The Genomic Shock Hypothesis: Genetic and Epigenetic Alterations of Transposable Elements after Interspecific Hybridization in Plants
Abstract
:1. Transposable Elements
2. Epigenetic Control of Transposable Elements
2.1. Epigenetic Silencing of TEs: Initiation
2.2. Epigenetic Silencing of TEs: Maintenance
2.3. Epigenetic Silencing of TEs: Loss
3. Changes in the Epigenetic Silencing of the TEs in Plant Interspecific Hybrids
3.1. Zea
3.2. Helianthus
3.3. Capsella
3.4. Aegilops
3.5. Arabidopsis
3.6. Arachis
3.7. Brassica
3.8. Camellia
3.9. Dactylorhiza
3.10. Gossypium
3.11. Lotus
3.12. Nicotiana
3.13. Oryza
3.14. Poa
3.15. Populus
3.16. Solanum
3.17. Triticum
3.18. Vitis
3.19. Yucca
3.20. Cajanus
3.21. Hieracium
3.22. Mimulus
3.23. Prunus
3.24. Spartina
3.25. Sorghum
| Species | Hybrid Type |
Genomic
Shock |
Transcription
Alterations |
DNA
Methylation 1 Alterations | Description | References |
|---|---|---|---|---|---|---|
| Capsella bursa-pastoris (C. grandiflora × C. orientalis) | Allotetraploid | Yes | - | - | Higher number of TEs only in gene-rich chromosome arms with no important global differences. | [65,66] |
| Helianthus spp. | Hybrid Natural & synthetic | Yes | Yes | - | Ancient hybrids have more DNA than parents due to the expansion of certain TE families that are transcriptionally active. Synthetic hybrids do not show increases in genome size. | [8,50,59,60,61,62,63,64] |
| Zea mays | Hybrid | Yes | - | - | Alterations in siRNAs and DNA methylation. | [56,57] |
| Hybrid | Yes | - | - | Differences in TE protein accumulation. | [55] | |
| Hybrid | Yes | Both | - | Most TE families do not show transcriptional differences in the hybrid, but some yes. | [54] | |
| Aegilops spp. | Hybrid | Limited | - | Higher | Increase in copy number of some families and increase in DNA methylation in the hybrid. | [67] |
| Aegilops geniculata × A. triuncialis | Hybrid | Limited | - | - | Activation of some retrotransposon families. | [68] |
| Aegilops markgrafii | Allotetraploid | Limited | - | - | Activation of some TE families. | [69] |
| Aegilops sharonensis × Triticum monococcum | Allohexaploid | Limited | Yes | - | Transcriptional activation of some retrotransposon families. | [114,115] |
| Aegilops speltoides | Hybrid | Limited | - | - | Activation of some TE families. | [70] |
| Arabidopsis suecica (A.thaliana × A. arenosa) | Allotetraploid Natural & synthetic | Limited | Yes | - | Limited higher transpositional activity of TEs in the hybrid. Changes in siRNA population. | [33,71,72,73] |
| Arachis duranensis × A. ipaensis | Allotetraploid | Limited | - | - | Mobilization of AhMITE1. | [82] |
| Brassica napus | Allotetraploid synthetic | Limited | Yes | Higher | Activation of some families is associated with changes in DNA methylation and siRNA contents in some cases and no activation in others. | [86,87,88,89,90,91,92,93,94] |
| Camellia azalea × C. amplexicaulis | Hybrid | Limited | Higher | - | Increase in TE transcription. | [95] |
| Dactylorhiza | Allotetraploids | Limited | - | - | Increase in genome size due to the activity of an MITE family. | [96] |
| Gossypium arboreum × G. raimondi | Hybrid | Undeter-mined | - | - | Many lncRNAs are activated in the hybrid corresponding to LINEs. | [97] |
| Lotus | RIL population | Limited | - | Lower | Mobilization of some retrotransposons associated with demethylation, but does not seem to affect all TE families. | [98] |
| Nicotiana arentsii, N. rustica, N. tabacum | Allotetraploid | Limited | - | - | Increase in copy number of some retrotransposon families. | [99,100] |
| Oryza sativa × Oenothera biennis | Hybrid | Limited | - | Altered | Mobilization of some TEs and changes in DNA methylation. | [105] |
| Oryza sativa × Zizania latifolia | Hybrid | Limited | Yes | Altered | Increase in some TEs copy numbers and transcription. Changes in DNA methylation. | [101,102,103,104] |
| Poa annua (P. infirma × P. supina) | Allotetraploid | Limited | - | - | Differences in TE content and distribution between subgenomes and between individuals. | [106] |
| Populus canadiensis (P. deltoides × P. nigra) | Allotetraploid | Limited | Higher | - | Differences in the presence of new copies and the transcription of certain retrotransposon families, but not a generalized activation of the TEs. | [107] |
| Solanum kurtzianum × S. microdontum | Hybrid | Limited | - | Lower | Tnt1 and Tto1 retrotransposons have moderate mobility and demethylation in the hybrid. | [112] |
| Solanum tuberosum × S. kurtzianum | Hybrid Allotetraploid | Limited | - | - | Activation of certain TE families. | [110] |
| Triticum aestivum × Secale cereale | Allohexaploid | Limited | - | - | DNA sequence rearrangements associated with TEs. | [122] |
| Triticum turgidum × Aegilops tauschii | Hybrid Allohexaploid | Limited | Yes | Altered | Changes in transcriptional activity and DNA methylation in some TE families. | [116,117,118,119,120,121] |
| Vitis | Hybrids | Limited | - | - | Increase in Gret1 LTR-retrotransposon copy number in hybrids. | [123] |
| Yucca aloifolia × Yucca filamentosa | Hybrid | Limited | Similar | - | No significant changes in TE abundance or transcription. Only one LTR retrotransposon family has more abundance in the hybrid. | [124] |
| Arabidopsis thaliana × Arabidopsis lyrata | Allotetraploid | No | - | Yes | No increases in TE mobility. | [34,74,75] |
| Arabidopsis thaliana Col-0 × Ler | Hybrid | No | - | Higher | No differences in small RNAs. | [76,77,78,79,80] |
| Arabidopsis thaliana Col-0 × met-1 mutant | Hybrid | No | Lower | Higher | Lower transcription and higher DNA methylation compared to mut1. | [81] |
| Brassica napus | Allotetraploid natural | No | - | - | No differences. | [83,84,85] |
| Cajanus cajan | Hybrid | No | - | Higher | DMRs. | [125] |
| Hieracium intybaceum × H. prenanthoides | Hybrid Triploid hybrid | No | - | - | No increase in the TE copy number. Overabundance of endogenous pararetrovirus in triploid hybrids. | [126] |
| Mimulus guttatus × Mimulus luteus | Allopolyploid Triploid hybrid | No | - | Lower | Lower DNA methylation in the F1 hybrid returns to the parental levels in few generations, but shows differences between subgenomes. | [127] |
| Prunus persica × P. dulcis | Hybrid | No | Similar | Similar | DMRs. | [128,129,135] |
| Spartina spp | Hybrid Allotetraploid | No | Some | Some | Few new insertions were detected, a limited TE transcriptional increase, and limited DNA methylation changes. Differential expression of TE-related small RNAs. | [130,131,132,133] |
| Solanum lycopersicum × S. pimpinellifolium | Hybrid | No | - | Lower | DNA methylation is lower in the hybrid. | [109] |
| Sorghum halepense (S. bicolor × S. ropinquum) | Allotetraploid | No | - | - | No differences in TE content. | [134] |
