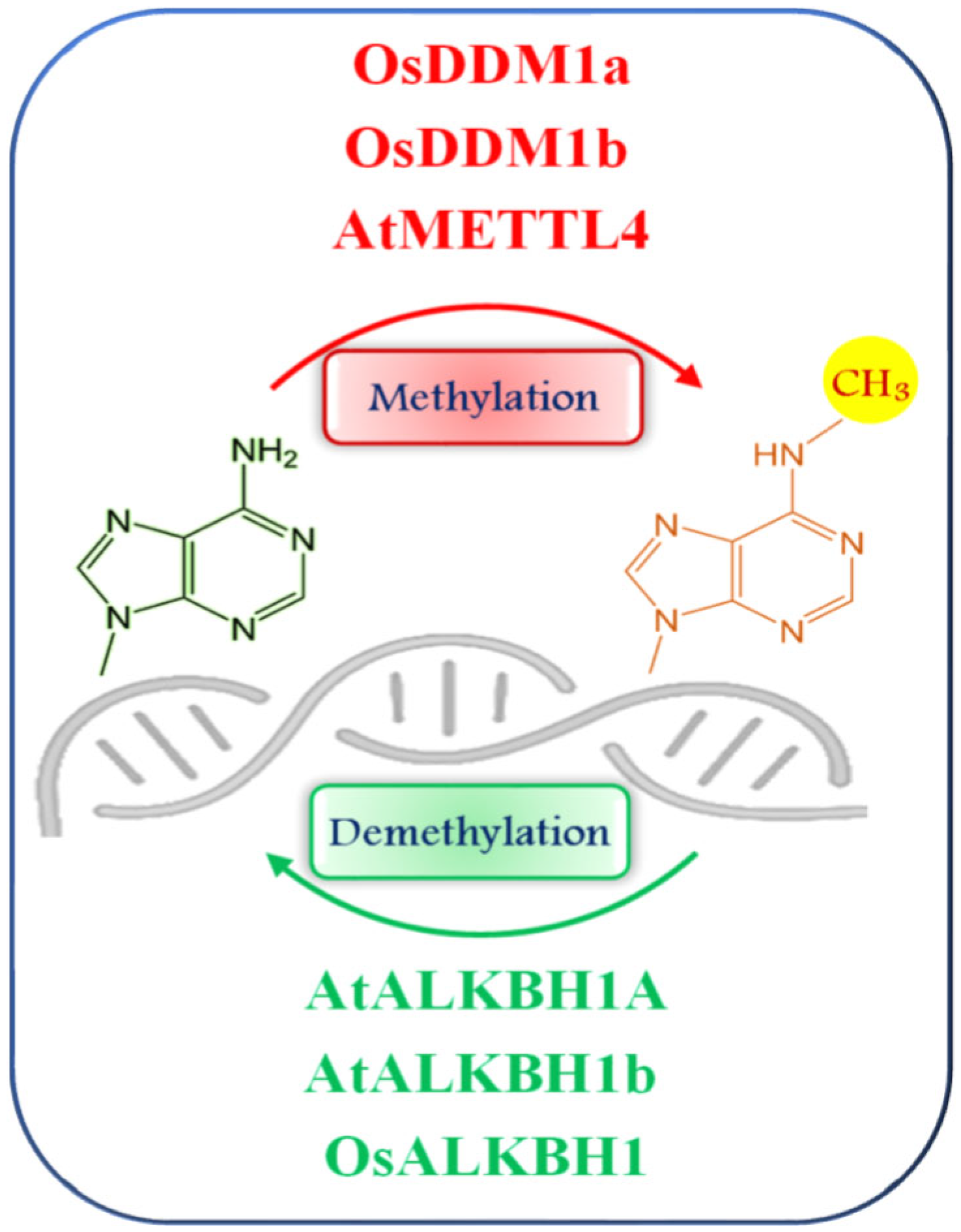
2. Plant DNA Methyltransferases and Demethylases: An Insight
3. DNA Methylation in Abiotic Stress
3.1. DNA Methylation During Salt Stress
3.2. DNA Methylation During Drought or Water-Deficit Stress
3.3. DNA Methylation During Heat or High Temperature Stress
3.4. DNA Methylation During Cold Stress
3.5. DNA Methylation During Heavy Metal Stress
4. Conclusions and Future Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Lodhi, N.; Ranjan, A.; Singh, M.; Srivastava, R.; Singh, S.P.; Chaturvedi, C.P.; Ansari, S.A.; Sawant, S.V.; Tuli, R. Interactions between upstream and core promoter sequences determine gene expression and nucleosome positioning in tobacco PR-1a promoter. Biochim. Biophys. Acta 2008, 1779, 634–644. [Google Scholar] [CrossRef]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- Pikaard, C.S.; Mittelsten Scheid, O. Epigenetic Regulation in Plants. Cold Spring Harb. Perspect. Biol. 2014, 6, a019315. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Rai, K.M.; Srivastava, R. Plant Biosynthetic Engineering Through Transcription Regulation: An Insight into Molecular Mechanisms During Environmental Stress. In Biosynthetic Technology and Environmental Challenges; Springer: Singapore, 2018; pp. 51–72. [Google Scholar] [CrossRef]
- Lodhi, N.; Srivastava, R. Histone Demethylases: Insights into Human. Encyclopedia. 2023. Available online: https://encyclopedia.pub/entry/45957 (accessed on 20 May 2025).
- Kumari, A.; Soni, A.; Srivastava, R.; Pandey, R.; Bhardwaj, A.R. Exploring the RNAi Vistas: From Discovery to Emerging Frontiers. In Non-Coding RNAs for Crop Improvement: Concepts and Applications; Goswami, K., Gelaw, T.A., Sanan-Mishra, N., Eds.; Springer Nature: Singapore, 2025; pp. 1–24. [Google Scholar]
- Srivastava, R.; Srivastava, R.; Singh, U.M. Understanding the patterns of gene expression during climate change. In Climate Change Effect on Crop Productivity; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group: Abingdon, UK, 2014; pp. 279–328. ISBN 978-1-4822-2920-2. [Google Scholar]
- Pandey, B.; Prakash, P.; Verma, P.C.; Srivastava, R. Regulated Gene Expression by Synthetic Modulation of the Promoter Architecture in Plants; Elsevier: Amsterdam, The Netherlands, 2019; pp. 235–255. [Google Scholar] [CrossRef]
- Chauhan, D.; Singh, D.; Pandey, H.; Srivastava, R.; Dhiman, V.K.; Dhiman, V.K. Impact of transcription factor in plant abiotic stress: A recent advancement for crop improvement. In Plant Transcription Factors; Academic Press: Cambridge, MA, USA, 2022; pp. 271–286. [Google Scholar] [CrossRef]
- Dubey, N.K.; Srivastava, R.; Singh, M.; Bhutani, S.; Ranjan, A. Genetic Engineering for Crop Improvement Against Stresses and Future Agriculture. In Omics and Genome Editing: Revolution in Crop Improvement for Sustainable Agriculture; Springer: Berlin/Heidelberg, Germany, 2025; pp. 55–67. [Google Scholar]
- Kiran, K.; Ansari, S.A.; Srivastava, R.; Lodhi, N.; Chaturvedi, C.P.; Sawant, S.V.; Tuli, R. The TATA-box sequence in the basal promoter contributes to determining light-dependent gene expression in plants. Plant Physiol. 2006, 142, 364–376. [Google Scholar] [CrossRef]
- Srivastava, R.; Rai, K.M.; Pandey, B.; Singh, S.P.; Sawant, S.V. Spt-Ada-Gcn5-Acetyltransferase (SAGA) Complex in Plants: Genome Wide Identification, Evolutionary Conservation and Functional Determination. PLoS ONE 2015, 10, e0134709. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Ansari, S.A.; Srivastava, R.; Mantri, S.; Asif, M.H.; Sawant, S.V.; Tuli, R. A T9G mutation in the prototype TATA-box TCACTATATATAG determines nucleosome formation and synergy with upstream activator sequences in plant promoters. Plant Physiol. 2009, 151, 2174–2186. [Google Scholar] [CrossRef]
- Srivastava, R.; Ahn, S.H. Modifications of RNA polymerase II CTD: Connections to the histone code and cellular function. Biotechnol. Adv. 2015, 33, 856–872. [Google Scholar] [CrossRef]
- Srivastava, R.; Rai, K.M.; Srivastava, M.; Kumar, V.; Pandey, B.; Singh, S.P.; Bag, S.K.; Singh, B.D.; Tuli, R.; Sawant, S.V. Distinct Role of Core Promoter Architecture in Regulation of Light Mediated Responses in Plant Genes. Mol. Plant 2014, 7, 626–641. [Google Scholar] [CrossRef]
- Srivastava, R.; Singh, U.M.; Dubey, N.K. Histone Modifications by different histone modifiers: Insights into histone writers and erasers during chromatin modification. J. Biol. Sci. Med. 2016, 2, 45–54. [Google Scholar]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef]
- D’Urso, A.; Brickner, J.H. Mechanisms of epigenetic memory. Trends Genet. 2014, 30, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Gallusci, P.; Agius, D.R.; Moschou, P.N.; Dobránszki, J.; Kaiserli, E.; Martinelli, F. Deep inside the epigenetic memories of stressed plants. Trends Plant Sci. 2023, 28, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, J.; Liu, B.; Xu, Z.-Y. Dynamic regulation of DNA methylation and histone modifications in response to abiotic stresses in plants. J. Integr. Plant Biol. 2022, 64, 2252–2274. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Talarico, E.; Zambelli, A.; Araniti, F.; Greco, E.; Chiappetta, A.; Bruno, L. Unravelling the Epigenetic Code: DNA Methylation in Plants and Its Role in Stress Response. Epigenomes 2024, 8, 30. [Google Scholar] [CrossRef]
- Xie, G.; Du, X.; Hu, H.; Du, J. Molecular Mechanisms Underlying the Establishment, Maintenance, and Removal of DNA Methylation in Plants. Annu. Rev. Plant Biol. 2025, 76, 143–170. [Google Scholar] [CrossRef]
- Srivastava, R.; Lodhi, N. DNA Methylation Malleability and Dysregulation in Cancer Progression: Understanding the Role of PARP1. Biomolecules 2022, 12, 417. [Google Scholar] [CrossRef]
- Chen, Z.X.; Riggs, A.D. DNA methylation and demethylation in mammals. J. Biol. Chem. 2011, 286, 18347–18353. [Google Scholar] [CrossRef]
- Cao, X.; Jacobsen, S.E. Role of the Arabidopsis DRM Methyltransferases in De Novo DNA Methylation and Gene Silencing. Curr. Biol. 2002, 12, 1138–1144. [Google Scholar] [CrossRef]
- He, X.-J.; Ma, Z.-Y.; Liu, Z.-W. Non-Coding RNA Transcription and RNA-Directed DNA Methylation in Arabidopsis. Mol. Plant 2014, 7, 1406–1414. [Google Scholar] [CrossRef] [PubMed]
