Epigenetic Regulation of Mammalian Cardiomyocyte Development
Abstract
1. Introduction
2. Cellular Events and Morphological Changes That Regulate the Contractile Activity of Cardiomyocytes
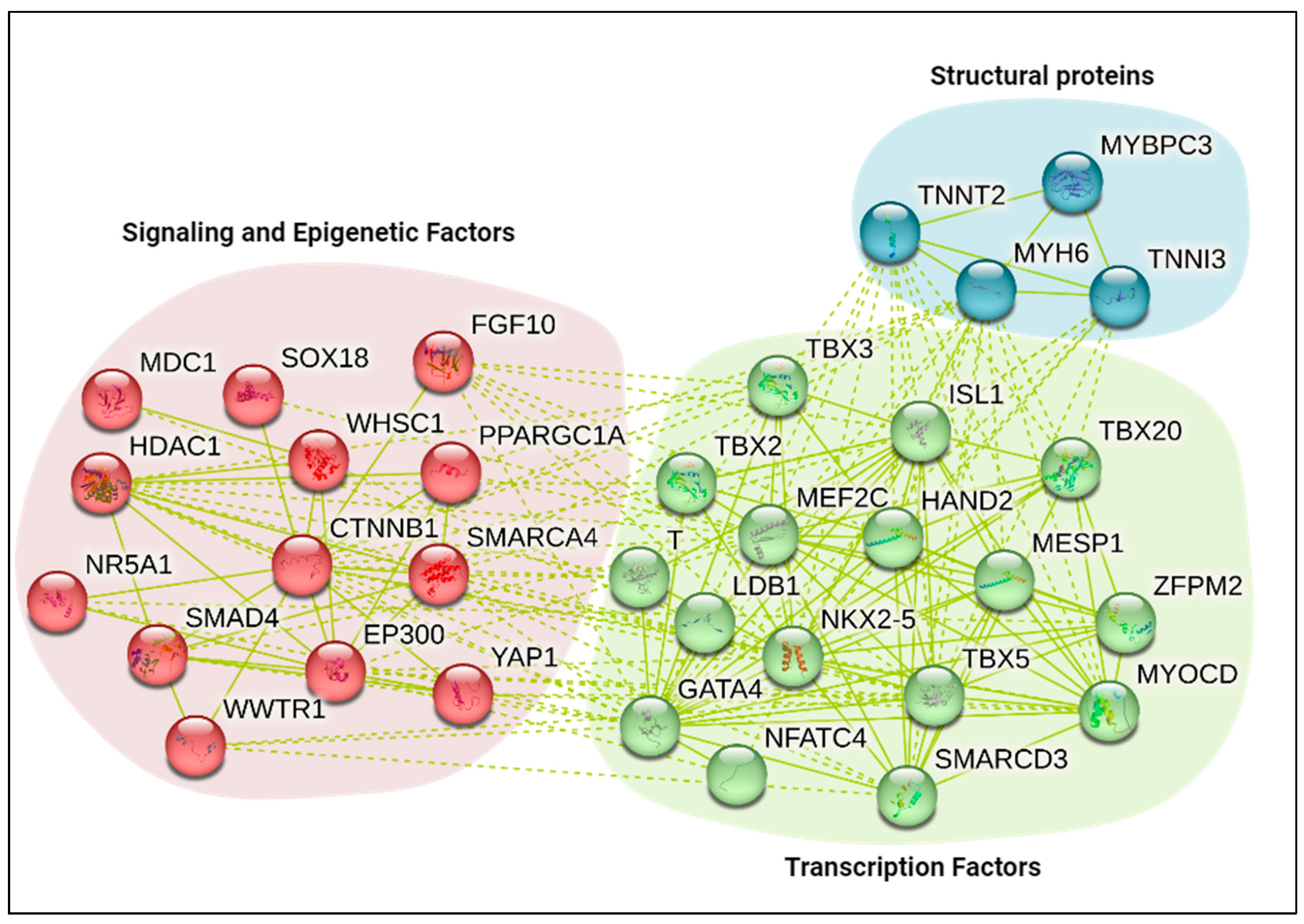
3. Transcriptional Regulation of Cardiomyocyte Development
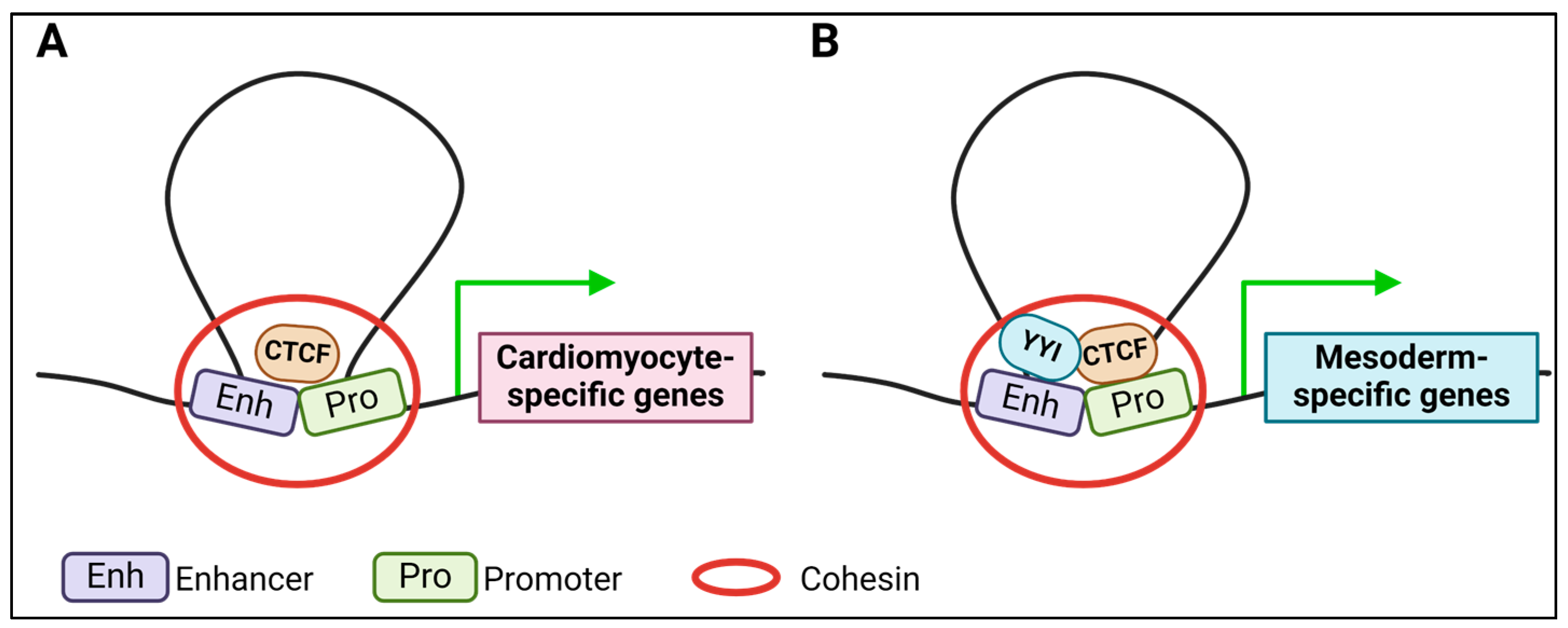
4. Insulator Activity of Transcription Factors
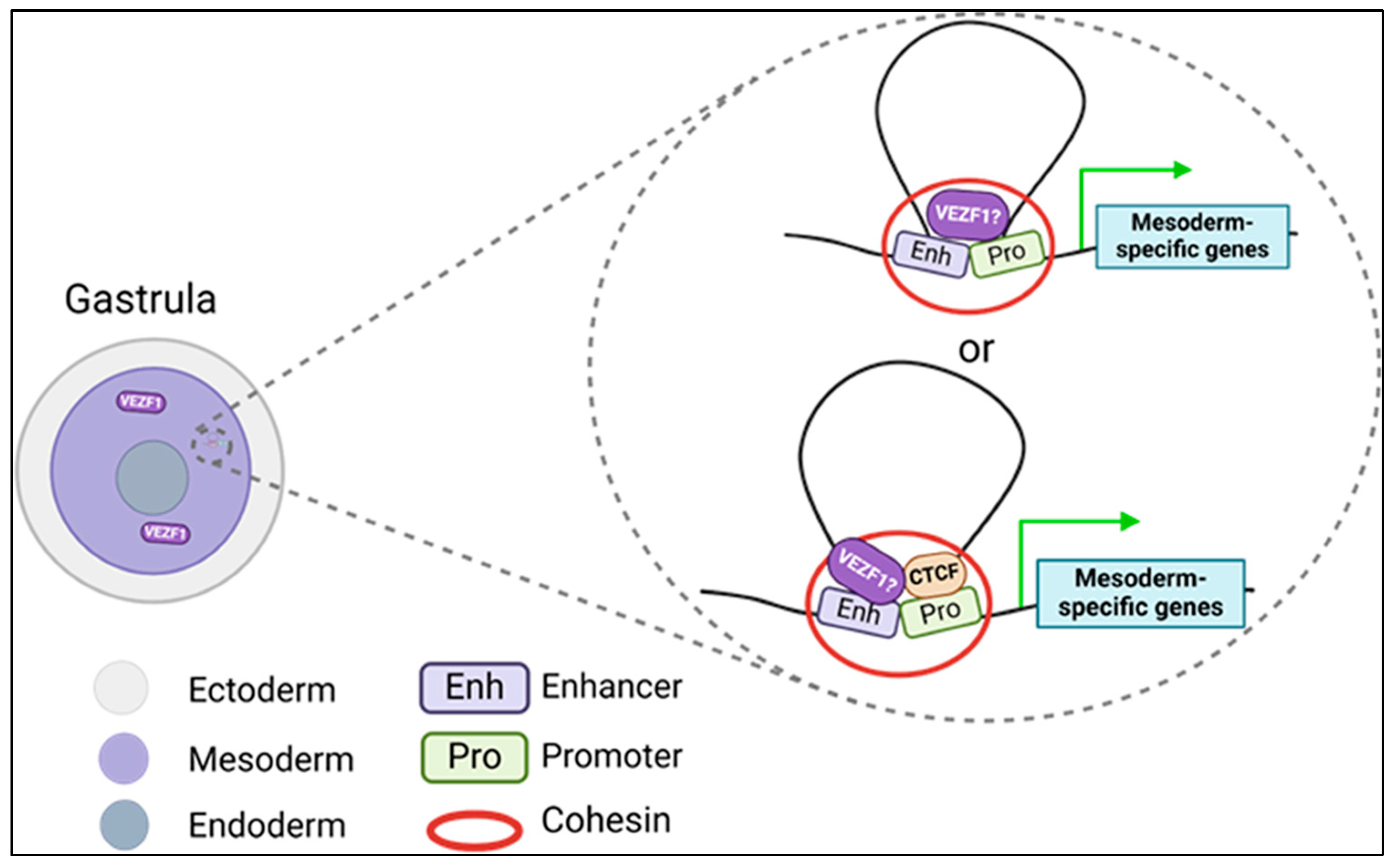
5. The Emerging Role of VEZF1 in Cardiac Development
6. Epigenetic Regulation of Cardiomyocyte Differentiation
7. DNA Methylation
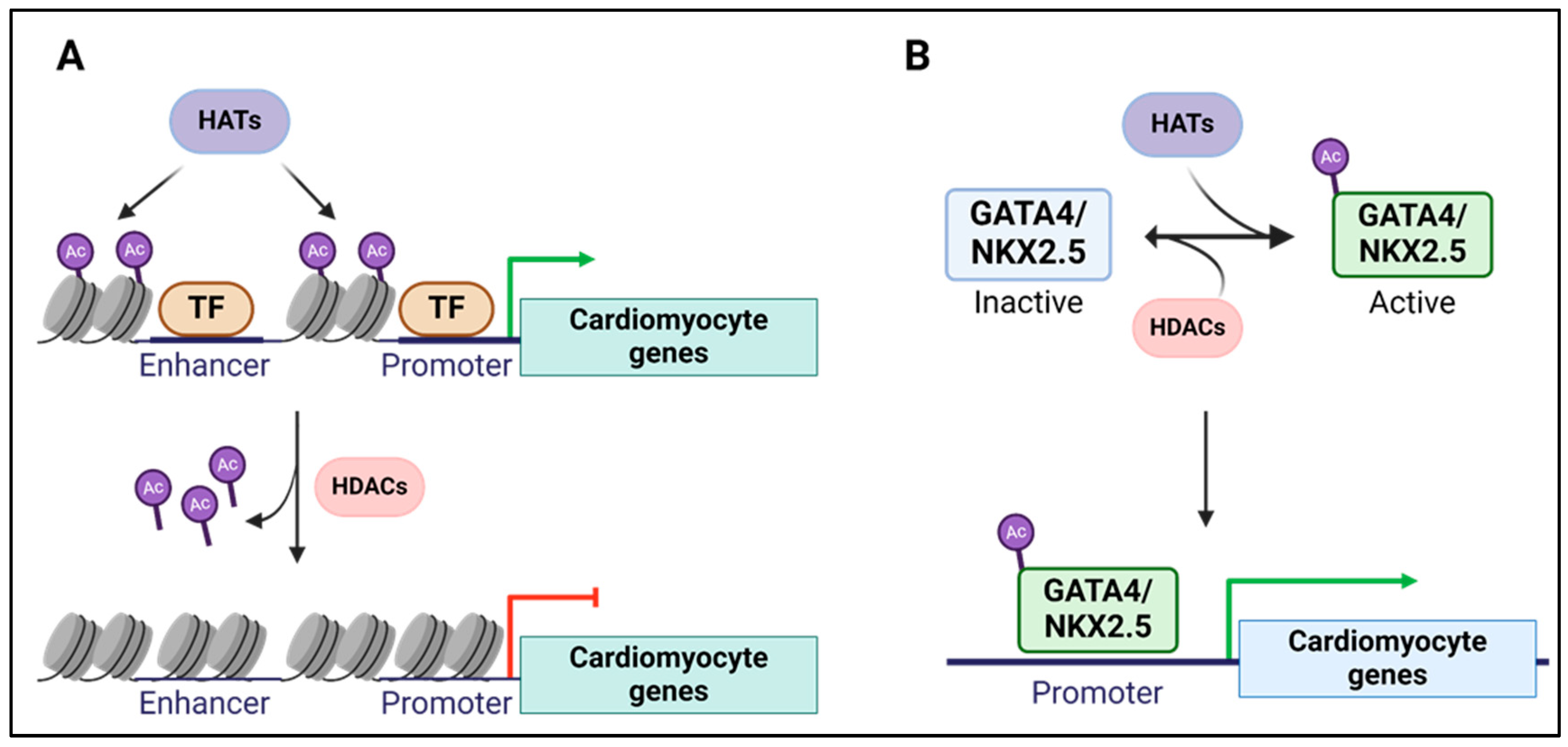
8. Histone Modifications
9. Noncoding RNAs in Cardiomyocyte Development
10. ATP-Dependent Chromatin Remodelers
11. Conclusions and Outcomes
Funding
Conflicts of Interest
References
- Rossant, J.; Tam, P.P.L. Early human embryonic development: Blastocyst formation to gastrulation. Dev. Cell 2022, 57, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Salehin, N.; Knowles, H.; Masamsetti, V.P.; Tam, P.P.L. Mammalian gastrulation: Signalling activity and transcriptional regulation of cell lineage differentiation and germ layer formation. Biochem. Soc. Trans. 2022, 50, 1619–1631. [Google Scholar] [CrossRef] [PubMed]
- Ivanovitch, K.; Soro-Barrio, P.; Chakravarty, P.; Jones, R.A.; Bell, D.M.; Mousavy Gharavy, S.N.; Stamataki, D.; Delile, J.; Smith, J.C.; Briscoe, J. Ventricular, atrial, and outflow tract heart progenitors arise from spatially and molecularly distinct regions of the primitive streak. PLoS Biol. 2021, 19, e3001200. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.S.K.; Shi, X.; Toyama, A.; Arpke, R.W.; Dandapat, A.; Iacovino, M.; Kang, J.; Le, G.; Hagen, H.R.; Garry, D.J.; et al. Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell 2013, 12, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Maas, R.G.C.; van den Dolder, F.W.; Yuan, Q.; van der Velden, J.; Wu, S.M.; Sluijter, J.P.G.; Buikema, J.W. Harnessing developmental cues for cardiomyocyte production. Development 2023, 150, dev201483. [Google Scholar] [CrossRef] [PubMed]
- Später, D.; Hansson, E.M.; Zangi, L.; Chien, K.R. How to make a cardiomyocyte. Development 2014, 141, 4418–4431. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.G.; Buckingham, M.E.; Moorman, A.F. Heart fields and cardiac morphogenesis. Cold Spring Harb. Perspect. Med. 2014, 4, a015750. [Google Scholar] [CrossRef] [PubMed]
- Brade, T.; Pane, L.S.; Moretti, A.; Chien, K.R.; Laugwitz, K.L. Embryonic heart progenitors and cardiogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a013847. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Meilhac, S.; Zaffran, S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 2005, 6, 826–835. [Google Scholar] [CrossRef]
- Tan, C.M.J.; Lewandowski, A.J. The Transitional Heart: From Early Embryonic and Fetal Development to Neonatal Life. Fetal Diagn. Ther. 2019, 47, 373–386. [Google Scholar] [CrossRef]
- Li, F.; Wang, X.; Capasso, J.M.; Gerdes, A.M. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J. Mol. Cell. Cardiol. 1996, 28, 1737–1746. [Google Scholar] [CrossRef] [PubMed]
- Bensley, J.G.; Stacy, V.K.; De Matteo, R.; Harding, R.; Black, M.J. Cardiac remodelling as a result of pre-term birth: Implications for future cardiovascular disease. Eur. Heart J. 2010, 31, 2058–2066. [Google Scholar] [CrossRef] [PubMed]
- Bertagnolli, M.; Huyard, F.; Cloutier, A.; Anstey, Z.; Huot-Marchand, J.; Fallaha, C.; Paradis, P.; Schiffrin, E.L.; Deblois, D.; Nuyt, A.M. Transient neonatal high oxygen exposure leads to early adult cardiac dysfunction, remodeling, and activation of the renin-angiotensin system. Hypertension 2014, 63, 143–150. [Google Scholar] [CrossRef] [PubMed]
- van Amerongen, M.J.; Engel, F.B. Features of cardiomyocyte proliferation and its potential for cardiac regeneration. J. Cell. Mol. Med. 2008, 12, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.E.; Tokuyama, T.; Anzai, T.; Chanthra, N.; Uosaki, H. Sarcomere maturation: Function acquisition, molecular mechanism, and interplay with other organelles. Philos. Trans. R. Soc. B Biol. Sci. 2022, 377, 20210325. [Google Scholar] [CrossRef] [PubMed]
- Correia, M.; Santos, F.; da Silva Ferreira, R.; Ferreira, R.; Bernardes de Jesus, B.; Nóbrega-Pereira, S. Metabolic Determinants in Cardiomyocyte Function and Heart Regenerative Strategies. Metabolites 2022, 12, 500. [Google Scholar] [CrossRef] [PubMed]
- Dhein, S.; Salameh, A. Remodeling of Cardiac Gap Junctional Cell-Cell Coupling. Cells 2021, 10, 2422. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.A.; Spinale, F.G. The structure and function of the cardiac myocyte: A review of fundamental concepts. J. Thorac. Cardiovasc. Surg. 1999, 118, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Pu, W.T. Cardiomyocyte Maturation. Circ. Res. 2020, 126, 1086–1106. [Google Scholar] [CrossRef]
- Rapila, R.; Korhonen, T.; Tavi, P. Excitation-contraction coupling of the mouse embryonic cardiomyocyte. J. Gen. Physiol. 2008, 132, 397–405. [Google Scholar] [CrossRef]
- Santulli, G.; Nakashima, R.; Yuan, Q.; Marks, A.R. Intracellular calcium release channels: An update. J. Physiol. 2017, 595, 3041–3051. [Google Scholar] [CrossRef] [PubMed]