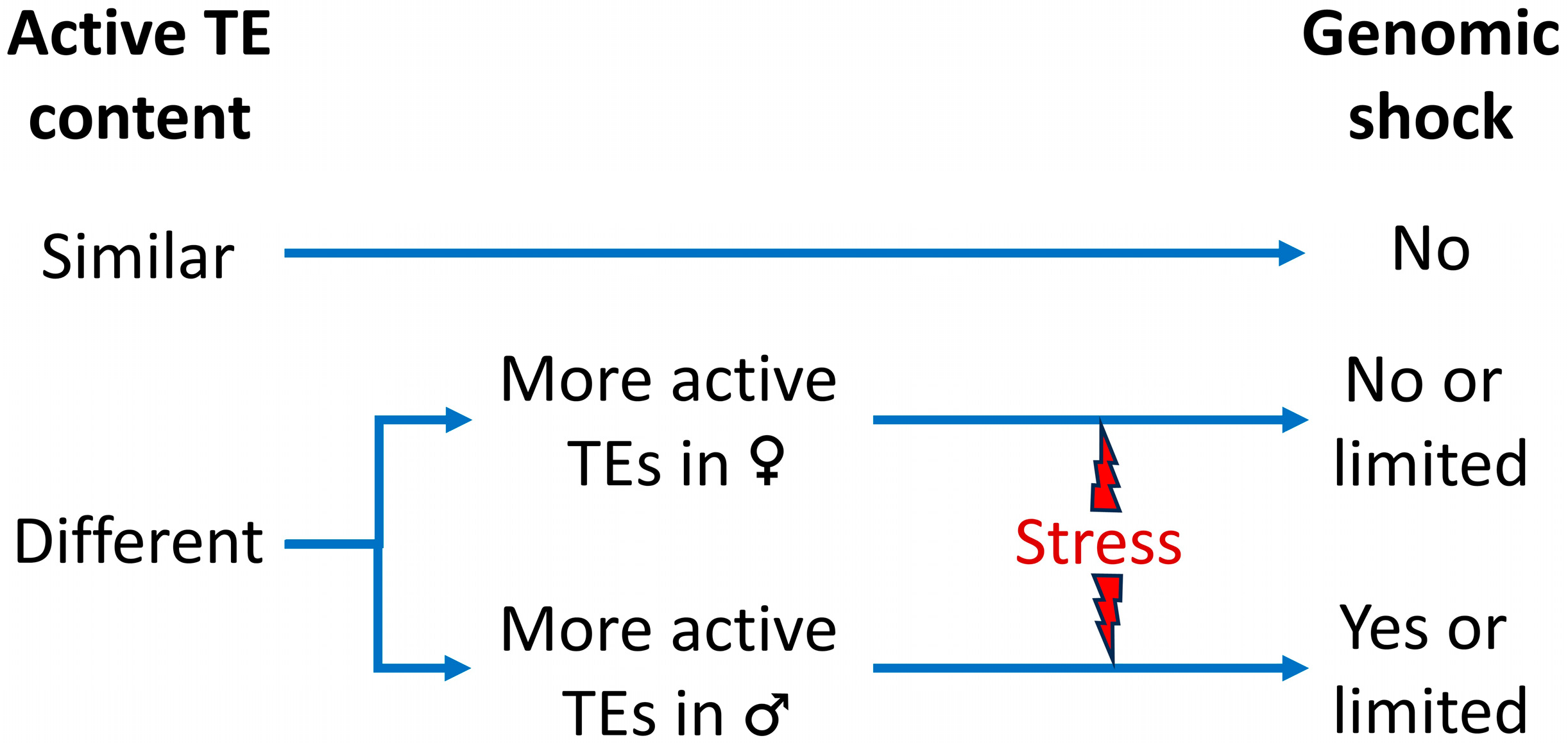
4. Conclusions: Genomic Shock?
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feschotte, C.; Jiang, N.; Wessler, S.R. Plant transposable elements: Where genetics meets genomics. Nat. Rev. Genet. 2002, 3, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Gundlach, H.; Spannagl, M.; Uauy, C.; Borrill, P.; Ramírez-González, R.H.; De Oliveira, R.; Mayer, K.F.X.; Paux, E.; Choulet, F. Impact of Transposable Elements on Genome Structure and Evolution in Bread Wheat. Genome Biol. 2018, 19, 103. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.I.; Kim, N.S. Transposable elements and genome size variations in plants. Genom. Inform. 2014, 12, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Piegu, B.; Guyot, R.; Picault, N.; Roulin, A.; Saniyal, A.; Kim, H.; Collura, K.; Brar, D.S.; Jackson, S.; Wing, R.A.; et al. Doubling Genome Size without Polyploidization: Dynamics of Retrotransposition-Driven Genomic Expansions in Oryza australiensis, a Wild Relative of Rice. Genome Res. 2006, 16, 1262–1269. [Google Scholar] [CrossRef] [PubMed]
- Vicient, C.M.; Suoniemi, A.; Anamthawat-Jónsson, K.; Tanskanen, J.; Beharav, A.; Nevo, E.; Schulman, A.H. Retrotransposon BARE-1 and Its Role in Genome Evolution in the Genus Hordeum. Plant Cell. 1999, 11, 1769–1784. [Google Scholar] [CrossRef] [PubMed]
- Vicient, C.M.; Casacuberta, J.M. Impact of transposable elements on polyploid plant genomes. Ann. Bot. 2017, 120, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Lisch, D. How important are transposons for plant evolution? Nat. Rev. Genet. 2013, 14, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Fambrini, M.; Usai, G.; Vangelisti, A.; Mascagni, F.; Pugliesi, C. The plastic genome: The impact of transposable elements on gene functionality and genomic structural variations. Genesis 2020, 58, e23399. [Google Scholar] [CrossRef]
- Mhiri, C.; Borges, F.; Grandbastien, M.A. Specificities and dynamics of transposable elements in land plants. Biology 2022, 11, 488. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. Helitrons on a roll: Eukaryotic rolling-circle transposons. Trends Genet. 2007, 23, 521–529. [Google Scholar] [CrossRef] [PubMed]
- De Tomás, C.; Vicient, C.M. Genome-wide identification of Reverse Transcriptase domains of recently inserted endogenous plant pararetrovirus (Caulimoviridae). Front. Plant Sci. 2022, 13, 1011565. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Cuerda-Gil, D.; Shahid, S.; Slotkin, R.K. The epigenetic control of the transposable element life cycle in plant genomes and beyond. Annu. Rev. Genet. 2022, 56, 63–87. [Google Scholar] [CrossRef] [PubMed]
- Pachamuthu, K.; Borges, F. Epigenetic control of transposons during plant reproduction: From meiosis to hybrid seeds. Curr. Opin. Plant Biol. 2023, 75, 102419. [Google Scholar] [CrossRef] [PubMed]
- Heard, E.; Martienssen, R.A. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell 2014, 157, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Matzke, M.A.; Kanno, T.; Matzke, A.J. RNA-directed DNA methylation: The evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 2015, 66, 243–267. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, R.K.; Freeling, M.; Lisch, D. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat. Genet. 2005, 37, 641–644. [Google Scholar] [CrossRef]
- Burgess, D.; Li, H.; Zhao, M.; Kim, S.Y.; Lisch, D. Silencing of mutator elements in maize involves distinct populations of small RNAs and distinct patterns of DNA methylation. Genetics 2020, 215, 379–391. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, J.; Zuo, T.; Zhao, M.; Lisch, D.; Peterson, T. Small RNA-mediated de novo silencing of Ac/ds transposons is initiated by alternative transposition in maize. Genetics 2020, 215, 393–406. [Google Scholar] [CrossRef]
- Zhai, J.; Bischof, S.; Wang, H.; Feng, S.; Lee, T.F.; Teng, C.; Chen, X.; Park, S.Y.; Liu, L.; Gallego-Bartolome, J.; et al. A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 2015, 163, 445–455. [Google Scholar] [CrossRef]
- Erdmann, R.M.; Picard, C.L. RNA-Directed DNA methylation. PLoS Genet. 2020, 16, e1009034. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Gent, J.I.; Zynda, G.; Song, J.; Makarevitch, I.; Hirsch, C.D.; Hirsch, C.N.; Dawe, R.K.; Madzima, T.F.; McGinnis, K.M.; et al. RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc. Natl. Acad. Sci. USA 2015, 112, 14728–14733. [Google Scholar] [CrossRef]
- Kim, E.Y.; Wang, L.; Lei, Z.; Li, H.; Fan, W.; Cho, J. Ribosome stalling and SGS3 phase separation prime the epigenetic silencing of transposons. Nativ. Plants 2021, 7, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Cuerda-Gil, D.; Slotkin, R.K. Non-canonical RNA-directed DNA methylation. Nativ. Plants 2016, 3, 16163. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhao, M. How transposable elements are recognized and epigenetically silenced in plants? Curr. Opin. Plant Biol. 2023, 75, 102428. [Google Scholar] [CrossRef]