- Matzke, M.A.; Kanno, T.; Matzke, A.J.M. RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants. Annu. Rev. Plant Biol. 2015, 66, 243–267. [Google Scholar] [CrossRef]
- Wambui Mbichi, R.; Wang, Q.-F.; Wan, T. RNA directed DNA methylation and seed plant genome evolution. Plant Cell Rep. 2020, 39, 983–996. [Google Scholar] [CrossRef] [PubMed]
- Stroud, H.; Do, T.; Du, J.; Zhong, X.; Feng, S.; Johnson, L.; Patel, D.J.; Jacobsen, S.E. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 2014, 21, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, R.M.; Picard, C.L. RNA-directed DNA Methylation. PLoS Genet. 2020, 16, e1009034. [Google Scholar] [CrossRef]
- Law, J.A.; Vashisht, A.A.; Wohlschlegel, J.A.; Jacobsen, S.E. SHH1, a Homeodomain Protein Required for DNA Methylation, As Well As RDR2, RDM4, and Chromatin Remodeling Factors, Associate with RNA Polymerase IV. PLoS Genet. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Zhou, M.; Palanca, A.M.S.; Law, J.A. Locus-specific control of the de novo DNA methylation pathway in Arabidopsis by the CLASSY family. Nat. Genet. 2018, 50, 865–873. [Google Scholar] [CrossRef]
- Kankel, M.; Ramsey, D.; Stokes, T.; Flowers, S.; Haag, J.; Jeddeloh, J.; Riddle, N.; Verbsky, M.; Richards, E. Arabidopsis MET1 Cytosine Methyltransferase Mutants. Genetics 2003, 163, 1109–1122. [Google Scholar] [CrossRef]
- Bewick, A.J.; Niederhuth, C.E.; Ji, L.; Rohr, N.A.; Griffin, P.T.; Leebens-Mack, J.; Schmitz, R.J. The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants. Genome Biol. 2017, 18, 65. [Google Scholar] [CrossRef]
- Du, X.; Yang, Z.; Xie, G.; Wang, C.; Zhang, L.; Yan, K.; Yang, M.; Li, S.; Zhu, J.K.; Du, J. Molecular basis of the plant ROS1-mediated active DNA demethylation. Nat. Plants 2023, 9, 271–279. [Google Scholar] [CrossRef]
- Meng, H.; Cao, Y.; Qin, J.; Song, X.; Zhang, Q.; Shi, Y.; Cao, L. DNA methylation, its mediators and genome integrity. Int. J. Biol. Sci. 2015, 11, 604–617. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Miki, D.; Zhang, H.; Liu, Y.; Zhang, X.; Tang, K.; Kan, Y.; La, H.; Li, X.; Li, S.; et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 2012, 336, 1445–1448. [Google Scholar] [CrossRef]
- Li, Y.; Kumar, S.; Qian, W. Active DNA demethylation: Mechanism and role in plant development. Plant Cell Rep. 2018, 37, 77–85. [Google Scholar] [CrossRef]
- Liang, Z.; Shen, L.; Cui, X.; Bao, S.; Geng, Y.; Yu, G.; Liang, F.; Xie, S.; Lu, T.; Gu, X.; et al. DNA N(6)-Adenine Methylation in Arabidopsis thaliana. Dev. Cell 2018, 45, 406–416.e403. [Google Scholar] [CrossRef]
- Zhang, Q.; Liang, Z.; Cui, X.; Ji, C.; Li, Y.; Zhang, P.; Liu, J.; Riaz, A.; Yao, P.; Liu, M.; et al. N6-Methyladenine DNA Methylation in Japonica and Indica Rice Genomes and Its Association with Gene Expression, Plant Development, and Stress Responses. Mol. Plant 2018, 11, 1492–1508. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, C.; Liu, H.; Zhou, Q.; Liu, Q.; Guo, Y.; Peng, T.; Song, J.; Zhang, J.; Chen, L.; et al. Identification and analysis of adenine N(6)-methylation sites in the rice genome. Nat. Plants 2018, 4, 554–563. [Google Scholar] [CrossRef]
- Xiao, C.L.; Zhu, S.; He, M.; Chen, D.; Zhang, Q.; Chen, Y.; Yu, G.; Liu, J.; Xie, S.Q.; Luo, F.; et al. N(6)-Methyladenine DNA Modification in the Human Genome. Mol. Cell 2018, 71, 306–318.e307. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, S.; Nelakanti, R.; Zhao, W.; Liu, G.; Li, Z.; Liu, X.; Wu, T.; Xiao, A.; Li, H. Mammalian ALKBH1 serves as an N(6)-mA demethylase of unpairing DNA. Cell Res. 2020, 30, 197–210. [Google Scholar] [CrossRef]
- Li, D.; Du, J.; Gao, M.; He, C. Identification of AtALKBH1A and AtALKBH1D as DNA N6-adenine demethylases in Arabidopsis thaliana. Plant Sci. 2024, 342, 112055. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Ma, A.; Lei, J.; He, C. METTL4-mediated N(6)-methyladenine DNA modification regulates thermotolerance in Arabidopsis thaliana. Plant Sci. 2024, 338, 111916. [Google Scholar] [CrossRef] [PubMed]
- Kooke, R.; Johannes, F.; Wardenaar, R.; Becker, F.; Etcheverry, M.; Colot, V.; Vreugdenhil, D.; Keurentjes, J.J. Epigenetic basis of morphological variation and phenotypic plasticity in Arabidopsis thaliana. Plant Cell 2015, 27, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Latzel, V.; Rendina González, A.P.; Rosenthal, J. Epigenetic Memory as a Basis for Intelligent Behavior in Clonal Plants. Front. Plant Sci. 2016, 7, 1354. [Google Scholar] [CrossRef]
- Jean Finnegan, E.; Kovac, K.A.; Jaligot, E.; Sheldon, C.C.; James Peacock, W.; Dennis, E.S. The downregulation of FLOWERING LOCUS C (FLC) expression in plants with low levels of DNA methylation and by vernalization occurs by distinct mechanisms. Plant J. 2005, 44, 420–432. [Google Scholar] [CrossRef]
- Song, Q.; Zhang, T.; Stelly, D.M.; Chen, Z.J. Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons. Genome Biol. 2017, 18, 99. [Google Scholar] [CrossRef]
- Baldini, A.; Battaglia, F.; Perrella, G. The generation of novel epialleles in plants: The prospective behind re-shaping the epigenome. Front. Plant Sci. 2025, 16, 1544744. [Google Scholar] [CrossRef]
- Wang, F.; Han, T.; Jeffrey Chen, Z. Circadian and photoperiodic regulation of the vegetative to reproductive transition in plants. Commun. Biol. 2024, 7, 579. [Google Scholar] [CrossRef] [PubMed]
- Dowen, R.H.; Pelizzola, M.; Schmitz, R.J.; Lister, R.; Dowen, J.M.; Nery, J.R.; Dixon, J.E.; Ecker, J.R. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 2012, 109, E2183–E2191. [Google Scholar] [CrossRef] [PubMed]
- Deleris, A.; Halter, T.; Navarro, L. DNA Methylation and Demethylation in Plant Immunity. Annu. Rev. Phytopathol. 2016, 54, 579–603. [Google Scholar] [CrossRef] [PubMed]
- López, M.-E.; Roquis, D.; Becker, C.; Denoyes, B.; Bucher, E. DNA methylation dynamics during stress response in woodland strawberry (Fragaria vesca). Hortic. Res. 2022, 9, uhac174. [Google Scholar] [CrossRef]
- Kumar, S.; Beena, A.S.; Awana, M.; Singh, A. Salt-Induced Tissue-Specific Cytosine Methylation Downregulates Expression of HKT Genes in Contrasting Wheat (Triticum aestivum L.) Genotypes. DNA Cell Biol. 2017, 36, 283–294. [Google Scholar] [CrossRef]