- Louch, W.E.; Koivumäki, J.T.; Tavi, P. Calcium signalling in developing cardiomyocytes: Implications for model systems and disease. J. Physiol. 2015, 593, 1047–1063. [Google Scholar] [CrossRef] [PubMed]
- Sasse, P.; Zhang, J.; Cleemann, L.; Morad, M.; Hescheler, J.; Fleischmann, B.K. Intracellular Ca2+ oscillations, a potential pacemaking mechanism in early embryonic heart cells. J. Gen. Physiol. 2007, 130, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, T.; Rapila, R.; Tavi, P. Mathematical model of mouse embryonic cardiomyocyte excitation-contraction coupling. J. Gen. Physiol. 2008, 132, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Eisner, D.A.; Caldwell, J.L.; Kistamás, K.; Trafford, A.W. Calcium and Excitation-Contraction Coupling in the Heart. Circ. Res. 2017, 121, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Fearnley, C.J.; Roderick, H.L.; Bootman, M.D. Calcium signaling in cardiac myocytes. Cold Spring Harb. Perspect. Biol. 2011, 3, a004242. [Google Scholar] [CrossRef] [PubMed]
- Werner, J.C.; Sicard, R.E.; Schuler, H.G. Palmitate oxidation by isolated working fetal and newborn pig hearts. Am. J. Physiol. 1989, 256, E315–E321. [Google Scholar] [CrossRef] [PubMed]
- Persad, K.L.; Lopaschuk, G.D. Energy Metabolism on Mitochondrial Maturation and Its Effects on Cardiomyocyte Cell Fate. Front. Cell Dev. Biol. 2022, 10, 886393. [Google Scholar] [CrossRef] [PubMed]
- Neary, M.T.; Ng, K.E.; Ludtmann, M.H.; Hall, A.R.; Piotrowska, I.; Ong, S.B.; Hausenloy, D.J.; Mohun, T.J.; Abramov, A.Y.; Breckenridge, R.A. Hypoxia signaling controls postnatal changes in cardiac mitochondrial morphology and function. J. Mol. Cell. Cardiol. 2014, 74, 340–352. [Google Scholar] [CrossRef]
- Lopaschuk, G.D.; Spafford, M.A.; Marsh, D.R. Glycolysis is predominant source of myocardial ATP production immediately after birth. Am. J. Physiol. 1991, 261, H1698–H1705. [Google Scholar] [CrossRef]
- Piquereau, J.; Ventura-Clapier, R. Maturation of Cardiac Energy Metabolism During Perinatal Development. Front. Physiol. 2018, 9, 959. [Google Scholar] [CrossRef] [PubMed]
- Crocini, C.; Gotthardt, M. Cardiac sarcomere mechanics in health and disease. Biophys. Rev. 2021, 13, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chahal, D.; Marquez, V.; Lee, S.S. Chapter 15—Cirrhotic cardiomyopathy. In Cardio-Hepatology; Taniguchi, T., Lee, S.S., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 217–246. [Google Scholar] [CrossRef]
- Lompre, A.M.; Schwartz, K.; d’Albis, A.; Lacombe, G.; Van Thiem, N.; Swynghedauw, B. Myosin isoenzyme redistribution in chronic heart overload. Nature 1979, 282, 105–107. [Google Scholar] [CrossRef]
- Anzai, T.; Yamagata, T.; Uosaki, H. Comparative Transcriptome Landscape of Mouse and Human Hearts. Front. Cell Dev. Biol. 2020, 8, 268. [Google Scholar] [CrossRef] [PubMed]
- Taegtmeyer, H.; Sen, S.; Vela, D. Return to the fetal gene program: A suggested metabolic link to gene expression in the heart. Ann. N. Y. Acad. Sci. 2010, 1188, 191–198. [Google Scholar] [CrossRef]
- Greaser, M.L.; Krzesinski, P.R.; Warren, C.M.; Kirkpatrick, B.; Campbell, K.S.; Moss, R.L. Developmental changes in rat cardiac titin/connectin: Transitions in normal animals and in mutants with a delayed pattern of isoform transition. J. Muscle Res. Cell Motil. 2005, 26, 325–332. [Google Scholar] [CrossRef]
- Sheng, J.J.; Jin, J.P. TNNI1, TNNI2 and TNNI3: Evolution, regulation, and protein structure-function relationships. Gene 2016, 576, 385–394. [Google Scholar] [CrossRef]
- Herrmann, B.G.; Kispert, A. The T genes in embryogenesis. Trends Genet. 1994, 10, 280–286. [Google Scholar] [CrossRef]
- Herrmann, B.G.; Labeit, S.; Poustka, A.; King, T.R.; Lehrach, H. Cloning of the T gene required in mesoderm formation in the mouse. Nature 1990, 343, 617–622. [Google Scholar] [CrossRef]
- Schulte-Merker, S.; Eeden, F.V.; Halpern, M.E.; Kimmel, C.B.; Nüsslein-Volhard, C. no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development 1994, 120, 1009–1015. [Google Scholar] [CrossRef]
- Schulte-Merker, S.; Ho, R.K.; Herrmann, B.G.; Nüsslein-Volhard, C. The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development 1992, 116, 1021–1032. [Google Scholar] [CrossRef]
- Hotta, K.; Takahashi, H.; Asakura, T.; Saitoh, B.; Takatori, N.; Satou, Y.; Satoh, N. Characterization of Brachyury-downstream notochord genes in the Ciona intestinalis embryo. Dev. Biol. 2000, 224, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Chesley, P. Development of the short-tailed mutant in the house mouse. J. Exp. Zool. 1935, 70, 429–459. [Google Scholar] [CrossRef]
- Showell, C.; Binder, O.; Conlon, F.L. T-box genes in early embryogenesis. Dev. Dyn. 2004, 229, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Schulte-Merker, S.; Smith, J.C. Mesoderm formation in response to Brachyury requires FGF signalling. Curr. Biol. 1995, 5, 62–67. [Google Scholar] [CrossRef]
- Isaacs, H.V.; Pownall, M.E.; Slack, J.M. eFGF regulates Xbra expression during Xenopus gastrulation. Embo J. 1994, 13, 4469–4481. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, Y.; Yasuo, H.; Satoh, N.; Nishida, H. Basic fibroblast growth factor induces notochord formation and the expression of As-T, a Brachyury homolog, during ascidian embryogenesis. Development 1996, 122, 2023–2031. [Google Scholar] [CrossRef]
