Abstract
Experimental alteration of DNA methylation is a suitable tool to infer the relationship between phenotypic and epigenetic variation in plants. A detailed analysis of the genome-wide effect of demethylating agents, such as 5-azacytidine (5azaC), and zebularine is only available for the model species Arabidopsis thaliana, which suggests that 5azaC may have a slightly larger effect. In this study, global methylation estimates obtained by high-performance liquid chromatography (HPLC) analyses were conducted to investigate the impact of 5azaC treatment on leaf and root tissue in Erodium cicutarium (Geraniaceae), which is an annual herb native to Mediterranean Europe that is currently naturalized in all continents, sometimes becoming invasive. We used seeds collected from two natural populations in SE Spain. Root tissue of the second generation (F2) greenhouse-grown seedlings had a significantly lower global cytosine methylation content than leaf tissue (13.0 vs. 17.7% of all cytosines). Leaf tissue consistently decreased methylation after treatment, but the response of root tissue varied according to seed provenance, suggesting that genetic background can mediate the response to experimental demethylation. We also found that both leaf number and leaf length were reduced in treated seedlings supporting a consistent phenotypic effect of the treatment regardless of seedling provenance. These findings suggest that, although the consequences of experimental demethylation may be tissue- and background-specific, this method is effective in altering early seedling development, and can thus be useful in ecological epigenetic studies that are aiming to investigate the links between epigenetic and phenotypic variation in non-model plant species.
Keywords:
DNA methylation; ecological epigenetics; Geraniaceae; HPLC; root; seed priming; 5-azacytidine 1. Introduction
Changes in DNA methylation, histone modification, and small RNAs impact on chromatin structure, modulate the expression of genes and transposable elements (TE), and determine the changes in phenotypic expression independent of genetic changes. These epigenetic changes are highly dynamic, are often under environmental or developmental influence, and their phenotypic consequences can be analyzed at cell, tissue, and organism levels [1,2,3,4]. In plants, epigenetic regulation has been related to individual and population responses to environmental stress and there is a growing interest in understanding how epigenetic regulation can contribute to transgenerational stress memories [5,6,7,8,9]. Cytosine DNA methylation is a key epigenetic process that is suitable for transgenerational transmission in flowering plants because, in contrast to animals, methylation marks are not extensively erased at gametogenesis [1,10]. At the molecular level, changes in the methylation status of cytosines after experimental stress are loci specific and its global sign and magnitude can vary across and within species (see [11] for a literature review). Thus, experimental studies in plants with different genomic and ecological features should contribute to a better understanding of the specificities of epigenetic responses to stress in terms of molecular and phenotypic changes that could be transmitted to progeny [12].
Experimental inhibition of the activity of DNA methyltransferases (DNMTs) is a powerful strategy to modulate DNA methylation that has been broadly assayed in medicine but to a much lower extent in plant science [13]. In plants, DNMT inhibitors (DNMTi hereafter), mainly 5-azacytidine, 5-aza-2′-deoxycytidine, and zebularine, have proven useful to obtain epimutants with characteristic phenotypes that, at least in some cases, remained stable across multiple generations (e.g., [14,15,16,17,18]). Zebularine is chemically more stable, forms a reversible complex with DNMTs, and exhibits a weaker inhibition activity and cytotoxicity than azacytosine analogs [13]. A comprehensive analysis of the molecular response to treatment with these DNMTi in plants is only available for the model Arabidopsis thaliana [19,20]. In this model species, seed exposure to either 5-azacytidine (5azaC) or zebularine imposes a dose-dependent loss of DNA methylation at the seedling stage, which, at higher concentrations (>50 µM), was larger for 5azaC [20]. Interestingly, demethylation induced by the two compounds occurred in all sequence contexts and altered both gene and TE expression [19,20].
The application of DNMTi is also a suitable tool to experimentally dissect the epigenetic contribution to phenotypic and functional responses to controlled stressful conditions. Nevertheless, different investigations have used different compounds and protocols (see e.g., [21,22,23,24]), and, occasionally, variations in the magnitude of the observed response are reported when different genetic lines or source populations are used (e.g., [21,22,23]). In addition to such a genotype-specific response, we should note that the magnitude of methylation changes in response to DNMTi treatment and/or experimental stress is usually analyzed in a single tissue, yet global methylation is known to differ between tissues and developmental stages [1], and changes in DNA methylation in response to stress can differ between tissues of treated individuals (see e.g., [3,25,26,27,28]). Above and below-ground plant organs are highly differentiated, and experience distinctive environments, thus, exploring variation in methylation between them might be particularly useful when trying to interpret plant responses to certain soil stress factors such as drought or salinity (see e.g., [25,26]).