- Al-Lawati, A.; Al-Bahry, S.; Victor, R.; Al-Lawati, A.H.; Yaish, M.W. Salt stress alters DNA methylation levels in alfalfa (Medicago spp.). Genet. Mol. Res. 2016, 15, 15018299. [Google Scholar] [CrossRef]
- Li, Z.; Hu, Y.; Chang, M.; Kashif, M.H.; Tang, M.; Luo, D.; Cao, S.; Lu, H.; Zhang, W.; Huang, Z.; et al. 5-azacytidine pre-treatment alters DNA methylation levels and induces genes responsive to salt stress in kenaf (Hibiscus cannabinus L.). Chemosphere 2021, 271, 129562. [Google Scholar] [CrossRef] [PubMed]
- Omar, S.A.; Feng, Y.; Yu, M.; Eldin, S.A.G.; Eldenary, M.E.; Shabala, S.; Allakhverdiev, S.I.; Abdelfattah, M.H. Exogenous application of 5-azacitidin, royal jelly and folic acid regulate plant redox state, expression level of DNA methyltransferases and alleviate adverse effects of salinity stress on Vicia faba L. plants. Heliyon 2024, 10, e30934. [Google Scholar] [CrossRef]
- Beyrne, C.C.; Iusem, N.D.; González, R.M. Effect of Salt Stress on Cytosine Methylation within GL2, An Arabidopsis thaliana Gene Involved in Root Epidermal Cell Differentiation. Absence of Inheritance in the Unstressed Progeny. Int. J. Mol. Sci. 2019, 20, 4446. [Google Scholar] [CrossRef]
- Awana, M.; Yadav, K.; Rani, K.; Gaikwad, K.; Praveen, S.; Kumar, S.; Singh, A. Insights into Salt Stress-Induced Biochemical, Molecular and Epigenetic Regulation of Spatial Responses in Pigeonpea (Cajanus cajan L.). J. Plant Growth Regul. 2019, 38, 1545–1561. [Google Scholar] [CrossRef]
- Hoseini, M.; Arzani, A.; Saeidi, G.; Araniti, F. Agro-Physiological and DNA Methylation Responses to Salinity Stress in Wheat (Triticum aestivum L.), Aegilops cylindrica Host, and Their Introgressed Lines. Plants 2024, 13, 2673. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, D. Transcriptome and DNA Methylome Analysis of Two Contrasting Rice Genotypes under Salt Stress during Germination. Int. J. Mol. Sci. 2023, 24, 3978. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.J.; Donoghue, M.T.A.; Barros, P.; Saibo, N.J.; Santos, A.P.; Oliveira, M.M. Uncovering Differentially Methylated Regions (DMRs) in a Salt-Tolerant Rice Variety under Stress: One Step towards New Regulatory Regions for Enhanced Salt Tolerance. Epigenomes 2019, 3, 4. [Google Scholar] [CrossRef]
- Skorupa, M.; Szczepanek, J.; Mazur, J.; Domagalski, K.; Tretyn, A.; Tyburski, J. Salt stress and salt shock differently affect DNA methylation in salt-responsive genes in sugar beet and its wild, halophytic ancestor. PLoS ONE 2021, 16, e0251675. [Google Scholar] [CrossRef]
- Yang, X.; Bai, Z.; He, Y.; Wang, N.; Sun, L.; Li, Y.; Yin, Z.; Wang, X.; Zhang, B.; Han, M.; et al. Genome-wide characterization of DNA methyltransferase family genes implies GhDMT6 improving tolerance of salt and drought on cotton. BMC Plant Biol. 2024, 24, 312. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, C.; Xu, X.; Kan, J.; Li, H.; Lin, J.; Cheng, Z.; Chang, Y. Comprehensive Analysis of the DNA Methyltransferase Genes and Their Association with Salt Response in Pyrus betulaefolia. Forests 2023, 14, 1751. [Google Scholar] [CrossRef]
- Fan, S.; Liu, H.; Liu, J.; Hua, W.; Xu, S.; Li, J. Systematic Analysis of the DNA Methylase and Demethylase Gene Families in Rapeseed (Brassica napus L.) and Their Expression Variations After Salt and Heat stresses. Int. J. Mol. Sci. 2020, 21, 953. [Google Scholar] [CrossRef]
- Miryeganeh, M.; Marlétaz, F.; Gavriouchkina, D.; Saze, H. De novo genome assembly and in natura epigenomics reveal salinity-induced DNA methylation in the mangrove tree Bruguiera gymnorhiza. New Phytol. 2022, 233, 2094–2110. [Google Scholar] [CrossRef]
- Moglia, A.; Gianoglio, S.; Acquadro, A.; Valentino, D.; Milani, A.M.; Lanteri, S.; Comino, C. Identification of DNA methyltransferases and demethylases in Solanum melongena L., and their transcription dynamics during fruit development and after salt and drought stresses. PLoS ONE 2019, 14, e0223581. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, X.; Di, Y.; Li, J.; Li, K.; Wei, H.; Zhang, F.; Su, Z. Systematic Analysis of DNA Demethylase Gene Families in Foxtail Millet (Setaria italica L.) and Their Expression Variations after Abiotic Stresses. Int. J. Mol. Sci. 2024, 25, 4464. [Google Scholar] [CrossRef]
- Bharti, P.; Mahajan, M.; Vishwakarma, A.K.; Bhardwaj, J.; Yadav, S.K. AtROS1 overexpression provides evidence for epigenetic regulation of genes encoding enzymes of flavonoid biosynthesis and antioxidant pathways during salt stress in transgenic tobacco. J. Exp. Bot. 2015, 66, 5959–5969. [Google Scholar] [CrossRef]
- Zhu, X.; Xie, S.; Tang, K.; Kalia, R.K.; Liu, N.; Ma, J.; Bressan, R.A.; Zhu, J.-K. Non-CG DNA methylation-deficiency mutations enhance mutagenesis rates during salt adaptation in cultured Arabidopsis cells. Stress Biol. 2021, 1, 12. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, L.; Peng, Y.; Lu, Z.; Zheng, M.; Wang, Z.; Liu, J.; He, Y.; Luo, J. ZmKTF1 promotes salt tolerance by mediating RNA-directed DNA methylation in maize. New Phytol. 2025, 245, 200–214. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Miki, D.; Tang, K.; Zhou, H.R.; Zheng, Z.; Chen, W.; Ma, Z.Y.; Yang, L.; Zhang, H.; Liu, R.; et al. A Pre-mRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis. PLoS Genet. 2013, 9, e1003779. [Google Scholar] [CrossRef]
- Baek, D.; Jiang, J.; Chung, J.S.; Wang, B.; Chen, J.; Xin, Z.; Shi, H. Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol. 2011, 52, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Garg, R. Unravelling Differential DNA Methylation Patterns in Genotype Dependent Manner under Salinity Stress Response in Chickpea. Int. J. Mol. Sci. 2023, 24, 1863. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, J.; Zhao, L.; Zhou, G.; Tang, Y. Genome-wide DNA methylation analysis of sorghum leaves following foreign GA3 exposure under salt stress. Genomics 2025, 117, 111000. [Google Scholar] [CrossRef]
- Srivastava, R.; Bajpai, R.; Khan, Z.; Singh, S.P.; Mehrotra, R.; Dubey, N.K. Insight into strigolactone hormone functions in plant parasitic weeds: A regulatory perspective. Indian J. Exp. Biol. 2022, 60, 659–666. [Google Scholar] [CrossRef]
- Lu, X.; Cui, J.; Qi, J.; Li, S.; Yu, W.; Li, C. The strigolactones-mediated DNA demethylation activates the phosphoinositide pathway in response to salt stress. Int. J. Biol. Macromol. 2025, 301, 139954. [Google Scholar] [CrossRef]