- van den Ameele, J.; Tiberi, L.; Bondue, A.; Paulissen, C.; Herpoel, A.; Iacovino, M.; Kyba, M.; Blanpain, C.; Vanderhaeghen, P. Eomesodermin induces Mesp1 expression and cardiac differentiation from embryonic stem cells in the absence of Activin. EMBO Rep. 2012, 13, 355–362. [Google Scholar] [CrossRef]
- Costello, I.; Pimeisl, I.M.; Dräger, S.; Bikoff, E.K.; Robertson, E.J.; Arnold, S.J. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat. Cell Biol. 2011, 13, 1084–1091. [Google Scholar] [CrossRef]
- David, R.; Jarsch, V.B.; Schwarz, F.; Nathan, P.; Gegg, M.; Lickert, H.; Franz, W.-M. Induction of MesP1 by Brachyury(T) generates the common multipotent cardiovascular stem cell. Cardiovasc. Res. 2011, 92, 115–122. [Google Scholar] [CrossRef]
- Saga, Y.; Kitajima, S.; Miyagawa-Tomita, S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc. Med. 2000, 10, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Bondue, A.; Lapouge, G.; Paulissen, C.; Semeraro, C.; Iacovino, M.; Kyba, M.; Blanpain, C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell 2008, 3, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Bondue, A.; Blanpain, C. Mesp1. Circ. Res. 2010, 107, 1414–1427. [Google Scholar] [CrossRef] [PubMed]
- Lindsley, R.C.; Gill, J.G.; Murphy, T.L.; Langer, E.M.; Cai, M.; Mashayekhi, M.; Wang, W.; Niwa, N.; Nerbonne, J.M.; Kyba, M. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell 2008, 3, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, R.J.; Olson, E.N. Building the heart piece by piece: Modularity of cis-elements regulating Nkx2-5 transcription. Development 1999, 126, 4187–4192. [Google Scholar] [CrossRef]
- McFadden, D.G.; Charité, J.; Richardson, J.A.; Srivastava, D.; Firulli, A.B.; Olson, E.N. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development 2000, 127, 5331–5341. [Google Scholar] [CrossRef] [PubMed]
- Durocher, D.; Charron, F.; Warren, R.; Schwartz, R.J.; Nemer, M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997, 16, 5687–5696. [Google Scholar] [CrossRef]
- Lee, Y.; Shioi, T.; Kasahara, H.; Jobe, S.M.; Wiese, R.J.; Markham, B.E.; Izumo, S. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol. Cell. Biol. 1998, 18, 3120–3129. [Google Scholar] [CrossRef]
- Molkentin, J.D.; Lin, Q.; Duncan, S.A.; Olson, E.N. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997, 11, 1061–1072. [Google Scholar] [CrossRef]
- Kuo, C.T.; Morrisey, E.E.; Anandappa, R.; Sigrist, K.; Lu, M.M.; Parmacek, M.S.; Soudais, C.; Leiden, J.M. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997, 11, 1048–1060. [Google Scholar] [CrossRef]
- Lyons, I.; Parsons, L.M.; Hartley, L.; Li, R.; Andrews, J.E.; Robb, L.; Harvey, R.P. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995, 9, 1654–1666. [Google Scholar] [CrossRef]
- Snyder, M.; Huang, X.Y.; Zhang, J.J. Stat3 directly controls the expression of Tbx5, Nkx2.5, and GATA4 and is essential for cardiomyocyte differentiation of P19CL6 cells. J. Biol. Chem. 2010, 285, 23639–23646. [Google Scholar] [CrossRef] [PubMed]
- Searcy, R.D.; Vincent, E.B.; Liberatore, C.M.; Yutzey, K.E. A GATA-dependent nkx-2. 5 regulatory element activates early cardiac gene expression in transgenic mice. Development 1998, 125, 4461–4470. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Dong, A.; Karasaki, K.; Sogabe, S.; Okamoto, D.; Saigo, M.; Ishida, M.; Yoshizumi, M.; Kokubo, H. Smad4 regulates the nuclear translocation of Nkx2-5 in cardiac differentiation. Sci. Rep. 2021, 11, 3588. [Google Scholar] [CrossRef] [PubMed]
- Malek Mohammadi, M.; Kattih, B.; Grund, A.; Froese, N.; Korf-Klingebiel, M.; Gigina, A.; Schrameck, U.; Rudat, C.; Liang, Q.; Kispert, A.; et al. The transcription factor GATA4 promotes myocardial regeneration in neonatal mice. EMBO Mol. Med. 2017, 9, 265–279. [Google Scholar] [CrossRef] [PubMed]
- von Both, I.; Silvestri, C.; Erdemir, T.; Lickert, H.; Walls, J.R.; Henkelman, R.M.; Rossant, J.; Harvey, R.P.; Attisano, L.; Wrana, J.L. Foxh1 Is Essential for Development of the Anterior Heart Field. Dev. Cell 2004, 7, 331–345. [Google Scholar] [CrossRef]
- Lin, L.; Cui, L.; Zhou, W.; Dufort, D.; Zhang, X.; Cai, C.-L.; Bu, L.; Yang, L.; Martin, J.; Kemler, R. β-Catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 9313–9318. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Li, Y.; Wang, Y.; Liu, Y.; Wang, W.; Jia, Z.; Chen, P.; Ma, K.; Zhou, C. Wnt-promoted Isl1 expression through a novel TCF/LEF1 binding site and H3K9 acetylation in early stages of cardiomyocyte differentiation of P19CL6 cells. Mol. Cell. Biochem. 2014, 391, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.-L.; Liang, X.; Shi, Y.; Chu, P.-H.; Pfaff, S.L.; Chen, J.; Evans, S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 2003, 5, 877–889. [Google Scholar] [CrossRef]
- Moretti, A.; Caron, L.; Nakano, A.; Lam, J.T.; Bernshausen, A.; Chen, Y.; Qyang, Y.; Bu, L.; Sasaki, M.; Martin-Puig, S. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 2006, 127, 1151–1165. [Google Scholar] [CrossRef]