In this study, we focused in Erodium cicutarium (L.) L’Hér. (Geraniaceae), a diploid annual herb native to Mediterranean Europe, north Africa, and western Asia, which is actually found globally in temperate areas with hot summers of both hemispheres [29,30]. A fast growing cycle, which allows early flowering and fruiting before seasonal summer drought, and autonomous seed-pollination have together contributed to its naturalization in diverse habitats from all of the continents, where it can displace native species [31,32,33,34]. These features make the species interesting for epigenetic analyses. We investigate the impact of 5azaC treatment at seed germination in early seedling size and DNA methylation level in leaves and roots. To gain generality, we used seeds that were collected from two natural populations located in Cazorla mountains (SE Spain) distant in straight-line ca. 10 km, with different inclination and soil depth. Application of DNMTi at seed germination is particularly appealing for basic and applied purposes because (i) demethylation at early developmental stages is expected to be amplified during subsequent cell divisions, and thus affect a high proportion of cells in the fully grown organism [2]; (ii) it resembles some seed priming protocols used to improve germination of stored seeds [35]; and, (iii) the transition from seed to seedling is key for individual establishment and plant population dynamics, and its success can be directly influenced by initial root development and early individual growth [36]. Our specific questions were (i) does 5azaC treatment affect early seedling size? (ii) Does 5azaC treatment affect global DNA cytosine methylation in seedlings’ leaf and root tissue and, if it does, is the effect similar in both tissues?, and (iii) are the effects consistent in seeds obtained from different provenance? Our results confirmed that addition of 5azaC at seed germination reduced early seedling size and global cytosine methylation in leaves. However, the response of root tissue varied according to seed provenance supporting the interest of analyzing above and below-ground tissues in trying to understand epigenetic regulation in plants.
2. Results
There was a significant 5azaC treatment effect on seedling phenotype (Supplementary Materials, Figure S1): treated seedlings produced fewer leaves than control ones (4.9 ± 0.4 vs. 7.7 ± 0.4; F1,36 = 11.6; p = 0.0016; Figure 1A) and their leaves were also shorter (3.0 ± 0.3 mm and 4.34 ± 0.3 mm, respectively; F1,36 = 22.7; p < 0.0001; Figure 1B). Seedlings from the two provenances did not differ in phenotype (F1,36 = 1.29; p = 0.26 for leaf number; F1,36 = 0.38; p = 0.54 for maximum leaf length), and responded similarly to the treatment (F1,36 = 0.02; p = 0.90; F1,36 = 0.34; p = 0.56 for the treatment * provenance interaction in leaf number and maximum leaf length, respectively).
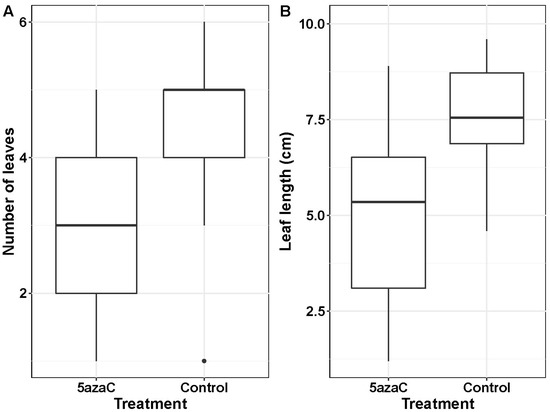
Figure 1.
Phenotypic consequences of the demethylation treatment at seed germination. Boxplots showing that seedlings treated with 5-azacytidine (5azaC) produced (A) fewer leaves than control seedlings and (B) their leaves were also shorter. Data from the two provenances combined.