- Fortini, E.A.; Batista, D.S.; Felipe, S.H.S.; Silva, T.D.; Correia, L.N.F.; Farias, L.M.; Faria, D.V.; Pinto, V.B.; Santa-Catarina, C.; Silveira, V.; et al. Physiological, epigenetic, and proteomic responses in Pfaffia glomerata growth in vitro under salt stress and 5-azacytidine. Protoplasma 2023, 260, 467–482. [Google Scholar] [CrossRef]
- Wang, L.; Cao, S.; Wang, P.; Lu, K.; Song, Q.; Zhao, F.J.; Chen, Z.J. DNA hypomethylation in tetraploid rice potentiates stress-responsive gene expression for salt tolerance. Proc. Natl. Acad. Sci. USA 2021, 118, e2023981118. [Google Scholar] [CrossRef] [PubMed]
- Amraee, L.; Rahmani, F.; Abdollahi Mandoulakani, B. 24-Epibrassinolide alters DNA cytosine methylation of Linum usitatissimum L. under salinity stress. Plant Physiol. Biochem. 2019, 139, 478–484. [Google Scholar] [CrossRef]
- Paul, A.; Dasgupta, P.; Roy, D.; Chaudhuri, S. Comparative analysis of Histone modifications and DNA methylation at OsBZ8 locus under salinity stress in IR64 and Nonabokra rice varieties. Plant Mol. Biol. 2017, 95, 63–88. [Google Scholar] [CrossRef]
- Movahedi, A.; Zhang, J.; Sun, W.; Mohammadi, K.; Almasi Zadeh Yaghuti, A.; Wei, H.; Wu, X.; Yin, T.; Zhuge, Q. Functional analyses of PtRDM1 gene overexpression in poplars and evaluation of its effect on DNA methylation and response to salt stress. Plant Physiol. Biochem. 2018, 127, 64–73. [Google Scholar] [CrossRef]
- Santos, A.P.; Ferreira, L.; Maroco, J.; Oliveira, M.M. Abiotic stress and induced DNA hypomethylation cause interphase chromatin structural changes in rice rDNA loci. Cytogenet. Genome Res. 2011, 132, 297–303. [Google Scholar] [CrossRef]
- Srivastava, R.; Srivastava, R.; Ahn, S.H. The Epigenetic Pathways to Ribosomal DNA Silencing. Microbiol. Mol. Biol. Rev. 2016, 80, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, G.; Guo, S.; Wang, Y.; Sun, J. SlSAMS1 enhances salt tolerance through regulation DNA methylation of SlGI in tomato. Plant Sci. 2023, 335, 111808. [Google Scholar] [CrossRef]
- Tobiasz-Salach, R.; Mazurek, M.; Jacek, B. Physiological, Biochemical, and Epigenetic Reaction of Maize (Zea mays L.) to Cultivation in Conditions of Varying Soil Salinity and Foliar Application of Silicon. Int. J. Mol. Sci. 2023, 24, 1141. [Google Scholar] [CrossRef] [PubMed]
- Victoria, D.; Aliki, K.; Venetia, K.; Georgios, M.; Zoe, H. Spatial and temporal expression of cytosine-5 DNA methyltransferase and DNA demethylase gene families of the Ricinus communis during seed development and drought stress. Plant Growth Regul. 2018, 84, 81–94. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, S.; Zhou, C.; Chen, L.; Fu, H.; Li, X.; Lin, Y.; Lai, Z.; Guo, Y. Genome-wide investigation and transcriptional analysis of cytosine-5 DNA methyltransferase and DNA demethylase gene families in tea plant (Camellia sinensis) under abiotic stress and withering processing. PeerJ 2020, 8, e8432. [Google Scholar] [CrossRef]
- Xu, J.; Zhou, S.; Gong, X.; Song, Y.; van Nocker, S.; Ma, F.; Guan, Q. Single-base methylome analysis reveals dynamic epigenomic differences associated with water deficit in apple. Plant Biotechnol. J. 2018, 16, 672–687. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.C.B.; Camillo, L.R.; Santos, D.B.; Amorim, M.S.; Gonçalves, L.P.; Barbosa, A.C.O.; Rocha Junior, D.S.; Alcântara, G.M.; Costa, M.G.C. Identification of DEMETER-like DNA demethylase gene family in citrus and their role in drought stress-adaptive responses. Comput. Biol. Chem. 2024, 112, 108128. [Google Scholar] [CrossRef] [PubMed]
- Kapazoglou, A.; Drosou, V.; Argiriou, A.; Tsaftaris, A.S. The study of a barley epigenetic regulator, HvDME, in seed development and under drought. BMC Plant Biol. 2013, 13, 172. [Google Scholar] [CrossRef]
- Drosou, V.; Kapazoglou, A.; Letsiou, S.; Tsaftaris, A.S.; Argiriou, A. Drought induces variation in the DNA methylation status of the barley HvDME promoter. J. Plant Res. 2021, 134, 1351–1362. [Google Scholar] [CrossRef]
- Wang, Y.; Xun, H.; Lv, J.; Ju, W.; Jiang, Y.; Wang, M.; Guo, R.; Zhang, M.; Ding, X.; Liu, B.; et al. A modulatory role of CG methylation on gene expression in soybean implicates its potential utility in breeding. Plant Biotechnol. J. 2025, 23, 1585–1600. [Google Scholar] [CrossRef]
- Sun, M.; Yang, Z.; Liu, L.; Duan, L. DNA Methylation in Plant Responses and Adaption to Abiotic Stresses. Int. J. Mol. Sci. 2022, 23, 6910. [Google Scholar] [CrossRef]
- Kaur, A.; Grewal, A.; Sharma, P. Comparative analysis of DNA methylation changes in two contrasting wheat genotypes under water deficit. Biol. Plant. 2018, 62, 471–478. [Google Scholar] [CrossRef]
- González, R.M.; Ricardi, M.M.; Iusem, N.D. Epigenetic marks in an adaptive water stress-responsive gene in tomato roots under normal and drought conditions. Epigenetics 2013, 8, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Sapna, H.; Ashwini, N.; Ramesh, S.; Nataraja, K.N. Assessment of DNA methylation pattern under drought stress using methylation-sensitive randomly amplified polymorphism analysis in rice. Plant Genet. Resour. Charact. Util. 2020, 18, 222–230. [Google Scholar] [CrossRef]
- Sallam, N.; Moussa, M.; Yacout, M.; El-Seedy, A. Differential DNA Methylation under Drought Stress in Maize. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 2527–2543. [Google Scholar] [CrossRef]
- Abid, G.; Mingeot, D.; Muhovski, Y.; Mergeai, G.; Aouida, M.; Abdelkarim, S.; Aroua, I.; El Ayed, M.; M’hamdi, M.; Sassi, K.; et al. Analysis of DNA methylation patterns associated with drought stress response in faba bean (Vicia faba L.) using methylation-sensitive amplification polymorphism (MSAP). Environ. Exp. Bot. 2017, 142, 34–44. [Google Scholar] [CrossRef]
- Liang, D.; Zhang, Z.; Wu, H.; Huang, C.; Shuai, P.; Ye, C.-Y.; Tang, S.; Wang, Y.; Yang, L.; Wang, J.; et al. Single-base-resolution methylomes of populus trichocarpa reveal the association between DNA methylation and drought stress. BMC Genet. 2014, 15, S9. [Google Scholar] [CrossRef]
- Wang, W.; Qin, Q.; Sun, F.; Wang, Y.; Xu, D.; Li, Z.; Fu, B. Genome-Wide Differences in DNA Methylation Changes in Two Contrasting Rice Genotypes in Response to Drought Conditions. Front. Plant Sci. 2016, 7, 1675. [Google Scholar] [CrossRef]