- Singh, J.; Freeling, M.; Lisch, D. A position effect on the heritability of epigenetic silencing. PLoS Genet. 2008, 4, e1000216. [Google Scholar] [CrossRef] [PubMed]
- Roquis, D.; Robertson, M.; Yu, L.; Thieme, M.; Julkowska, M.; Bucher, E. Genomic impact of stress-induced transposable element mobility in Arabidopsis. Nucleic Acids Res. 2021, 49, 10431–10447. [Google Scholar] [CrossRef]
- Guo, W.; Wang, D.; Lisch, D. RNA-directed DNA methylation prevents rapid and heritable reversal of transposon silencing under heat stress in Zea mays. PLoS Genet. 2021, 17, e1009326. [Google Scholar] [CrossRef]
- Ito, H.; Kim, J.M.; Matsunaga, W.; Saze, H.; Matsui, A.; Endo, T.A.; Harukawa, Y.; Takagi, H.; Yaegashi, H.; Masuta, Y.; et al. A stress-activated transposon in arabidopsis induces transgenerational abscisic acid insensitivity. Sci. Rep. 2016, 6, 23181. [Google Scholar] [CrossRef]
- Klein, S.P.; Anderson, S.N. The evolution and function of transposons in epigenetic regulation in response to the environment. Curr. Opin. Plant Biol. 2022, 69, 102277. [Google Scholar] [CrossRef]
- Hirochika, H.; Sugimoto, K.; Otsuki, Y.; Tsugawa, H.; Kanda, M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 1966, 93, 7783–7788. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.J.; Schultz, M.D.; Lewsey, M.G.; O’Malley, R.C.; Urich, M.A.; Libiger, O.; Schork, N.J.; Ecker, J.R. Transgenerational epigenetic instability is a source of novel methylation variants. Science 2011, 334, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Josefsson, C.; Dilkes, B.; Comai, L. Parent-dependent loss of gene silencing during interspecies hybridization. Curr. Biol. 2006, 16, 1322–1328. [Google Scholar] [CrossRef] [PubMed]
- Göbel, U.; Arce, A.L.; He, F.; Rico, A.; Schmitz, G.; de Meaux, J. Robustness of transposable element regulation but no genomic shock observed in interspecific Arabidopsis hybrids. Genome Biol. Evol. 2018, 10, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Yañez-Santos, A.M.; Paz, R.C.; Paz-Sepúlveda, P.B.; Urdampilleta, J.D. Full-length LTR retroelements in Capsicum annuum revealed a few species-specific family bursts with insertional preferences. Chrom. Res. 2021, 29, 261–284. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Kim, M.Y.; Hsieh, P.H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef]
- Sigman, M.J.; Slotkin, R.K. The first rule of plant transposable element silencing: Location, location, location. Plant Cell 2016, 28, 304–313. [Google Scholar] [CrossRef]
- Becker, Y. Evolution of viruses by acquisition of cellular RNA or DNA nucleotide sequences and genes: An introduction. Virus Genes. 2000, 21, 7–12. [Google Scholar] [CrossRef]
- Agol, V.I.; Gmyl, A.P. Viral security proteins: Counteracting host defences. Nat. Rev. Microbiol. 2010, 8, 867–878. [Google Scholar] [CrossRef]
- Sasaki, T.; Kato, K.; Hosaka, A.; Fu, Y.; Toyoda, A.; Fujiyama, A.; Tarutani, Y.; Kakutani, T. Arms race between anti-silencing and RdDM in noncoding regions of transposable elements. EMBO Rep. 2023, 24, e56678. [Google Scholar] [CrossRef]
- Fedoroff, N.V. Molecular genetics and epigenetics of CACTA elements. Methods Mol. Biol. 2013, 1057, 177–192. [Google Scholar] [PubMed]
- Duan, C.G.; Wang, X.; Xie, S.; Pan, L.; Miki, D.; Tang, K.; Hsu, C.C.; Lei, M.; Zhong, Y.; Hou, Y.J.; et al. A pair of transposon-derived proteins function in a histone acetyltransferase complex for active DNA demethylation. Cell Res. 2017, 27, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Vicient, C.M.; Casacuberta, J.M. Additional ORFs in Plant LTR-Retrotransposons. Front. Plant Sci. 2020, 11, 555. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Orte, E.; Vicient, C.M.; Martínez-Izquierdo, J.A. Grande retrotransposons contain an accessory gene in the unusually long 3’-internal region that encodes a nuclear protein transcribed from its own promoter. Plant Mol. Biol. 2013, 81, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Ellstrand, N.C. Is gene flow the most important evolutionary force in plants? Am. J. Bot. 2014, 101, 737–753. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.S.; Batley, J. Creating new interspecific hybrid and polyploid crops. Trends Biotechnol. 2015, 33, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Donoso, J.M.; Picañol, R.; Serra, O.; Howad, W.; Alegre, S.; Arús, P.; Eduardo, I. Exploring almond genetic variability useful for peach improvement: Mapping major genes and QTLs in two interspecific almond × peach populations. Mol. Breed. 2016, 36, 16. [Google Scholar] [CrossRef]