Leaf DNA was consistently more methylated than root DNA across samples (Figure 2). On average, 17.68% (±0.17) of all cytosines were methylated in leaf DNA, in comparison to only 13.02% (±0.17) in roots. When all of the samples were combined, a global effect of the demethylation treatment was found, although it has only marginal significance (Table 1). Note however that a significant 3-way treatment * tissue * provenance interaction in the analysis of global cytosine methylation suggests some complex effects of the experimental treatment (Table 1). In leaves, 5azaC treatment decreased methylation consistently in individuals from the two populations (Figure 3; F1,36 = 7.69, p = 0.009 and F1,36 = 0.67, p = 0.79 for the treatment and interaction effects, respectively). However, in roots, a significant interaction between treatment and provenance was found (F1,36 = 9.73, p = 0.036), which arose because the 5azaC treatment decreased methylation in root DNA only in individuals from Nava de las Correhuelas (CH) site whereas in tended to increase in those obtained from Puerto del Tejo (PT) site (Figure 3).
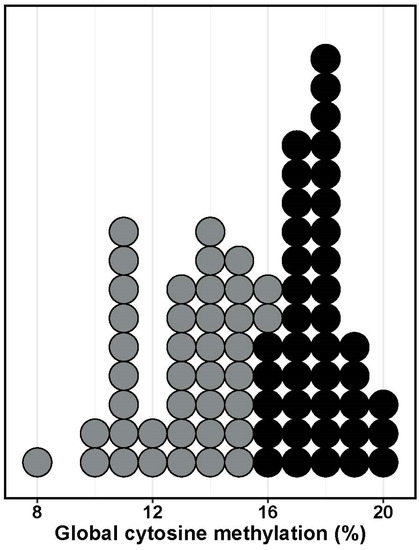
Figure 2.
Density histogram showing the range of variation in percent of global cytosine methylated recorded in leaves (black) and roots (grey) of Erodium cicutarium seedlings. Each dot represents the average value of the two replicates processed (N = 40 individuals).

Table 1.
Summary of the ANOVA results of the full factorial design conducted to analyze the variation in global cytosine methylation percentage of Erodium cicutarium seedlings. Two replicate measurements per plant were taken and plants were included in the model as a random effect.
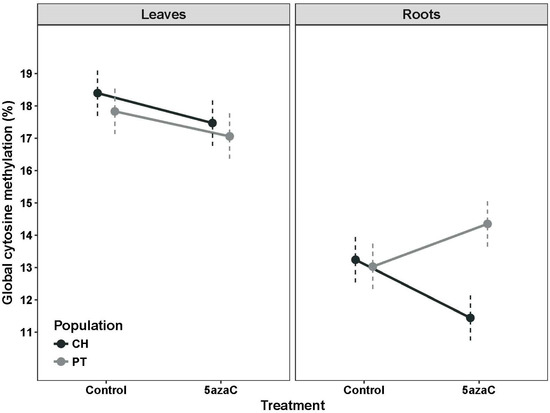
Figure 3.
Interaction plot for the effects of provenance and treatment in percent of global cytosine methylated in leaves and roots of Erodium cicutarium seedlings. The two provenances are Nava de las Correhuelas [CH] and Puerto del Tejo [PT] natural populations in SE Spain. Dots and dashed lines represent leas-squeared means and their 95% confidence level, respectively.
3. Discussion
Analyses of natural and induced epimutants have improved our understanding of epigenetics in ecological adaptation and evolutionary change [37,38,39,40]. In particular, experimental demethylation with DNMTi has contributed to reveal the broad range of plant traits and developmental processes that are regulated through DNA cytosine methylation, because such experiments have been able to, among others, alter sex expression [15], modify growth, leaf traits, flowering time or fruit ripening (e.g., [17,18,21,41,42,43,44]), and induce dwarfism at maturity [14,17]. The phenotypic consequences of the cell reprogramming associated to genome-wide demethylation, however, are not necessarily the same across species, genetic backgrounds, and cell lines [13]. For instance, the application of 5azaC induced metastable early flowering lines in flax [41], delayed flowering in some but not all of A. thaliana genetic lines assayed [21] and both early and late flowering lines in strawberry [42]. In this study, we documented a reduction in leaf size and leaf production during early seedling development in E. cicutarium that was treated with 5azaC. Reduced size at early development stages has usually a negative impact for seedling survival in wild plant populations, particularly in seasonal environments with summer drought (e.g., [45]), suggesting that altering DNA methylation could affect E. cicutarium establishment. We further revealed tissue and provenance-specific molecular responses to this treatment.