- Ding, G.; Cao, L.; Zhou, J.; Li, Z.; Lai, Y.; Liu, K.; Luo, Y.; Bai, L.; Wang, X.; Wang, T.; et al. DNA Methylation Correlates with the Expression of Drought-Responsive Genes and Drought Resistance in Rice. Agronomy 2022, 12, 1445. [Google Scholar] [CrossRef]
- Bhardwaj, J.; Mahajan, M.; Yadav, S.K. Comparative analysis of DNA methylation polymorphism in drought sensitive (HPKC2) and tolerant (HPK4) genotypes of horse Gram (Macrotyloma uniflorum). Biochem. Genet. 2013, 51, 493–502. [Google Scholar] [CrossRef]
- Lyu, Z.; Zhang, G.; Song, Y.; Diao, S.; He, C.; Zhang, J. Transcriptome and DNA methylome provide insights into the molecular regulation of drought stress in sea buckthorn. Genomics 2022, 114, 110345. [Google Scholar] [CrossRef]
- Luo, D.; Shan, C.; Zengqiang, L.; Meiqiong, T.; Caijin, W.; Enerand, M.; Zhen, H.; Jiao, P.; Xia, W.; Qijing, W.; et al. Methyl-Sensitive Amplification Polymorphism (MSAP) Analysis Provides Insights into the DNA Methylation Underlying Heterosis in Kenaf (Hibiscus Cannabinus L.) Drought Tolerance. J. Nat. Fibers 2022, 19, 13665–13680. [Google Scholar] [CrossRef]
- Naderi, S.; Maali-Amiri, R.; Sadeghi, L.; Hamidi, A. Physio-biochemical and DNA methylation analysis of the defense response network of wheat to drought stress. Plant Physiol. Biochem. 2024, 209, 108516. [Google Scholar] [CrossRef]
- Mao, H.; Wang, H.; Liu, S.; Li, Z.; Yang, X.; Yan, J.; Li, J.; Tran, L.-S.P.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015, 6, 8326. [Google Scholar] [CrossRef] [PubMed]
- Surdonja, K.; Eggert, K.; Hajirezaei, M.-R.; Harshavardhan, V.T.; Seiler, C.; Von Wirén, N.; Sreenivasulu, N.; Kuhlmann, M. Increase of DNA Methylation at the HvCKX2.1 Promoter by Terminal Drought Stress in Barley. Epigenomes 2017, 1, 9. [Google Scholar] [CrossRef]
- Wang, W.S.; Pan, Y.J.; Zhao, X.Q.; Dwivedi, D.; Zhu, L.H.; Ali, J.; Fu, B.Y.; Li, Z.K. Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J. Exp. Bot. 2011, 62, 1951–1960. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, L.; Li, M.; Lou, Q.; Xia, H.; Wang, P.; Li, T.; Liu, H.; Luo, L. Transgenerational variations in DNA methylation induced by drought stress in two rice varieties with distinguished difference to drought resistance. PLoS ONE 2013, 8, e80253. [Google Scholar] [CrossRef]
- Kou, S.; Gu, Q.; Duan, L.; Liu, G.; Yuan, P.; Li, H.; Wu, Z.; Liu, W.; Huang, P.; Liu, L. Genome-Wide Bisulphite Sequencing Uncovered the Contribution of DNA Methylation to Rice Short-Term Drought Memory Formation. J. Plant Growth Regul. 2022, 41, 2903–2917. [Google Scholar] [CrossRef]
- Zi, N.; Ren, W.; Guo, H.; Yuan, F.; Liu, Y.; Fry, E. DNA Methylation Participates in Drought Stress Memory and Response to Drought in Medicago ruthenica. Genes 2024, 15, 1286. [Google Scholar] [CrossRef]
- Shi, N.; Fan, Y.; Zhang, W.; Zhang, Z.; Pu, Z.; Li, Z.; Hu, L.; Bi, Z.; Yao, P.; Liu, Y.; et al. Genome-Wide Identification and Drought-Responsive Functional Analysis of the GST Gene Family in Potato (Solanum tuberosum L.). Antioxidants 2025, 14, 239. [Google Scholar] [CrossRef] [PubMed]
- Fei, Y.; Xue, Y.; Du, P.; Yang, S.; Deng, X. Expression analysis and promoter methylation under osmotic and salinity stress of TaGAPC1 in wheat (Triticum aestivum L). Protoplasma 2017, 254, 987–996. [Google Scholar] [CrossRef]
- Niu, C.; Jiang, L.; Cao, F.; Liu, C.; Guo, J.; Zhang, Z.; Yue, Q.; Hou, N.; Liu, Z.; Li, X.; et al. Methylation of a MITE insertion in the MdRFNR1-1 promoter is positively associated with its allelic expression in apple in response to drought stress. Plant Cell 2022, 34, 3983–4006. [Google Scholar] [CrossRef]
- Li, P.; Yang, H.; Wang, L.; Liu, H.; Huo, H.; Zhang, C.; Liu, A.; Zhu, A.; Hu, J.; Lin, Y.; et al. Physiological and Transcriptome Analyses Reveal Short-Term Responses and Formation of Memory Under Drought Stress in Rice. Front. Genet. 2019, 10, 55. [Google Scholar] [CrossRef]
- Fan, H.; Li, T.; Guan, L.; Li, Z.; Guo, N.; Cai, Y.; Lin, Y. Effects of exogenous nitric oxide on antioxidation and DNA methylation of Dendrobium huoshanense grown under drought stress. Plant Cell Tissue Organ Cult. 2012, 109, 307–314. [Google Scholar] [CrossRef]
- Mittler, R.; Finka, A.; Goloubinoff, P. How do plants feel the heat? Trends Biochem. Sci. 2012, 37, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Sun, Y.; Du, X.; Chu, Z.; Zhong, X.; Chen, X. Plant-specific histone deacetylases associate with ARGONAUTE4 to promote heterochromatin stabilization and plant heat tolerance. New Phytol. 2023, 238, 252–269. [Google Scholar] [CrossRef] [PubMed]
- Korotko, U.; Chwiałkowska, K.; Sańko-Sawczenko, I.; Kwasniewski, M. DNA Demethylation in Response to Heat Stress in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 1555. [Google Scholar] [CrossRef] [PubMed]
- Malabarba, J.; Windels, D.; Xu, W.; Verdier, J. Regulation of DNA (de)Methylation Positively Impacts Seed Germination during Seed Development under Heat Stress. Genes 2021, 12, 457. [Google Scholar] [CrossRef]
- Qian, Y.; Hu, W.; Liao, J.; Zhang, J.; Ren, Q. The Dynamics of DNA methylation in the maize (Zea mays L.) inbred line B73 response to heat stress at the seedling stage. Biochem. Biophys. Res. Commun. 2019, 512, 742–749. [Google Scholar] [CrossRef]
- Ci, D.; Song, Y.; Tian, M.; Zhang, D. Methylation of miRNA genes in the response to temperature stress in Populus simonii. Front. Plant Sci. 2015, 6, 921. [Google Scholar] [CrossRef]
- Gong, Z.; Zheng, J.; Yang, N.; Li, X.; Qian, S.; Sun, F.; Geng, S.; Liang, Y.; Wang, J. Whole-Genome Bisulfite Sequencing (WGBS) Analysis of Gossypium hirsutum under High-Temperature Stress Conditions. Genes 2024, 15, 1241. [Google Scholar] [CrossRef]
- Liu, T.; Li, Y.; Duan, W.; Huang, F.; Hou, X. Cold acclimation alters DNA methylation patterns and confers tolerance to heat and increases growth rate in Brassica rapa. J. Exp. Bot. 2017, 68, 1213–1224. [Google Scholar] [CrossRef]