- Bashir, T.; Sailer, C.; Gerber, F.; Loganathan, N.; Bhoopalan, H.; Eichenberger, C.; Grossniklaus, U.; Baskar, R. Hybridization alters spontaneous mutation rates in a parent-of-origin-dependent fashion in Arabidopsis. Plant Physiol. 2014, 165, 424–437. [Google Scholar] [CrossRef]
- Nieto-Feliner, G.; Casacuberta, J.; Wendel, J.F. Genomics of evolutionary novelty in hybrids and polyploids. Front. Genet. 2020, 11, 792. [Google Scholar] [CrossRef]
- Baack, E.J.; Whitney, K.D.; Rieseberg, L.H. Hybridization and genome size evolution: Timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytol. 2005, 167, 623–630. [Google Scholar] [CrossRef]
- Michalak, P. Epigenetic, transposon and small RNA determinants of hybrid dysfunctions. Heredity 2009, 102, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Comai, L.; Madlung, A.; Josefsson, C.; Tyagi, A. Do the different parental ‘heteromes’ cause genomic shock in newly formed allopolyploids? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003, 358, 1149–1155. [Google Scholar] [CrossRef]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.N.; Stitzer, M.C.; Zhou, P.; Ross-Ibarra, J.; Hirsch, C.D.; Springer, N.M. Dynamic Patterns of Transcript Abundance of Transposable Element Families in Maize. G3 2019, 5, 3673–3682. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Chen, Y.; Zhang, G.; Xing, J.; Hu, Z.; Feng, W.; Yao, Y.; Peng, H.; Du, J.; Zhang, Y.; et al. Comparative proteomic analysis of embryos between a maize hybrid and its parental lines during early stages of seed germination. PLoS ONE 2013, 8, e65867. [Google Scholar] [CrossRef] [PubMed]
- Barber, W.T.; Zhang, W.; Win, H.; Varala, K.K.; Dorweiler, J.E.; Hudson, M.E.; Moose, S.P. Repeat associated small RNAs vary among parents and following hybridization in maize. Proc. Natl. Acad. Sci. USA 2012, 109, 10444–10449. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yang, D.; Wang, D.; Liang, C.; Wang, J.; Lisch, D.; Zhao, M. Heritable changes of epialleles in maize can be triggered in the absence of DNA methylation. bioRxiv, 2023; preprint. [Google Scholar]
- Ungerer, M.C.; Strakosh, S.C.; Zhen, Y. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr. Biol. 2006, 16, R872–R873. [Google Scholar] [CrossRef]
- Ungerer, M.C.; Strakosh, S.C.; Stimpson, K.M. Proliferation of Ty3/gypsy-like retrotransposons in hybrid sunflower taxa inferred from phylogenetic data. BMC Biol. 2009, 7, 40. [Google Scholar] [CrossRef]
- Staton, S.E.; Ungerer, M.C.; Moore, R.C. The genomic organization of Ty3/gypsy-like retrotransposons in Helianthus (Asteraceae) homoploid hybrid species. Am. J. Bot. 2009, 96, 1646–1655. [Google Scholar] [CrossRef]
- Kawakami, T.; Strakosh, S.C.; Zhen, Y.; Ungerer, M.C. Different scales of Ty1/copia-like retrotransposon proliferation in the genomes of three diploid hybrid sunflower species. Heredity 2010, 104, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, T.; Dhakal, P.; Katterhenry, A.N.; Heatherington, C.A.; Ungerer, M.C. Transposable element proliferation and genome expansion are rare in contemporary sunflower hybrid populations despite widespread transcriptional activity of LTR retrotransposons. Genome Biol. Evol. 2011, 3, 156–167. [Google Scholar] [CrossRef]
- Ungerer, M.C.; Kawakami, T. Transcriptional dynamics of LTR retrotransposons in early generation and ancient sunflower hybrids. Genome Biol. Evol. 2013, 5, 329–337. [Google Scholar] [CrossRef]
- Renaut, S.; Rowe, H.C.; Ungerer, M.C.; Rieseberg, L.H. Genomics of homoploid hybrid speciation: Diversity and transcriptional activity of long terminal repeat retrotransposons in hybrid sunflowers. Phil. Tran. R. Soc. Lond. Biol. Sci. 2014, 369, 20130345. [Google Scholar] [CrossRef] [PubMed]
- Ågren, J.A.; Huang, H.R.; Wright, S.I. Transposable element evolution in the allotetraploid Capsella bursa-pastoris. Am. J. Bot. 2016, 103, 1197–1202. [Google Scholar] [CrossRef] [PubMed]
- Duan, T.; Sicard, A.; Glémin, S.; Lascoux, M. Expression pattern of resynthesized allotetraploid Capsella is determined by hybridization, not whole-genome duplication. New Phytol. 2023, 237, 339–353. [Google Scholar] [CrossRef]
- Senerchia, N.; Felber, F.; Parisod, C. Genome reorganization in F1 hybrids uncovers the role of retrotransposons in reproductive isolation. Proc. R. Soc. B. Biol. Sci. 2015, 282, 20142874. [Google Scholar]
- Senerchia, N.; Felber, F.; North, B.; Sarr, A.; Guadagnuolo, R.; Parisod, C. Differential introgression and reorganization of retrotransposons in hybrid zones between wild wheats. Mol. Ecol. 2016, 25, 2518–2528. [Google Scholar] [CrossRef]