- Bu, L.; Jiang, X.; Martin-Puig, S.; Caron, L.; Zhu, S.; Shao, Y.; Roberts, D.J.; Huang, P.L.; Domian, I.J.; Chien, K.R. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature 2009, 460, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Dodou, E.; Verzi, M.P.; Anderson, J.P.; Xu, S.-M.; Black, B.L. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development 2004, 131, 3931–3942. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.; Qian, L.; Cheng, P.; Nigam, V.; Arnold, J.; Srivastava, D. A regulatory pathway involving Notch1/β-catenin/Isl1 determines cardiac progenitor cell fate. Nat. Cell Biol. 2009, 11, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Miao, D.; Li, Y.; Gao, R. Spotlight on Isl1: A Key Player in Cardiovascular Development and Diseases. Front. Cell Dev. Biol. 2021, 9, 793605. [Google Scholar] [CrossRef]
- Lin, Q.; Schwarz, J.; Bucana, C.; Olson, E.N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 1997, 276, 1404–1407. [Google Scholar] [CrossRef]
- Materna, S.C.; Sinha, T.; Barnes, R.M.; Lammerts van Bueren, K.; Black, B.L. Cardiovascular development and survival require Mef2c function in the myocardial but not the endothelial lineage. Dev. Biol. 2019, 445, 170–177. [Google Scholar] [CrossRef]
- Edmondson, D.G.; Lyons, G.E.; Martin, J.F.; Olson, E.N. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development 1994, 120, 1251–1263. [Google Scholar] [CrossRef] [PubMed]
- Black, B.L.; Olson, E.N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 1998, 14, 167–196. [Google Scholar] [CrossRef]
- Bhavsar, P.K.; Dellow, K.A.; Yacoub, M.H.; Brand, N.J.; Barton, P.J.R. Identification of cis-acting DNA elements required for expression of the human cardiac troponin I gene promoter. J. Mol. Cell. Cardiol. 2000, 32, 95–108. [Google Scholar] [CrossRef]
- Morin, S.; Charron, F.; Robitaille, L.; Nemer, M. GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J. 2000, 19, 2046–2055. [Google Scholar] [CrossRef]
- Vanpoucke, G.; Goossens, S.; De Craene, B.; Gilbert, B.; van Roy, F.; Berx, G. GATA-4 and MEF2C transcription factors control the tissue-specific expression of the alphaT-catenin gene CTNNA3. Nucleic Acids Res. 2004, 32, 4155–4165. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Paige, S.L.; Plonowska, K.; Xu, A.; Wu, S.M. Molecular regulation of cardiomyocyte differentiation. Circ. Res. 2015, 116, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Burridge, P.W.; Sharma, A.; Wu, J.C. Genetic and Epigenetic Regulation of Human Cardiac Reprogramming and Differentiation in Regenerative Medicine. Annu. Rev. Genet. 2015, 49, 461–484. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.C.; West, A.G.; Felsenfeld, G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 1999, 98, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, I.; Gambetta, M.C. The Role of Insulation in Patterning Gene Expression. Genes 2019, 10, 767. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Corces, V.G. Chromatin insulators: A role in nuclear organization and gene expression. Adv. Cancer Res. 2011, 110, 43–76. [Google Scholar] [CrossRef]
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.T.; Corces, V.G. CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014, 15, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.B.; Pan, H.; Hannenhalli, S.; Cheng, Y.; Ma, J.; Fedoriw, A.; Lobanenkov, V.; Latham, K.E.; Schultz, R.M.; Bartolomei, M.S. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development 2008, 135, 2729–2738. [Google Scholar] [CrossRef]
- Moore, J.M.; Rabaia, N.A.; Smith, L.E.; Fagerlie, S.; Gurley, K.; Loukinov, D.; Disteche, C.M.; Collins, S.J.; Kemp, C.J.; Lobanenkov, V.V.; et al. Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PLoS ONE 2012, 7, e34915. [Google Scholar] [CrossRef]
- Gomez-Velazquez, M.; Badia-Careaga, C.; Lechuga-Vieco, A.V.; Nieto-Arellano, R.; Tena, J.J.; Rollan, I.; Alvarez, A.; Torroja, C.; Caceres, E.F.; Roy, A.R.; et al. CTCF counter-regulates cardiomyocyte development and maturation programs in the embryonic heart. PLoS Genet. 2017, 13, e1006985. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.P.; Tan, W.L.W.; Anene-Nzelu, C.G.; Lee, C.J.M.; Li, P.Y.; Luu, T.D.A.; Chan, C.X.; Tiang, Z.; Ng, S.L.; Huang, X.; et al. Robust CTCF-Based Chromatin Architecture Underpins Epigenetic Changes in the Heart Failure Stress–Gene Response. Circulation 2019, 139, 1937–1956. [Google Scholar] [CrossRef] [PubMed]
- Schwalie, P.C.; Ward, M.C.; Cain, C.E.; Faure, A.J.; Gilad, Y.; Odom, D.T.; Flicek, P. Co-binding by YY1 identifies the transcriptionally active, highly conserved set of CTCF-bound regions in primate genomes. Genome Biol. 2013, 14, R148. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, A.S.; Li, C.H.; Zamudio, A.V.; Sigova, A.A.; Hannett, N.M.; Day, D.S.; Abraham, B.J.; Cohen, M.A.; Nabet, B.; Buckley, D.L.; et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 2017, 171, 1573–1588.e28. [Google Scholar] [CrossRef] [PubMed]