- Li, J.; Huang, Q.; Sun, M.; Zhang, T.; Li, H.; Chen, B.; Xu, K.; Gao, G.; Li, F.; Yan, G.; et al. Global DNA methylation variations after short-term heat shock treatment in cultured microspores of Brassica napus cv. Topas. Sci. Rep. 2016, 6, 38401. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Li, J.; Li, H.; Li, F.; Xu, K.; Yan, G.; Chen, B.; Qiao, J.; Wu, X. Comparison of the heat stress induced variations in DNA methylation between heat-tolerant and heat-sensitive rapeseed seedlings. Breed. Sci. 2014, 64, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, S.; Wang, T.; Tao, Z.; Huang, S.; Lin, N.; Zhao, Y.; Wang, C.; Li, P. Cooperative condensation of RNA-DIRECTED DNA METHYLATION 16 splicing isoforms enhances heat tolerance in Arabidopsis. Nat. Commun. 2025, 16, 433. [Google Scholar] [CrossRef] [PubMed]
- Folsom, J.J.; Begcy, K.; Hao, X.; Wang, D.; Walia, H. Rice fertilization-Independent Endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol. 2014, 165, 238–248. [Google Scholar] [CrossRef]
- Jiang, J.; Gwee, J.; Fang, J.; Leichter, S.M.; Sanders, D.; Ji, X.; Song, J.; Zhong, X. Substrate specificity and protein stability drive the divergence of plant-specific DNA methyltransferases. Sci. Adv. 2024, 10, eadr2222. [Google Scholar] [CrossRef]
- Shen, X.; De Jonge, J.; Forsberg, S.K.; Pettersson, M.E.; Sheng, Z.; Hennig, L.; Carlborg, Ö. Natural CMT2 variation is associated with genome-wide methylation changes and temperature seasonality. PLoS Genet. 2014, 10, e1004842. [Google Scholar] [CrossRef]
- Nozawa, K.; Chen, J.; Jiang, J.; Leichter, S.M.; Yamada, M.; Suzuki, T.; Liu, F.; Ito, H.; Zhong, X. DNA methyltransferase CHROMOMETHYLASE3 prevents ONSEN transposon silencing under heat stress. PLoS Genet. 2021, 17, e1009710. [Google Scholar] [CrossRef] [PubMed]
- Popova, O.V.; Dinh, H.Q.; Aufsatz, W.; Jonak, C. The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol. Plant 2013, 6, 396–410. [Google Scholar] [CrossRef]
- Naydenov, M.; Baev, V.; Apostolova, E.; Gospodinova, N.; Sablok, G.; Gozmanova, M.; Yahubyan, G. High-temperature effect on genes engaged in DNA methylation and affected by DNA methylation in Arabidopsis. Plant Physiol. Biochem. 2015, 87, 102–108. [Google Scholar] [CrossRef]
- Tang, X.; Wang, Q.; Yuan, H.; Huang, X. Chilling-induced DNA Demethylation is associated with the cold tolerance of Hevea brasiliensis. BMC Plant Biol. 2018, 18, 70. [Google Scholar] [CrossRef] [PubMed]
- Gutschker, S.; Corral, J.M.; Schmiedl, A.; Ludewig, F.; Koch, W.; Fiedler-Wiechers, K.; Czarnecki, O.; Harms, K.; Keller, I.; Martins Rodrigues, C.; et al. Multi-omics data integration reveals link between epigenetic modifications and gene expression in sugar beet (Beta vulgaris subsp. vulgaris) in response to cold. BMC Genom. 2022, 23, 144. [Google Scholar] [CrossRef]
- Yu, G.; Zhang, B.; Chen, Q.; Huang, Z.; Zhang, B.; Wang, K.; Han, J. Dynamic DNA methylation modifications in the cold stress response of cassava. Genomics 2024, 116, 110871. [Google Scholar] [CrossRef]
- Song, Y.; Liu, L.; Feng, Y.; Wei, Y.; Yue, X.; He, W.; Zhang, H.; An, L. Chilling- and Freezing-Induced Alterations in Cytosine Methylation and Its Association with the Cold Tolerance of an Alpine Subnival Plant, Chorispora bungeana. PLoS ONE 2015, 10, e0135485. [Google Scholar] [CrossRef]
- Fan, H.H.; Wei, J.; Li, T.C.; Li, Z.P.; Guo, N.; Cai, Y.P.; Lin, Y. DNA methylation alterations of upland cotton (Gossypium hirsutum) in response to cold stress. Acta Physiol. Plant. 2013, 35, 2445–2453. [Google Scholar] [CrossRef]
- Shan, X.; Wang, X.; Yang, G.; Wu, Y.; Su, S.; Li, S.; Liu, H.; Yuan, Y. Analysis of the DNA methylation of maize (Zea mays L.) in response to cold stress based on methylation-sensitive amplified polymorphisms. J. Plant Biol. 2013, 56, 32–38. [Google Scholar] [CrossRef]
- Kenchanmane Raju, S.K.; Shao, M.R.; Wamboldt, Y.; Mackenzie, S. Epigenomic plasticity of Arabidopsis msh1 mutants under prolonged cold stress. Plant Direct 2018, 2, e00079. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, L.; Zhao, L.; Li, J.; He, S.; Zhou, K.; Yang, F.; Huang, M.; Jiang, L.; Li, L. Trichostatin A Selectively Suppresses the Cold-Induced Transcription of the ZmDREB1 Gene in Maize. PLoS ONE 2011, 6, e22132. [Google Scholar] [CrossRef]
- Steward, N.; Ito, M.; Yamaguchi, Y.; Koizumi, N.; Sano, H. Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J. Biol. Chem. 2002, 277, 37741–37746. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lang, C.; Wu, Y.; Meng, D.; Yang, T.; Li, D.; Jin, T.; Zhou, X. ROS1-mediated decrease in DNA methylation and increase in expression of defense genes and stress response genes in Arabidopsis thaliana due to abiotic stresses. BMC Plant Biol. 2022, 22, 104. [Google Scholar] [CrossRef]
- Chan, Z.; Wang, Y.; Cao, M.; Gong, Y.; Mu, Z.; Wang, H.; Hu, Y.; Deng, X.; He, X.J.; Zhu, J.K. RDM4 modulates cold stress resistance in Arabidopsis partially through the CBF-mediated pathway. New Phytol. 2016, 209, 1527–1539. [Google Scholar] [CrossRef]
- Kidokoro, S.; Kim, J.S.; Ishikawa, T.; Suzuki, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DREB1A/CBF3 Is Repressed by Transgene-Induced DNA Methylation in the Arabidopsis ice1-1 Mutant. Plant Cell 2020, 32, 1035–1048. [Google Scholar] [CrossRef]
- Li, Z.; Wang, W.; Yu, X.; Zhao, P.; Li, W.; Zhang, X.; Peng, M.; Li, S.; Ruan, M. Integrated analysis of DNA methylome and transcriptome revealing epigenetic regulation of CRIR1-promoted cold tolerance. BMC Plant Biol. 2024, 24, 631. [Google Scholar] [CrossRef]
- Huang, J.; Wang, Y.; Yu, J.; Li, F.; Yi, L.; Li, Y.; Xie, N.; Wu, Q.; Samarina, L.; Tong, W.; et al. Evolutionary Landscape of Tea Circular RNAs and Its Contribution to Chilling Tolerance of Tea Plant. Int. J. Mol. Sci. 2023, 24, 1478. [Google Scholar] [CrossRef] [PubMed]
- Hashida, S.N.; Uchiyama, T.; Martin, C.; Kishima, Y.; Sano, Y.; Mikami, T. The temperature-dependent change in methylation of the Antirrhinum transposon Tam3 is controlled by the activity of its transposase. Plant Cell 2006, 18, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Steward, N.; Kusano, T.; Sano, H. Expression of ZmMET1, a gene encoding a DNA methyltransferase from maize, is associated not only with DNA replication in actively proliferating cells, but also with altered DNA methylation status in cold-stressed quiescent cells. Nucleic Acids Res. 2000, 28, 3250–3259. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.S.; Zhang, X.; Zhang, W.; Shen, D.; Wang, H.; Xia, Y.; Qiu, Y.; Song, J.; Wang, C.; Li, X. The association of changes in DNA methylation with temperature-dependent sex determination in cucumber. J. Exp. Bot. 2017, 68, 2899–2912. [Google Scholar] [CrossRef]