- Danilova, T.V.; Akhunova, A.R.; Akhunov, E.D.; Friebe, B.; Gill, B.S. Major structural genomic alterations can be associated with hybrid speciation in Aegilops markgrafii (Triticeae). Plant J. 2017, 92, 317–330. [Google Scholar] [CrossRef]
- Shams, I.; Raskina, O. Intraspecific and intraorganismal copy number dynamics of retrotransposons and tandem repeat in Aegilops speltoides Tausch (Poaceae, Triticeae). Protoplasma 2018, 255, 1023–1038. [Google Scholar] [CrossRef]
- Madlung, A.; Tyagi, A.P.; Watson, B.; Jiang, H.; Kagochi, T.; Doerge, R.W.; Martienssen, R.; Comai, L. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 2005, 41, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Madlung, A.; Henkhaus, N.; Jurevic, L.; Kahsai, E.A.; Bernhard, J. Natural variation and persistent developmental instabilities in geographically diverse accessions of the allopolyploid Arabidopsis suecica. Physiol. Plant. 2012, 144, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Lu, J.; Tian, L.; Ramachandran, V.; Kasschau, K.D.; Chapman, E.J.; Carrington, J.C.; Chen, X.; Wang, X.J.; Chen, Z.J. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. USA 2009, 106, 17835–17840. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, J.; Jean, M.; Belzile, F. The allotetraploid Arabidopsis thaliana-Arabidopsis lyrata subsp. petraea as an alternative model system for the study of polyploidy in plants. Mol. Genet. Genom. 2009, 281, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Hu, B.; Becker, C.; Doan, E.S.; Berendzen, K.W.; Weigel, D.; Liu, C. Altered chromatin compaction and histone methylation drive non-additive gene expression in an interspecific Arabidopsis hybrid. Genome Biol. 2017, 18, 157. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, M.W.; Tanurdzić, M.; Lippman, Z.; Jiang, H.; Carrasquillo, R.; Rabinowicz, P.D.; Dedhia, N.; McCombie, W.R.; Agier, N.; Bulski, A.; et al. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007, 5, e174. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Varala, K.; Moose, S.P.; Hudson, M.E. The inheritance pattern of 24 nt siRNA clusters in arabidopsis hybrids is influenced by proximity to transposable elements. PLoS ONE 2012, 7, e47043. [Google Scholar] [CrossRef]
- Greaves, I.K.; Groszmann, M.; Ying, H.; Taylor, J.M.; Peacock, W.J.; Dennis, E.S. Trans chromosomal methylation in Arabidopsis hybrids. Proc. Natl. Acad. Sci. USA 2012, 109, 3570–3575. [Google Scholar] [CrossRef]
- Groszmann, M.; Greaves, I.K.; Albertyn, Z.I.; Scofield, G.N.; Peacock, W.J.; Dennis, E.S. Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc. Natl. Acad. Sci. USA 2011, 108, 2617–2622. [Google Scholar] [CrossRef]
- Shen, H.; He, H.; Li, J.; Chen, W.; Wang, X.; Guo, L.; Peng, Z.; He, G.; Zhong, S.; Qi, Y.; et al. Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 2012, 24, 875–892. [Google Scholar] [CrossRef]
- Rigal, M.; Becker, C.; Pélissier, T.; Pogorelcnik, R.; Devos, J.; Ikeda, Y.; Weigel, D.; Mathieu, O. Epigenome confrontation triggers immediate reprogramming of DNA methylation and transposon silencing in Arabidopsis thaliana F1 epihybrids. Proc. Natl. Acad. Sci. USA 2016, 113, 2083–2092. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, X.; Hu, C.; Qiu, X.; Li, J.; Li, X.; Zhu, H.; Wang, J.; Sui, J.; Qiao, L. Identification and characterization of transposable element AhMITE1 in the genomes of cultivated and two wild peanuts. BMC Genom. 2022, 23, 500. [Google Scholar] [CrossRef] [PubMed]
- Alix, K.; Heslop-Harrison, J.S. The diversity of retroelements in diploid and allotetraploid Brassica species. Plant Mol. Biol. 2004, 54, 895–909. [Google Scholar] [CrossRef] [PubMed]
- Alix, K.; Joets, J.; Ryder, C.D.; Moore, J.; Barker, G.C.; Bailey, J.P.; King, G.J.; Pat Heslop-Harrison, J.S. The CACTA transposon Bot1 played a major role in Brassica genome divergence and gene proliferation. Plant J. 2008, 56, 1030–1044. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.; Zou, J.; Hubertus Behrens, F.; Chen, L.; Ye, C.; Dai, S.; Li, R.; Ni, M.; Jiang, X.; Qiu, J.; et al. Identification, evolution, and expression partitioning of miRNAs in allopolyploid Brassica napus. J. Exp. Bot. 2015, 66, 7241–7253. [Google Scholar] [CrossRef] [PubMed]
- Lukens, L.N.; Pires, J.C.; Leon, E.; Vogelzang, R.; Oslach, L.; Osborn, T. Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 2006, 140, 336–348. [Google Scholar] [CrossRef]
- Zou, J.; Fu, D.; Gong, H.; Qian, W.; Xia, W.; Pires, J.C.; Li, R.; Long, Y.; Mason, A.S.; Yang, T.J.; et al. De novo genetic variation associated with retrotransposon activation, genomic rearrangements and trait variation in a recombinant inbred line population of Brassica napus derived from interspecific hybridization with Brassica rapa. Plant J. 2011, 68, 212–224. [Google Scholar] [CrossRef]