- Beketaev, I.; Zhang, Y.; Kim, E.Y.; Yu, W.; Qian, L.; Wang, J. Critical role of YY1 in cardiac morphogenesis. Dev. Dyn. 2015, 244, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Gregoire, S.; Karra, R.; Passer, D.; Deutsch, M.A.; Krane, M.; Feistritzer, R.; Sturzu, A.; Domian, I.; Saga, Y.; Wu, S.M. Essential and unexpected role of Yin Yang 1 to promote mesodermal cardiac differentiation. Circ. Res. 2013, 112, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Gregoire, S.; Li, G.; Sturzu, A.C.; Schwartz, R.J.; Wu, S.M. YY1 Expression Is Sufficient for the Maintenance of Cardiac Progenitor Cell State. Stem Cells 2017, 35, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Koyano-Nakagawa, N.; Nishida, J.; Baldwin, D.; Arai, K.; Yokota, T. Molecular cloning of a novel human cDNA encoding a zinc finger protein that binds to the interleukin-3 promoter. Mol. Cell. Biol. 1994, 14, 5099–5107. [Google Scholar] [CrossRef]
- Aitsebaomo, J.; Kingsley-Kallesen, M.L.; Wu, Y.; Quertermous, T.; Patterson, C. Vezf1/DB1 Is an Endothelial Cell-specific Transcription Factor That Regulates Expression of the Endothelin-1 Promoter. J. Biol. Chem. 2001, 276, 39197–39205. [Google Scholar] [CrossRef]
- Gowher, H.; Brick, K.; Camerini-Otero, R.D.; Felsenfeld, G. Vezf1 protein binding sites genome-wide are associated with pausing of elongating RNA polymerase II. Proc. Natl. Acad. Sci. USA 2012, 109, 2370–2375. [Google Scholar] [CrossRef]
- Dickson, J.; Gowher, H.; Strogantsev, R.; Gaszner, M.; Hair, A.; Felsenfeld, G.; West, A.G. VEZF1 elements mediate protection from DNA methylation. PLoS Genet. 2010, 6, e1000804. [Google Scholar] [CrossRef] [PubMed]
- Gowher, H.; Stuhlmann, H.; Felsenfeld, G. Vezf1 regulates genomic DNA methylation through its effects on expression of DNA methyltransferase Dnmt3b. Genes Dev. 2008, 22, 2075–2084. [Google Scholar] [CrossRef] [PubMed]
- Ghirlando, R.; Giles, K.; Gowher, H.; Xiao, T.; Xu, Z.; Yao, H.; Felsenfeld, G. Chromatin domains, insulators, and the regulation of gene expression. Biochim. Biophys. Acta 2012, 1819, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Recillas-Targa, F.; Pikaart, M.J.; Burgess-Beusse, B.; Bell, A.C.; Litt, M.D.; West, A.G.; Gaszner, M.; Felsenfeld, G. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl. Acad. Sci. USA 2002, 99, 6883–6888. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.W.; Leahy, A.; Lee, H.H.; Stuhlmann, H. Vezf1: A Zn finger transcription factor restricted to endothelial cells and their precursors. Dev. Biol. 1999, 206, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Kuhnert, F.; Campagnolo, L.; Xiong, J.W.; Lemons, D.; Fitch, M.J.; Zou, Z.; Kiosses, W.B.; Gardner, H.; Stuhlmann, H. Dosage-dependent requirement for mouse Vezf1 in vascular system development. Dev. Biol. 2005, 283, 140–156. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Gupta, V.; Bjorge, J.; Shi, X.; Gong, W.; Garry, M.G.; Garry, D.J. ETV2 and VEZF1 interaction and regulation of the hematoendothelial lineage during embryogenesis. Front. Cell Dev. Biol. 2023, 11, 1109648. [Google Scholar] [CrossRef] [PubMed]
- AlAbdi, L.; He, M.; Yang, Q.; Norvil, A.B.; Gowher, H. The transcription factor Vezf1 represses the expression of the antiangiogenic factor Cited2 in endothelial cells. J. Biol. Chem. 2018, 293, 11109–11118. [Google Scholar] [CrossRef]
- Gerald, D.; Adini, I.; Shechter, S.; Perruzzi, C.; Varnau, J.; Hopkins, B.; Kazerounian, S.; Kurschat, P.; Blachon, S.; Khedkar, S.; et al. RhoB controls coordination of adult angiogenesis and lymphangiogenesis following injury by regulating VEZF1-mediated transcription. Nat. Commun. 2013, 4, 2824. [Google Scholar] [CrossRef]
- Bruderer, M.; Alini, M.; Stoddart, M.J. Role of HOXA9 and VEZF1 in endothelial biology. J. Vasc. Res. 2013, 50, 265–278. [Google Scholar] [CrossRef]
- Paavola, J.; Alakoski, T.; Ulvila, J.; Kilpiö, T.; Sirén, J.; Perttunen, S.; Narumanchi, S.; Wang, H.; Lin, R.; Porvari, K.; et al. Vezf1 regulates cardiac structure and contractile function. EBioMedicine 2020, 51, 102608. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.-Y.; Xie, M.-S.; Guo, Y.-H.; Yang, C.-X.; Gu, J.-N.; Qiao, Q.; Di, R.-M.; Qiu, X.-B.; Xu, Y.-J.; Yang, Y.-Q. VEZF1 loss-of-function mutation underlying familial dilated cardiomyopathy. Eur. J. Med. Genet. 2023, 66, 104705. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K. Role of epigenetics in developmental biology and transgenerational inheritance. Birth Defects Res. C Embryo Today 2011, 93, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Andersson, R.; Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020, 21, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Mensah, I.K.; Norvil, A.B.; AlAbdi, L.; McGovern, S.; Petell, C.J.; He, M.; Gowher, H. Misregulation of the expression and activity of DNA methyltransferases in cancer. NAR Cancer 2021, 3, zcab045. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.; Hendrich, B.; Reik, W.; Dean, W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002, 241, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Haas, J.; Frese, K.S.; Park, Y.J.; Keller, A.; Vogel, B.; Lindroth, A.M.; Weichenhan, D.; Franke, J.; Fischer, S.; Bauer, A.; et al. Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol. Med. 2013, 5, 413–429. [Google Scholar] [CrossRef] [PubMed]
- Movassagh, M.; Choy, M.K.; Knowles, D.A.; Cordeddu, L.; Haider, S.; Down, T.; Siggens, L.; Vujic, A.; Simeoni, I.; Penkett, C.; et al. Distinct epigenomic features in end-stage failing human hearts. Circulation 2011, 124, 2411–2422. [Google Scholar] [CrossRef]