- Xie, H.J.; Li, H.; Liu, D.; Dai, W.M.; He, J.Y.; Lin, S.; Duan, H.; Liu, L.L.; Chen, S.G.; Song, X.L.; et al. ICE1 demethylation drives the range expansion of a plant invader through cold tolerance divergence. Mol. Ecol. 2015, 24, 835–850. [Google Scholar] [CrossRef]
- Xie, H.; Sun, Y.; Cheng, B.; Xue, S.; Cheng, D.; Liu, L.; Meng, L.; Qiang, S. Variation in ICE1 Methylation Primarily Determines Phenotypic Variation in Freezing Tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2019, 60, 152–165. [Google Scholar] [CrossRef]
- Guo, H.; Wu, T.; Li, S.; He, Q.; Yang, Z.; Zhang, W.; Gan, Y.; Sun, P.; Xiang, G.; Zhang, H.; et al. The Methylation Patterns and Transcriptional Responses to Chilling Stress at the Seedling Stage in Rice. Int. J. Mol. Sci. 2019, 20, 5089. [Google Scholar] [CrossRef]
- Yue, R.; Li, Y.; Qi, Y.; Liang, X.; Zheng, Z.; Ye, Z.; Tong, W.; Si, X.; Zhang, Y.; Xia, E.; et al. Divergent MYB paralogs determine spatial distribution of linalool mediated by JA and DNA demethylation participating in aroma formation and cold tolerance of tea plants. Plant Biotechnol. J. 2025, 23, 1455–1475. [Google Scholar] [CrossRef] [PubMed]
- Xin, C.; Wang, J.; Chi, J.; Xu, Y.; Liang, R.; Jian, L.; Wang, L.; Guo, J. Intragenic cytosine methylation modification regulates the response of SUCLα1 to lower temperature in Solanaceae. Plant Sci 2025, 350, 112320. [Google Scholar] [CrossRef]
- Qian, C.; Shao, Y.; Cai, Z.; Zhang, B.; Sohail, H.; Liu, J.; Kan, J.; Zhang, M.; Xiao, L.; Yang, X.; et al. Melatonin Reduces Lignin Biosynthesis by Fostering Epigenetic Modifications in Water Bamboo Shoots under Cold Storage. J. Agric. Food Chem. 2025, 73, 7504–7516. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Shi, Y.; Huang, Y.; Lu, J.; Zhang, M.; Cao, X.; Hu, R.; Li, D.; Chen, W.; Zhu, C.; et al. XYLEM NAC DOMAIN 1 (EjXND1) relieves cold-induced lignification by negatively regulating the EjHB1-EjPRX12 module in loquat fruit. J. Adv. Res. 2024, 73, 93–104. [Google Scholar] [CrossRef]
- Huang, Y.; Liang, Z.; Lu, J.; Zhang, M.; Cao, X.; Hu, R.; Li, D.; Grierson, D.; Chen, W.; Zhu, C.; et al. The transcription factor EjNAC5 regulates loquat fruit chilling lignification. J. Exp. Bot. 2024, 75, 6625–6643. [Google Scholar] [CrossRef]
- Sicilia, A.; Scialò, E.; Puglisi, I.; Lo Piero, A.R. Anthocyanin Biosynthesis and DNA Methylation Dynamics in Sweet Orange Fruit [Citrus sinensis L. (Osbeck)] under Cold Stress. J. Agric. Food Chem. 2020, 68, 7024–7031. [Google Scholar] [CrossRef]
- Song, Y.; Jia, Z.; Hou, Y.; Ma, X.; Li, L.; Jin, X.; An, L. Roles of DNA Methylation in Cold Priming in Tartary Buckwheat. Front. Plant Sci. 2020, 11, 608540. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Cui, J.; Zheng, G.; Dong, X.; Wu, Z.; Fang, Y.; Sa, E.; Zhu, S.; Li, B.; Wei, H.; et al. BnaHSFA2, a heat shock transcription factor interacting with HSP70 and MPK11, enhances freezing tolerance in transgenic rapeseed. Plant Physiol. Biochem. 2025, 219, 109423. [Google Scholar] [CrossRef]
- Conde, D.; Le Gac, A.L.; Perales, M.; Dervinis, C.; Kirst, M.; Maury, S.; González-Melendi, P.; Allona, I. Chilling-responsive DEMETER-LIKE DNA demethylase mediates in poplar bud break. Plant Cell Environ. 2017, 40, 2236–2249. [Google Scholar] [CrossRef]
- Conde, D.; Moreno-Cortés, A.; Dervinis, C.; Ramos-Sánchez, J.M.; Kirst, M.; Perales, M.; González-Melendi, P.; Allona, I. Overexpression of DEMETER, a DNA demethylase, promotes early apical bud maturation in poplar. Plant Cell Environ. 2017, 40, 2806–2819. [Google Scholar] [CrossRef]
- Trap-Gentil, M.V.; Hébrard, C.; Lafon-Placette, C.; Delaunay, A.; Hagège, D.; Joseph, C.; Brignolas, F.; Lefebvre, M.; Barnes, S.; Maury, S. Time course and amplitude of DNA methylation in the shoot apical meristem are critical points for bolting induction in sugar beet and bolting tolerance between genotypes. J. Exp. Bot. 2011, 62, 2585–2597. [Google Scholar] [CrossRef]
- Duan, W.; Zhang, H.; Zhang, B.; Wu, X.; Shao, S.; Li, Y.; Hou, X.; Liu, T. Role of vernalization-mediated demethylation in the floral transition of Brassica rapa. Planta 2017, 245, 227–233. [Google Scholar] [CrossRef]
- Khan, A.R.; Enjalbert, J.; Marsollier, A.C.; Rousselet, A.; Goldringer, I.; Vitte, C. Vernalization treatment induces site-specific DNA hypermethylation at the VERNALIZATION-A1 (VRN-A1) locus in hexaploid winter wheat. BMC Plant Biol. 2013, 13, 209. [Google Scholar] [CrossRef] [PubMed]
- Mao, F.; Xie, H.; Shi, Y.; Jiang, S.; Wang, S.; Wu, Y. The Global Changes of N6-methyldeoxyadenosine in Response to Low Temperature in Arabidopsis thaliana and Rice. Plants 2023, 12, 2373. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Tao, X.; Fahim, A.M.; Xu, Y.; Zhang, Y.; Li, S.; Yang, G.; Pu, Y.; Wang, W.; Liu, L.; et al. Novel insights into the unique characterization of N6-methyladenosine RNA modification and regulating cold tolerance in winter Brassica rapa. Int. J. Biol. Macromol. 2025, 303, 140460. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Mishra, N.; Singh, U.M.; Srivastava, R. Genotoxicity: Mechanisms and its impact on human diseases. Octa J. Biosci. 2016, 4, 67–70. [Google Scholar]
- Ou, X.; Zhang, Y.; Xu, C.; Lin, X.; Zang, Q.; Zhuang, T.; Jiang, L.; von Wettstein, D.; Liu, B. Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice (Oryza sativa L.). PLoS ONE 2012, 7, e41143. [Google Scholar] [CrossRef]
- Miao, Y.; Cong, W.; Mu, J.; Fu, T.; Zhuang, T.; Yan, Y.; Kang, Y.; Yu, L.; Zhao, W.; Li, H.; et al. Various potentially toxic element tolerances in different rice genotypes correlate with distinct physiological responses and alterations in DNA methylation. Chemosphere 2022, 292, 133462. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, B. Identification of a rice metal tolerance protein OsMTP11 as a manganese transporter. PLoS ONE 2017, 12, e0174987. [Google Scholar] [CrossRef]
- Liu, X.S.; Li, H.; Feng, S.J.; Yang, Z.M. A transposable element-derived siRNAs involve DNA hypermethylation at the promoter of OsGSTZ4 for cadmium tolerance in rice. Gene 2024, 892, 147900. [Google Scholar] [CrossRef]