- Sarilar, V.; Palacios, P.M.; Rousselet, A.; Ridel, C.; Falque, M.; Eber, F.; Chèvre, A.M.; Joets, J.; Brabant, P.; Alix, K. Allopolyploidy has a moderate impact on restructuring at three contrasting transposable element insertion sites in resynthesized Brassica napus allotetraploids. New Phytol. 2013, 198, 593–604. [Google Scholar] [CrossRef]
- An, Z.; Tang, Z.; Ma, B.; Mason, A.S.; Guo, Y.; Yin, J.; Gao, C.; Wei, L.; Li, J.; Fu, D. Transposon variation by order during allopolyploidisation between Brassica oleracea and Brassica rapa. Plant Biol. 2014, 16, 825–835. [Google Scholar] [CrossRef]
- Zhang, J.; Li, G.; Li, H.; Pu, X.; Jiang, J.; Chai, L.; Zheng, B.; Cui, C.; Yang, Z.; Zhu, Y.; et al. Transcriptome Analysis of Interspecific Hybrid between Brassica napus and B. rapa Reveals Heterosis for Oil Rape Improvement. Int. J. Genom. 2015, 2015, 230985. [Google Scholar]
- Xu, Y.; Zhong, L.; Wu, X.; Fang, X.; Wang, J. Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta 2009, 229, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ge, X.; Shao, Y.; Sun, G.; Li, Z. Genomic change, retrotransposon mobilization and extensive cytosine methylation alteration in Brassica napus introgressions from two intertribal hybridizations. PLoS ONE 2013, 8, e56346. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Sun, S.; Hua, S.; Shen, E.; Ye, C.Y.; Cai, D.; Timko, M.P.; Zhu, Q.H.; Fan, L. Analysis of transcriptional and epigenetic changes in hybrid vigor of allopolyploid Brassica napus uncovers key roles for small RNAs. Plant J. 2017, 91, 874–893. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, L.; Cui, Y.; Yang, Y.; Wu, J.; Liang, J.; Li, X.; Zhang, X.; Zhang, Y.; Guo, Z.; et al. The lack of negative association between TE load and subgenome dominance in synthesized Brassica allotetraploids. Proc. Natl. Acad. Sci. USA 2023, 120, e2305208120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, X.K.; Fan, W.; Yan, D.F.; Zhong, N.S.; Gao, J.Y.; Zhang, W.J. Transcriptome analysis reveals hybridization-induced genome shock in an interspecific F1 hybrid from Camellia. Genome 2018, 61, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M.C.; Mandáková, T.; McCann, J.; Temsch, E.M.; Chase, M.W.; Hedrén, M.; Weiss-Schneeweiss, H.; Paun, O. Repeat dynamics across timescales: A perspective from sibling allotetraploid marsh orchids (Dactylorhiza majalis s.l.). Mol. Biol. Evol. 2022, 39, msac167. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Tao, X.; Feng, S.; Wang, L.; Hong, H.; Ma, W.; Shang, G.; Guo, S.; He, Y.; Zhou, B.; et al. LncRNAs in polyploid cotton interspecific hybrids are derived from transposon neofunctionalization. Genome Biol. 2018, 19, 195. [Google Scholar] [CrossRef] [PubMed]
- Fukai, E.; Yoshikawa, M.; Shah, N.; Sandal, N.; Miyao, A.; Ono, S.; Hirakawa, H.; Akyol, T.Y.; Umehara, Y.; Nonomura, K.I.; et al. Widespread and transgenerational retrotransposon activation in inter- and intra-species recombinant inbred populations of Lotus japonicus. Plant J. 2022, 111, 1397–1410. [Google Scholar] [CrossRef]
- Petit, M.; Guidat, C.; Daniel, J.; Denis, E.; Montoriol, E.; Bui, Q.T.; Lim, K.Y.; Kovarik, A.; Leitch, A.R.; Grandbastien, M.A.; et al. Mobilization of retrotransposons in synthetic allotetraploid tobacco. New Phytol. 2010, 186, 135–147. [Google Scholar] [CrossRef]
- Mhiri, C.; Parisod, C.; Daniel, J.; Petit, M.; Lim, K.Y.; de Borne, F.D.; Kovarik, A.; Leitch, A.R.; Grandbastien, M.A. Parental transposable element loads influence their dynamics in young Nicotiana hybrids and allotetraploids. New Phytol. 2019, 221, 1619–1633. [Google Scholar] [CrossRef]
- Liu, B.; Wendel, J.F. Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 2000, 43, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Liu, Z.; Dong, Z.; Wang, Y.; Chen, Y.; Lin, X.; Long, L.; Han, F.; Dong, Y.; Liu, B. Mobilization of the active MITE transposons mPing and Pong in rice by introgression from wild rice (Zizania latifolia Griseb.). Mol. Biol. Evol. 2005, 22, 976–990. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Y.; Wang, Y.M.; Zhang, Z.J.; Shen, Y.; Lin, X.Y.; Ou, X.F.; Han, F.P.; Liu, B. Extent and pattern of DNA methylation alteration in rice lines derived from introgressive hybridization of rice and Zizania latifolia Griseb. Theor. Appl. Genet. 2006, 113, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wang, H.; Wang, H.; Zhang, D.; Wu, Y.; Ou, X.; Liu, S.; Dong, Z.; Liu, B. Transpositional reactivation of the Dart transposon family in rice lines derived from introgressive hybridization with Zizania latifolia. BMC Plant Biol. 2010, 10, 190. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chai, Y.; Chu, X.; Zhao, Y.; Wu, Y.; Zhao, J.; Ngezahayo, F.; Xu, C.; Liu, B. Molecular characterization of a rice mutator-phenotype derived from an incompatible cross-pollination reveals transgenerational mobilization of multiple transposable elements and extensive epigenetic instability. BMC Plant Biol. 2009, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Benson, C.W.; Sheltra, M.R.; Maughan, P.J.; Jellen, E.N.; Robbins, M.D.; Bushman, B.S.; Patterson, E.L.; Hall, N.D.; Huff, D.R. Homoeologous evolution of the allotetraploid genome of Poa annua L. BMC Genom. 2023, 24, 350. [Google Scholar]