Some early epigenetic studies reported differences in global cytosine methylation among specific tissues/organs, as well as along tissue maturation stages (e.g., [1,46,47,48,49] and references therein). More recently, Whole Genome Bisulfite Sequencing in three Brassicaceae species found lower methylation levels in roots than shoots, with changes between the two tissues being more frequent at repetitive sites and transposons, although across-species variation in cytosine methylation patterns was larger [3]. Here, we found that, in E. cicutarium, methylation was also lower in roots than in above ground leaves (see [47,50] for similar results in chicory and cotton, respectively). Scarcity of studies, disparity of the analytical methods that were used to quantify cytosine methylation, and some contrasting results between the few species analyzed in more detail (see e.g., [51]) precludes us to ascertain whether, apart from a reduced methylation level in endosperm, methylation varies among plant tissues in some definite way [52]. The high-performance liquid chromatography (HPLC) method applied here could help to provide precise and genome reference-free global measure of DNA cytosine methylation, useful as a molecular phenotype for tissue-specific comparisons among and within species (see e.g., [47,50]) that could improve our current understanding of epigenetic regulation during plant development in a broader range of plant species.
In the present study on E. cicutarium, imbibition during 48 h of scarified seeds in 0.5 mM 5azaC reduced global DNA cytosine methylation in seedling leaves, difference being similar in individuals from the two study provenances (average difference 0.92% and 0.76%, for CH and PT site, respectively). The effect of treatment on roots, however, varied greatly depending on seed provenance: demethylation was larger than in leaves in seedlings from CH site (average difference 1.8%), whereas the roots of seedlings from PT site treated with 5azaC exhibited a higher global methylation than control siblings (average difference 1.42%). The methylation increase after the DNMTi treatment was unexpected. We are not aware of any previous study reporting similar findings but a differential response between leaves and roots after experimental stress has been previously documented for rice [28] and barley [53]. In particular, leaf and root barley DNA were methylated at similar level but under water-deficiency treatment more demethylation events were observed in leaf DNA, whereas novel methylations dominated in root DNA [53]. Further, methylation changes in response to salt stress were variable between leaves and roots in rice, but genotype-specific methylation changes were also highly relevant [28]. Actually, the relative relevance of genotype and tissue/organ-specific cytosine methylation patterns is not yet fully understood [51]. We can only speculate that increased methylation in E. cicutarium roots of PT after 5azaC treatment could have arisen as a consequence of the up-and-down variation in methylation level during early growth in roots [47], if the two provenances exhibited slightly different developmental stages that were not evident at collection date; as different sensitivity to other side cytotoxic effects associated to deeper cell reprogramming in root cells of plants from PT site; or due to demethylation-induced expression of a root-specific DNA methyltransferase operating in PT plants. A better understanding of specific functional effects of DNMTi could be gained by recording the phenotypic consequences and methylation levels at both above and below ground organs at different developmental stages. Altogether, current evidence highlights the added-value of including several tissues/organs and provenances/genotypes in experimental designs which include DNMTi. Its application together with other stress factors should help to better weigh the epigenetic component behind the functional phenotypic responses experienced by stressed individuals and its transgenerational transmission (see e.g., [22,23,24]).
In addition to simultaneously considering the demethylating impact of DNMTi on different plant parts, future studies should make efforts to converge on more similar experimental protocols. To date, broad variation across investigations in DNMTi application modes limits the reproducibility of the molecular results, precluding, thus, sound interpretation of species and genotype-specific phenotypic responses observed and even the assessment of the long-term biological impact of the effects of different compounds [20]. A non-exhaustive literature search illustrates the range of dosage (concentration and time of exposure) used for experimental inhibition of DNMT with different compounds at seed germination (Table 2). In A. thaliana, seed exposure to either 5azaC or zebularine imposes a dose-dependent loss of DNA methylation that tended to be higher for 5azaC at concentrations >50 µM [19,20]. Application of 50 µM 5azaC to seeds up to germination (336 h) increased mortality of germinated seedlings, delayed flowering, and reduced individual size and fruit production, although the magnitude of the response varied among genotypes [21]. Furthermore, zebularine in concentration up to 80 µM causes global DNA demethylation in A. thaliana and a delayed seedling growth, although both molecular and phenotypic effects can be overcome at adult stage by treatment release and subsequent growth in the absence of the drug [19]. In flax, a short duration treatment with a high 5azaC concentration (1.5 mM) generated more stable early flowering epimutants [16,41,54]. In strawberry [42], an increased phenotypic variance was obtained at intermediate concentrations (20–50 mM), whereas the highest one (100 mM) was entirely detrimental. Altogether, current evidence suggests that 5azaC may have a slightly larger and more stable effect in DNA although the most practical concentration to induce phenotypic variance without reducing survival can vary among species. More research is still required to fully understand the stability and transgeneration transmission of the epigenetic marks altered by DNMTi even for model species [20].