- Jin, B.; Li, Y.; Robertson, K.D. DNA methylation: Superior or subordinate in the epigenetic hierarchy? Genes Cancer 2011, 2, 607–617. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Gowher, H.; Jeltsch, A. Mammalian DNA methyltransferases: New discoveries and open questions. Biochem. Soc. Trans. 2018, 46, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, A.A.; Lin, M.; Lister, R.L.; Maslov, A.A.; Wang, Y.; Suzuki, M.; Wu, B.; Greally, J.M.; Zheng, D.; Zhou, B. DNA methylation is developmentally regulated for genes essential for cardiogenesis. J. Am. Heart Assoc. 2014, 3, e000976. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Liu, G.-H.; Plongthongkum, N.; Benner, C.; Yi, F.; Qu, J.; Suzuki, K.; Yang, J.; Zhang, W.; Li, M.; et al. Global DNA methylation and transcriptional analyses of human ESC-derived cardiomyocytes. Protein Cell 2014, 5, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Poulsen, R.; Zhao, L.; Wang, J.; Rivkees, S.A.; Wendler, C.C. Knockdown of DNA methyltransferase 1 reduces DNA methylation and alters expression patterns of cardiac genes in embryonic cardiomyocytes. FEBS Open Bio 2021, 11, 2364–2382. [Google Scholar] [CrossRef] [PubMed]
- Madsen, A.; Höppner, G.; Krause, J.; Hirt, M.N.; Laufer, S.D.; Schweizer, M.; Tan, W.L.W.; Mosqueira, D.; Anene-Nzelu, C.G.; Lim, I.; et al. An Important Role for DNMT3A-Mediated DNA Methylation in Cardiomyocyte Metabolism and Contractility. Circulation 2020, 142, 1562–1578. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Huang, B.O.; Xu, Y.M.; Li, J.; Huang, L.F.; Lin, J.; Zhang, J.; Min, Q.H.; Yang, W.M.; et al. Mechanism of alternative splicing and its regulation. Biomed. Rep. 2015, 3, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Shayevitch, R.; Askayo, D.; Keydar, I.; Ast, G. The importance of DNA methylation of exons on alternative splicing. RNA 2018, 24, 1351–1362. [Google Scholar] [CrossRef]
- Flores, K.; Wolschin, F.; Corneveaux, J.J.; Allen, A.N.; Huentelman, M.J.; Amdam, G.V. Genome-wide association between DNA methylation and alternative splicing in an invertebrate. BMC Genom. 2012, 13, 480. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, J.D.; Jung, M.; Chen, C.-y.; Lin, Z.; Ye, J.; Godatha, S.; Lizhar, E.; Wu, X.; Hsu, D.; Couture, L.A.; et al. Mapping Human Pluripotent-to-Cardiomyocyte Differentiation: Methylomes, Transcriptomes, and Exon DNA Methylation “Memories”. EBioMedicine 2016, 4, 74–85. [Google Scholar] [CrossRef]
- Gilsbach, R.; Preissl, S.; Grüning, B.A.; Schnick, T.; Burger, L.; Benes, V.; Würch, A.; Bönisch, U.; Günther, S.; Backofen, R.; et al. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat. Commun. 2014, 5, 5288. [Google Scholar] [CrossRef]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Henikoff, S. Histone variants on the move: Substrates for chromatin dynamics. Nat. Rev. Mol. Cell. Biol. 2017, 18, 115–126. [Google Scholar] [CrossRef]
- Tan, M.; Luo, H.; Lee, S.; Jin, F.; Yang, J.S.; Montellier, E.; Buchou, T.; Cheng, Z.; Rousseaux, S.; Rajagopal, N. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011, 146, 1016–1028. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Henikoff, S. The Yin and Yang of Histone Marks in Transcription. Annu. Rev. Genom. Hum. Genet. 2021, 22, 147–170. [Google Scholar] [CrossRef]
- Kuo, M.H.; Allis, C.D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 1998, 20, 615–626. [Google Scholar] [CrossRef]
- Peters, A.H.; Kubicek, S.; Mechtler, K.; O’Sullivan, R.J.; Derijck, A.A.; Perez-Burgos, L.; Kohlmaier, A.; Opravil, S.; Tachibana, M.; Shinkai, Y.; et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 2003, 12, 1577–1589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Reinberg, D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes Dev. 2001, 15, 2343–2360. [Google Scholar] [CrossRef]
- Zhang, Q.J.; Liu, Z.P. Histone methylations in heart development, congenital and adult heart diseases. Epigenomics 2015, 7, 321–330. [Google Scholar] [CrossRef]
- Chen, T.; Dent, S.Y. Chromatin modifiers and remodellers: Regulators of cellular differentiation. Nat. Rev. Genet. 2014, 15, 93–106. [Google Scholar] [CrossRef]
- Schueler, M.; Zhang, Q.; Schlesinger, J.; Tönjes, M.; Sperling, S.R. Dynamics of Srf, p300 and histone modifications during cardiac maturation in mouse. Mol. Biosyst. 2012, 8, 495–503. [Google Scholar] [CrossRef]
- Yao, T.-P.; Oh, S.P.; Fuchs, M.; Zhou, N.-D.; Ch’ng, L.-E.; Newsome, D.; Bronson, R.T.; Li, E.; Livingston, D.M.; Eckner, R. Gene dosage–dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 1998, 93, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Partanen, A.; Motoyama, J.; Hui, C.C. Developmentally regulated expression of the transcriptional cofactors/histone acetyltransferases CBP and p300 during mouse embryogenesis. Int. J. Dev. Biol. 2004, 43, 487–494. [Google Scholar]
- Sun, H.; Yang, X.; Zhu, J.; Lv, T.; Chen, Y.; Chen, G.; Zhong, L.; Li, Y.; Huang, X.; Huang, G.; et al. Inhibition of p300-HAT results in a reduced histone acetylation and down-regulation of gene expression in cardiac myocytes. Life Sci. 2010, 87, 707–714. [Google Scholar] [CrossRef]