- Usai, G.; Mascagni, F.; Vangelisti, A.; Giordani, T.; Ceccarelli, M.; Cavallini, A.; Natali, L. Interspecific hybridisation and LTR retrotransposon mobilisation-related structural variation in plants: A case study. Genomics 2020, 122, 1611–1621. [Google Scholar] [CrossRef]
- Mascagni, F.; Usai, G.; Natali, L.; Cavallini, A.; Giordani, T. A comparison of methods for LTR-retrotransposon insertion time profiling in the Populus trichocarpa genome. Caryologia 2018, 71, 85–92. [Google Scholar] [CrossRef]
- Raza, M.A.; Yu, N.; Wang, D.; Cao, L.; Gan, S.; Chen, L. Differential DNA methylation and gene expression in reciprocal hybrids between Solanum lycopersicum and S. pimpinellifolium. DNA Res. 2017, 24, 597–607. [Google Scholar] [CrossRef]
- Gantuz, M.; Morales, A.; Bertoldi, M.V.; Ibañez, V.N.; Duarte, P.F.; Marfil, C.F.; Masuelli, R.W. Hybridization and polyploidization effects on LTR-retrotransposon activation in potato genome. J. Plant Res. 2021, 135, 81–92. [Google Scholar] [CrossRef]
- Marfil, C.F.; Masuelli, R.W.; Davison, J.; Comai, L. Genomic instability in Solanum tuberosum × Solanum kurtzianum interspecific hybrids. Genome 2006, 49, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Paz, R.C.; Rendina González, A.P.; Ferrer, M.S.; Masuelli, R.W. Short-term hybridisation activates Tnt1 and Tto1 Copia retrotransposons in wild tuber-bearing Solanum species. Plant Biol. 2015, 1, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Shaked, H.; Kashkush, K.; Ozkan, H.; Feldman, M.; Levy, A.A. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 2001, 13, 1749–1759. [Google Scholar] [CrossRef] [PubMed]
- Kashkush, K.; Feldman, M.; Levy, A.A. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 2002, 160, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Kashkush, K.; Feldman, M.; Levy, A.A. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 2003, 33, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Banouh, M.; Armisen, D.; Bouguennec, A.; Huneau, C.; Sow, M.D.; Pont, C.; Salse, J.; Civáň, P. Low impact of polyploidization on the transcriptome of synthetic allohexaploid wheat. BMC Genom. 2023, 24, 255. [Google Scholar] [CrossRef] [PubMed]
- Kraitshtein, Z.; Yaakov, B.; Khasdan, V.; Kashkush, K. Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics 2010, 186, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Yaakov, B.; Kashkush, K. Massive alterations of the methylation patterns around DNA transposons in the first four generations of a newly formed wheat allohexaploid. Genome 2011, 54, 42–49. [Google Scholar] [CrossRef]
- Kenan-Eichler, M.; Leshkowitz, D.; Tal, L.; Noor, E.; Melamed-Bessudo, C.; Feldman, M.; Levy, A.A. Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 2011, 188, 263–272. [Google Scholar] [CrossRef]
- Kirov, I.; Dudnikov, M.; Merkulov, P.; Shingaliev, A.; Omarov, M.; Kolganova, E.; Sigaeva, A.; Karlov, G.; Soloviev, A. Nanopore RNA Sequencing Revealed Long Non-Coding and LTR Retrotransposon-Related RNAs Expressed at Early Stages of Triticale SEED Development. Plants 2020, 9, E1794. [Google Scholar] [CrossRef]
- Li, A.; Liu, D.; Wu, J.; Zhao, X.; Hao, M.; Geng, S.; Yan, J.; Jiang, X.; Zhang, L.; Wu, J.; et al. mRNA and Small RNA Transcriptomes Reveal Insights into Dynamic Homoeolog Regulation of Allopolyploid Heterosis in Nascent Hexaploid Wheat. Plant Cell 2014, 26, 1878–1900. [Google Scholar] [CrossRef] [PubMed]
- Bento, M.; Pereira, H.S.; Rocheta, M.; Gustafson, P.; Viegas, W.; Silva, M. Polyploidization as a retraction force in plant genome evolution: Sequence rearrangements in triticale. PLoS ONE 2008, 3, e1402. [Google Scholar] [CrossRef] [PubMed]
- Cadle-Davidson, M.M.; Owens, C.L. Genomic amplification of the Gret1 retroelement in white-fruited accessions of wild vitis and interspecific hybrids. Theor. Appl. Genet. 2008, 116, 1079–1094. [Google Scholar] [CrossRef] [PubMed]
- Heyduk, K.; McAssey, E.V.; Grimwood, J.; Shu, S.; Schmutz, J.; McKain, M.R.; Leebens-Mack, J. Hybridization History and Repetitive Element Content in the Genome of a Homoploid Hybrid, Yucca gloriosa (Asparagaceae). Front. Plant Sci. 2021, 11, 573767. [Google Scholar] [CrossRef] [PubMed]