- Feng, S.J.; Liu, X.S.; Tao, H.; Tan, S.K.; Chu, S.S.; Oono, Y.; Zhang, X.D.; Chen, J.; Yang, Z.M. Variation of DNA methylation patterns associated with gene expression in rice (Oryza sativa) exposed to cadmium. Plant Cell Environ. 2016, 39, 2629–2649. [Google Scholar] [CrossRef]
- Xin, C.; Chi, J.; Zhao, Y.; He, Y.; Guo, J. Cadmium stress alters cytosine methylation status and expression of a select set of genes in Nicotiana benthamiana. Plant Sci 2019, 284, 16–24. [Google Scholar] [CrossRef]
- Bai, Y.; Wang, X.; Ali, S.; Liu, Y.; Zhou, J.; Liu, M.; Liu, S.; Tang, Y. A 24-nt miR9560 modulates the transporter gene BrpHMA2 expression in Brassica parachinensis. Plant Genome 2025, 18, e70013. [Google Scholar] [CrossRef]
- Lancíková, V.; Kačírová, J.; Hricová, A. Identification and gene expression analysis of cytosine-5 DNA methyltransferase and demethylase genes in Amaranthus cruentus L. under heavy metal stress. Front. Plant Sci. 2022, 13, 1092067. [Google Scholar] [CrossRef]
- Chen, L.; Li, X.; Liu, H.; He, F.; Li, M.; Long, R.; Wang, X.; Kang, J.; Yang, Q. Comprehensive analysis of epigenetic modifications in alfalfa under cadmium stress. J. Hazard. Mater. 2025, 482, 136545. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Lu, H.; Wang, C.; Mubeen, S.; Cao, S.; Yue, J.; Pan, J.; Wu, X.; Wu, Q.; Zhang, H.; et al. Physiological and DNA methylation analysis provides epigenetic insights into kenaf cadmium tolerance heterosis. Plant Sci. 2023, 331, 111663. [Google Scholar] [CrossRef]
- Pacenza, M.; Muto, A.; Chiappetta, A.; Mariotti, L.; Talarico, E.; Picciarelli, P.; Picardi, E.; Bruno, L.; Bitonti, M.B. In Arabidopsis thaliana Cd differentially impacts on hormone genetic pathways in the methylation defective ddc mutant compared to wild type. Sci. Rep. 2021, 11, 10965. [Google Scholar] [CrossRef]
- Chakrabarti, M.; Mukherjee, A. Investigating the underlying mechanism of cadmium-induced plant adaptive response to genotoxic stress. Ecotoxicol. Environ. Saf. 2021, 209, 111817. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Gao, J.; Qin, C.; Ma, H.; Huang, H.; Song, P.; Luo, X.; Lin, H.; Shen, Y.; Pan, G.; et al. The Dynamics of DNA Methylation in Maize Roots under Pb Stress. Int. J. Mol. Sci. 2014, 15, 23537–23554. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Xu, L.; Wang, Y.; Dong, J.; Zhang, X.; Wang, K.; Ying, J.; Li, C.; Liu, L. Melatonin-induced DNA demethylation of metal transporters and antioxidant genes alleviates lead stress in radish plants. Hortic. Res. 2021, 8, 124. [Google Scholar] [CrossRef]
- Tang, M.; Li, R.; Chen, P. Exogenous glutathione can alleviate chromium toxicity in kenaf by activating antioxidant system and regulating DNA methylation. Chemosphere 2023, 337, 139305. [Google Scholar] [CrossRef]
- Guarino, F.; Cicatelli, A.; Nissim, W.G.; Colzi, I.; Gonnelli, C.; Basso, M.F.; Vergata, C.; Contaldi, F.; Martinelli, F.; Castiglione, S. Epigenetic changes induced by chronic and acute chromium stress treatments in Arabidopsis thaliana identified by the MSAP-Seq. Chemosphere 2024, 362, 142642. [Google Scholar] [CrossRef] [PubMed]
- Niedziela, A. The influence of Al(3+) on DNA methylation and sequence changes in the triticale (× Triticosecale Wittmack) genome. J. Appl. Genet. 2018, 59, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Taspinar, M.S.; Aydin, M.; Sigmaz, B.; Yagci, S.; Arslan, E.; Agar, G. Aluminum-Induced Changes on DNA Damage, DNA Methylation and LTR Retrotransposon Polymorphism in Maize. Arab. J. Sci. Eng. 2018, 43, 123–131. [Google Scholar] [CrossRef]
- Choi, C.S.; Sano, H. Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol. Genet. Genom. 2007, 277, 589–600. [Google Scholar] [CrossRef]
- Kashino-Fujii, M.; Yokosho, K.; Yamaji, N.; Yamane, M.; Saisho, D.; Sato, K.; Ma, J.F. Retrotransposon Insertion and DNA Methylation Regulate Aluminum Tolerance in European Barley Accessions. Plant Physiol. 2018, 178, 716–727. [Google Scholar] [CrossRef]
- Gullì, M.; Marchi, L.; Fragni, R.; Buschini, A.; Visioli, G. Epigenetic modifications preserve the hyperaccumulator Noccaea caerulescens from Ni geno-toxicity. Environ. Mol. Mutagen. 2018, 59, 464–475. [Google Scholar] [CrossRef]
- Aina, R.; Sgorbati, S.; Santagostino, A.; Labra, M.; Ghiani, A.; Citterio, S. Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hemp. Physiol. Plant. 2004, 121, 472–480. [Google Scholar] [CrossRef]
- Ding, S.; Zhang, H.; Zhou, C.; Bao, Y.; Xu, X.; Chen, Y.; Shen, Z.; Chen, C. Transcriptomic, epigenomic and physiological comparisons reveal key factors for different manganese tolerances in three Chenopodium ambrosioides L. populations. Plant Physiol. Biochem. 2023, 201, 107883. [Google Scholar] [CrossRef]
- Jing, M.; Zhang, H.; Wei, M.; Tang, Y.; Xia, Y.; Chen, Y.; Shen, Z.; Chen, C. Reactive Oxygen Species Partly Mediate DNA Methylation in Responses to Different Heavy Metals in Pokeweed. Front. Plant Sci. 2022, 13, 845108. [Google Scholar] [CrossRef]
- Cong, W.; Li, N.; Miao, Y.; Huang, Y.; Zhao, W.; Kang, Y.; Zhang, B.; Wang, J.; Zhang, J.; Lv, Y.; et al. DNA hypomethylation-associated transcriptional rewiring enables resistance to heavy metal mercury (Hg) stress in rice. J. Hazard. Mater. 2024, 461, 132649. [Google Scholar] [CrossRef] [PubMed]
- Zemanová, V.; Popov, M.; Pavlíková, D.; Kotrba, P.; Hnilička, F.; Česká, J.; Pavlík, M. Effect of arsenic stress on 5-methylcytosine, photosynthetic parameters and nutrient content in arsenic hyperaccumulator Pteris cretica (L.) var. Albo-lineata. BMC Plant Biol. 2020, 20, 130. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lodhi, N.; Srivastava, R. Dynamics and Malleability of Plant DNA Methylation During Abiotic Stresses. Epigenomes 2025, 9, 31. https://doi.org/10.3390/epigenomes9030031
Lodhi N, Srivastava R. Dynamics and Malleability of Plant DNA Methylation During Abiotic Stresses. Epigenomes. 2025; 9(3):31. https://doi.org/10.3390/epigenomes9030031
Chicago/Turabian StyleLodhi, Niraj, and Rakesh Srivastava. 2025. "Dynamics and Malleability of Plant DNA Methylation During Abiotic Stresses" Epigenomes 9, no. 3: 31. https://doi.org/10.3390/epigenomes9030031
APA StyleLodhi, N., & Srivastava, R. (2025). Dynamics and Malleability of Plant DNA Methylation During Abiotic Stresses. Epigenomes, 9(3), 31. https://doi.org/10.3390/epigenomes9030031