- Li, T.; Li, Y.-M.; Jia, Z.-Q.; Chen, P.; Ma, K.-T.; Zhou, C.-Y. Carboxyl terminus of Nkx2.5 impairs its interaction with p300. J. Mol. Biol. 2007, 370, 976–992. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.S.; Markham, B.E. p300 Functions as a coactivator of transcription factor GATA-4. J. Biol. Chem. 2001, 276, 37178–37185. [Google Scholar] [CrossRef]
- Montgomery, R.L.; Davis, C.A.; Potthoff, M.J.; Haberland, M.; Fielitz, J.; Qi, X.; Hill, J.A.; Richardson, J.A.; Olson, E.N. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007, 21, 1790–1802. [Google Scholar] [CrossRef]
- Karamboulas, C.; Swedani, A.; Ward, C.; Al-Madhoun, A.S.; Wilton, S.; Boisvenue, S.; Ridgeway, A.G.; Skerjanc, I.S. HDAC activity regulates entry of mesoderm cells into the cardiac muscle lineage. J. Cell Sci. 2006, 119, 4305–4314. [Google Scholar] [CrossRef]
- Backs, J.; Worst, B.C.; Lehmann, L.H.; Patrick, D.M.; Jebessa, Z.; Kreusser, M.M.; Sun, Q.; Chen, L.; Heft, C.; Katus, H.A.; et al. Selective repression of MEF2 activity by PKA-dependent proteolysis of HDAC4. J. Cell Biol. 2011, 195, 403–415. [Google Scholar] [CrossRef]
- Hohl, M.; Wagner, M.; Reil, J.-C.; Müller, S.-A.; Tauchnitz, M.; Zimmer, A.M.; Lehmann, L.H.; Thiel, G.; Böhm, M.; Backs, J. HDAC4 controls histone methylation in response to elevated cardiac load. J. Clin. Investig. 2013, 123, 1359–1370. [Google Scholar] [CrossRef]
- Trivedi, C.M.; Zhu, W.; Wang, Q.; Jia, C.; Kee, H.J.; Li, L.; Hannenhalli, S.; Epstein, J.A. Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev. Cell 2010, 19, 450–459. [Google Scholar] [CrossRef]
- Zaidi, S.; Choi, M.; Wakimoto, H.; Ma, L.; Jiang, J.; Overton, J.D.; Romano-Adesman, A.; Bjornson, R.D.; Breitbart, R.E.; Brown, K.K. De novo mutations in histone-modifying genes in congenital heart disease. Nature 2013, 498, 220–223. [Google Scholar] [CrossRef]
- Lee, S.; Lee, J.W.; Lee, S.-K. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev. Cell 2012, 22, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Nimura, K.; Ura, K.; Shiratori, H.; Ikawa, M.; Okabe, M.; Schwartz, R.J.; Kaneda, Y. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf–Hirschhorn syndrome. Nature 2009, 460, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Olguín, P.; Huang, Y.; Li, X.; Christodoulou, D.; Seidman, C.E.; Seidman, J.G.; Tarakhovsky, A.; Bruneau, B.G. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat. Genet. 2012, 44, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Mattiroli, F.; Penengo, L. Histone Ubiquitination: An Integrative Signaling Platform in Genome Stability. Trends Genet. 2021, 37, 566–581. [Google Scholar] [CrossRef]
- Oss-Ronen, L.; Sarusi, T.; Cohen, I. Histone Mono-Ubiquitination in Transcriptional Regulation and Its Mark on Life: Emerging Roles in Tissue Development and Disease. Cells 2022, 11, 2404. [Google Scholar] [CrossRef]
- Peng, X.; Feng, G.; Zhang, Y.; Sun, Y. PRC1 Stabilizes Cardiac Contraction by Regulating Cardiac Sarcomere Assembly and Cardiac Conduction System Construction. Int. J. Mol. Sci. 2021, 22, 11368. [Google Scholar] [CrossRef]
- Xie, W.; Nagarajan, S.; Baumgart, S.J.; Kosinsky, R.L.; Najafova, Z.; Kari, V.; Hennion, M.; Indenbirken, D.; Bonn, S.; Grundhoff, A.; et al. RNF40 regulates gene expression in an epigenetic context-dependent manner. Genome Biol. 2017, 18, 32. [Google Scholar] [CrossRef]
- Robson, A.; Makova, S.Z.; Barish, S.; Zaidi, S.; Mehta, S.; Drozd, J.; Jin, S.C.; Gelb, B.D.; Seidman, C.E.; Chung, W.K.; et al. Histone H2B monoubiquitination regulates heart development via epigenetic control of cilia motility. Proc. Natl. Acad. Sci. USA 2019, 116, 14049–14054. [Google Scholar] [CrossRef]
- VanDusen, N.J.; Lee, J.Y.; Gu, W.; Butler, C.E.; Sethi, I.; Zheng, Y.; King, J.S.; Zhou, P.; Suo, S.; Guo, Y.; et al. Massively parallel in vivo CRISPR screening identifies RNF20/40 as epigenetic regulators of cardiomyocyte maturation. Nat. Commun. 2021, 12, 4442. [Google Scholar] [CrossRef]
- Ryu, H.Y.; Hochstrasser, M. Histone sumoylation and chromatin dynamics. Nucleic Acids Res. 2021, 49, 6043–6052. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Chen, X.; Su, Q.; Lu, W.; Wang, Q.; Yuan, H.; Zhang, Z.; Wang, X.; Wu, H.; Qi, Y. The Function of SUMOylation and Its Critical Roles in Cardiovascular Diseases and Potential Clinical Implications. Int. J. Mol. Sci. 2021, 22, 10618. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.W.; Lee, S.; Furtado, M.B.; Xin, L.; Sparrow, D.B.; Martinez, C.G.; Dunwoodie, S.L.; Kurtenbach, E.; Mohun, T.; Rosenthal, N.; et al. Complex SUMO-1 regulation of cardiac transcription factor Nkx2-5. PLoS ONE 2011, 6, e24812. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.K.; McLendon, P.M.; Gulick, J.; James, J.; Khalili, K.; Robbins, J. UBC9-Mediated Sumoylation Favorably Impacts Cardiac Function in Compromised Hearts. Circ. Res. 2016, 118, 1894–1905. [Google Scholar] [CrossRef] [PubMed]
- Kaikkonen, M.U.; Lam, M.T.; Glass, C.K. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 2011, 90, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, S.; Moukette, B.; Hayasaka, T.; Haskell, A.K.; Mah, J.; Sepúlveda, M.N.; Tang, Y.; Kim, I.M. Noncoding RNAs as Key Regulators for Cardiac Development and Cardiovascular Diseases. J. Cardiovasc. Dev. Dis. 2023, 10, 166. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Krishnan, J. Non-coding RNAs in Cardiac Regeneration. Front. Physiol. 2021, 12, 650566. [Google Scholar] [CrossRef]