- Junaid, A.; Kumar, H.; Rao, A.R.; Patil, A.N.; Singh, N.K.; Gaikwad, K. Unravelling the epigenomic interactions between parental inbreds resulting in an altered hybrid methylome in pigeonpea. DNA Res. 2018, 25, 361–373. [Google Scholar] [CrossRef]
- Zagorski, D.; Hartmann, M.; Bertrand, Y.J.K.; Paštová, L.; Slavíková, R.; Josefiová, J.; Fehrer, J. Characterization and Dynamics of Repeatomes in Closely Related Species of Hieracium (Asteraceae) and Their Synthetic and Apomictic Hybrids. Front. Plant Sci. 2020, 11, 591053. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; Smith, R.; McKain, M.R.; Cooley, A.M.; Vallejo-Marin, M.; Yuan, Y.; Bewick, A.J.; Ji, L.; Platts, A.E.; Bowman, M.J.; et al. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year-old naturally established neo-allopolyploid Monkeyflower. Plant Cell 2017, 29, 2150–2167. [Google Scholar] [CrossRef]
- De Tomás, C.; Bardil, A.; Castanera, R.; Casacuberta, J.M.; Vicient, C.M. Absence of major epigenetic and transcriptomic changes accompanying an interspecific cross between peach and almond. Hortic. Res. 2022, 9, uhac127. [Google Scholar] [CrossRef]
- D’Amico-Willman, K.M.; Sideli, G.M.; Allen, B.J.; Anderson, E.S.; Gradziel, T.M.; Fresnedo-Ramírez, J. Identification of Putative Markers of Non-infectious Bud Failure in Almond (Prunus dulcis (Mill.) D.A. Webb) through Genome Wide DNA Methylation Profiling and Gene Expression Analysis in an Almond × Peach Hybrid Population. Front. Plant Sci. 2022, 13, 804145. [Google Scholar] [CrossRef]
- Parisod, C.; Salmon, A.; Zerjal, T.; Tenaillon, M.; Grandbastien, M.A.; Ainouche, M. Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol. 2009, 184, 1003–1015. [Google Scholar] [CrossRef]
- Giraud, D.; Lima, O.; Rousseau-Gueutin, M.; Salmon, A.; Aïnouche, M. Gene and Transposable Element Expression Evolution Following Recent and Past Polyploidy Events in Spartina (Poaceae). Front. Genet. 2021, 12, 589160. [Google Scholar] [CrossRef] [PubMed]
- Baumel, A.; Ainouche, M.; Kalendar, R.; Schulman, A.H. Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C.E. Hubbard (Poaceae). Mol. Biol. Evol. 2002, 19, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Cavé-Radet, A.; Giraud, D.; Lima, O.; El Amrani, A.; Aïnouche, M.; Salmon, A. Evolution of small RNA expression following hybridization and allopolyploidization: Insights from Spartina species (Poaceae, Chloridoideae). Plant Mol. Biol. 2019, 102, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.T.; Ishii, T.; Fuchs, J.; Hsieh, W.H.; Houben, A.; Lin, Y.R. The Evolutionary Dynamics of Repetitive DNA and Its Impact on the Genome Diversification in the Genus Sorghum. Front. Plant Sci. 2021, 12, 729734. [Google Scholar] [CrossRef] [PubMed]
- De Tomás, C. Transposons, a rich source of genetic variability in crops. Ph.D. Thesis, Facultat de Biociències, Universitat Autònoma de Barcelona, Barcelona, España, 2023. [Google Scholar]
- Drouin, M.; Hénault, M.; Hallin, J.; Landry, C.R. Testing the Genomic Shock Hypothesis Using Transposable Element Expression in Yeast Hybrids. Front. Fungal Biol. 2021, 2, 729264. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S. Epigenomics of Plant Responses to Environmental Stress. Epigenomes 2018, 2, 6. [Google Scholar] [CrossRef]
- Qiu, Y.; O’Connor, C.H.; Della Coletta, R.; Renk, J.S.; Monnahan, P.J.; Noshay, J.M.; Liang, Z.; Gilbert, A.; Anderson, S.N.; McGaugh, S.E.; et al. Whole-genome variation of transposable element insertions in a maize diversity panel. G3 2021, 11, jkab238. [Google Scholar] [CrossRef]
- Lee, Q.H.; Wright, S.; Bureau, T. Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 7376–7381. [Google Scholar] [CrossRef]
- Deniz, Ö.; Frost, J.M.; Branco, M.R. Regulation of transposable elements by DNA modifications. Nat. Rev. Genet. 2019, 20, 417–431. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Tomás, C.; Vicient, C.M. The Genomic Shock Hypothesis: Genetic and Epigenetic Alterations of Transposable Elements after Interspecific Hybridization in Plants. Epigenomes 2024, 8, 2. https://doi.org/10.3390/epigenomes8010002
de Tomás C, Vicient CM. The Genomic Shock Hypothesis: Genetic and Epigenetic Alterations of Transposable Elements after Interspecific Hybridization in Plants. Epigenomes. 2024; 8(1):2. https://doi.org/10.3390/epigenomes8010002
Chicago/Turabian Stylede Tomás, Carlos, and Carlos M. Vicient. 2024. "The Genomic Shock Hypothesis: Genetic and Epigenetic Alterations of Transposable Elements after Interspecific Hybridization in Plants" Epigenomes 8, no. 1: 2. https://doi.org/10.3390/epigenomes8010002
APA Stylede Tomás, C., & Vicient, C. M. (2024). The Genomic Shock Hypothesis: Genetic and Epigenetic Alterations of Transposable Elements after Interspecific Hybridization in Plants. Epigenomes, 8(1), 2. https://doi.org/10.3390/epigenomes8010002