- Devaux, Y.; Zangrando, J.; Schroen, B.; Creemers, E.E.; Pedrazzini, T.; Chang, C.-P.; Dorn, G.W.; Thum, T.; Heymans, S.; The Cardiolinc, n. Long noncoding RNAs in cardiac development and ageing. Nat. Rev. Cardiol. 2015, 12, 415–425. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Zhao, Y.; Samal, E.; Srivastava, D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005, 436, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Cianflone, E.; Scalise, M.; Marino, F.; Salerno, L.; Salerno, N.; Urbanek, K.; Torella, D. The negative regulation of gene expression by microRNAs as key driver of inducers and repressors of cardiomyocyte differentiation. Clin. Sci. 2022, 136, 1179–1203. [Google Scholar] [CrossRef] [PubMed]
- Kay, M.; Soltani, B.M.; Aghdaei, F.H.; Ansari, H.; Baharvand, H. Hsa-miR-335 regulates cardiac mesoderm and progenitor cell differentiation. Stem Cell Res. Ther. 2019, 10, 191. [Google Scholar] [CrossRef] [PubMed]
- Poon, E.N.; Hao, B.; Guan, D.; Jun Li, M.; Lu, J.; Yang, Y.; Wu, B.; Wu, S.C.; Webb, S.E.; Liang, Y.; et al. Integrated transcriptomic and regulatory network analyses identify microRNA-200c as a novel repressor of human pluripotent stem cell-derived cardiomyocyte differentiation and maturation. Cardiovasc. Res. 2018, 114, 894–906. [Google Scholar] [CrossRef]
- Ouyang, Z.; Wei, K. miRNA in cardiac development and regeneration. Cell Regen. 2021, 10, 14. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell. Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Anderson, K.M.; Anderson, D.M.; McAnally, J.R.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 2016, 539, 433–436. [Google Scholar] [CrossRef]
- Han, X.; Zhang, J.; Liu, Y.; Fan, X.; Ai, S.; Luo, Y.; Li, X.; Jin, H.; Luo, S.; Zheng, H.; et al. The lncRNA Hand2os1/Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development 2019, 146, dev176198. [Google Scholar] [CrossRef] [PubMed]
- Ritter, N.; Ali, T.; Kopitchinski, N.; Schuster, P.; Beisaw, A.; Hendrix, D.A.; Schulz, M.H.; Müller-McNicoll, M.; Dimmeler, S.; Grote, P. The lncRNA Locus Handsdown Regulates Cardiac Gene Programs and Is Essential for Early Mouse Development. Dev. Cell 2019, 50, 644–657.e8. [Google Scholar] [CrossRef] [PubMed]
- Hazra, R.; Brine, L.; Garcia, L.; Benz, B.; Chirathivat, N.; Shen, M.M.; Wilkinson, J.E.; Lyons, S.K.; Spector, D.L. Platr4 is an early embryonic lncRNA that exerts its function downstream on cardiogenic mesodermal lineage commitment. Dev. Cell 2022, 57, 2450–2468.e7. [Google Scholar] [CrossRef]
- Kay, M.; Soltani, B.M.; Nemir, M.; Aghagolzadeh, P.; Pezzuto, I.; Chouvardas, P.; Ruberto, F.; Movahedi, F.; Ansari, H.; Baharvand, H.; et al. The conserved long non-coding RNA CARMA regulates cardiomyocyte differentiation. Cardiovasc. Res. 2022, 118, 2339–2353. [Google Scholar] [CrossRef] [PubMed]
- Taliani, V.; Buonaiuto, G.; Desideri, F.; Setti, A.; Santini, T.; Galfrè, S.; Schirone, L.; Mariani, D.; Frati, G.; Valenti, V.; et al. The long noncoding RNA Charme supervises cardiomyocyte maturation by controlling cell differentiation programs in the developing heart. eLife 2023, 12, e81360. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell. Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Chang, C.-P.; Bruneau, B.G. Epigenetics and cardiovascular development. Annu. Rev. Physiol. 2012, 74, 41–68. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.K.; Bruneau, B.G. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 2009, 459, 708–711. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Hota, S.K.; Zhou, Y.-Q.; Novak, S.; Miguel-Perez, D.; Christodoulou, D.; Seidman, C.E.; Seidman, J.G.; Gregorio, C.C.; Henkelman, R.M.; et al. Cardiac-enriched BAF chromatin-remodeling complex subunit Baf60c regulates gene expression programs essential for heart development and function. Biol. Open 2018, 7, bio029512. [Google Scholar] [CrossRef]
- Takeuchi, J.K.; Lou, X.; Alexander, J.M.; Sugizaki, H.; Delgado-Olguín, P.; Holloway, A.K.; Mori, A.D.; Wylie, J.N.; Munson, C.; Zhu, Y. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat. Commun. 2011, 2, 187. [Google Scholar] [CrossRef]
- Hang, C.T.; Yang, J.; Han, P.; Cheng, H.L.; Shang, C.; Ashley, E.; Zhou, B.; Chang, C.P. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 2010, 466, 62–67. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mensah, I.K.; Gowher, H. Epigenetic Regulation of Mammalian Cardiomyocyte Development. Epigenomes 2024, 8, 25. https://doi.org/10.3390/epigenomes8030025
Mensah IK, Gowher H. Epigenetic Regulation of Mammalian Cardiomyocyte Development. Epigenomes. 2024; 8(3):25. https://doi.org/10.3390/epigenomes8030025
Chicago/Turabian StyleMensah, Isaiah K., and Humaira Gowher. 2024. "Epigenetic Regulation of Mammalian Cardiomyocyte Development" Epigenomes 8, no. 3: 25. https://doi.org/10.3390/epigenomes8030025
APA StyleMensah, I. K., & Gowher, H. (2024). Epigenetic Regulation of Mammalian Cardiomyocyte Development. Epigenomes, 8(3), 25. https://doi.org/10.3390/epigenomes8030025