Table 2.
Experimental conditions used for the application of different DNA methyltransferases inhibitors (DNMTi), namely 5-azacytidine (5azaC), 5-aza-2′-deoxycytidine (5azadC) and zebularine (Zebu), at seed germination stage and methods used to measure demethylation: bidimensional thin-layer chromatography (2D-TLC); High-Performance Liquid Chromatography (HPLC); Methyl-Sensitive Amplified Fragment Length Polymorphism (MSAP) and Next Generation Sequencing (NGS). In all cases DNA from either a single above ground vegetative organ or full seedlings was analyzed.
To sum up, experimental demethylation with DNMTi solutions is effective in altering early seedling development and young plant phenotypes and, thus, could help to investigate the links between epigenetic and phenotypic variation at intraspecific level. However, the magnitude and stability of decrease in global DNA methylation is compound-, tissue-, and genotype-specific, and we still have limited information about the relative relevance of such variations on different plant species. Methodological improvements by convergence of experimental protocols towards, as suggested by Fieldes and Amyot [16], a short duration with a high concentration DNMTi treatment, and complementary analyses of phenotype and DNA methylation in different tissues/organs, should contribute to better understand epigenetic regulation in plants and its role in their responses to environmental factors. The genome reference-free HPLC method here applied could contribute to this aim.
4. Materials and Methods
4.1. Experimental Design
To assess the effects of DNMTi on global methylation levels of leaves and roots, the experiment was conducted with seedlings grown in the greenhouse for two generations aiming to minimize the effects of heterogeneous natural growing conditions of maternal plants in their offspring phenotype and DNA methylation [54]. In spring 2015, open pollinated fruits were collected from individual plants growing at two E. cicutarium natural populations located in Cazorla mountains (Jaén province, Spain): Puerto del Tejo (PT, 1590 m asl) and Nava de las Correhuelas (CH, 1625 m asl) distant in straight-line ca. 10 km. Puerto del Tejo site has higher inclination and more rocky substrate. Some phenotypic differences could be observed between the two populations: individuals at PT site tended to produce less leaves than those at CH site (28.2 ± 4.0 vs. 35.0 ± 2.6; F1,47 = 3.0, p = 0.09), although they were larger in average rosette diameter (11.04 cm ± 1.0 vs. 8.66 cm ± 0.6; F1,47 = 6.02, p = 0.018). Further, leaves of flowering plants at PT site have higher global methylation percentage than those at CH site (18.10 ± 0.38% vs. 17.21 ± 0.25%; F1,49 = 5.322, p = 0.025). Roots were not studied in natural populations. Collected fruits were stored in darkness at room temperature for 4–5 months. In autumn 2015, the seeds were removed from fruits, scarified, and germinated in universal substrate (COMPO SANA®, Münster, Germany) mixed in 3:1 with perlite (substrate hereafter). These first generation (F1) seedlings were subsequently transferred to 1 L pots with the same substrate, regularly watered and monitored until the end of reproduction (ca. six months). Autonomously-pollinated fruits were collected in paper bags, and stored at room temperature for six months.
For the second generation, four seeds of five of these F1 maternal family lines from each field site were haphazardly selected. The seed coat of every seed, was slightly scarified with sandpaper, and was subsequently stored in labeled microplates at 4 °C until the beginning of our experiment. For the demethylation treatment, a stock solution of 5-Azacytidine (Sigma, St. Louis, MO, USA) in Dimethyl Sulfoxide (DMSO; Sigma) 100 mM was prepared, stored at −20 °C, and diluted in water to 0.5 mM just before treatment (5azaC Treatment). As a control a mock solution of DMSO in water (3:97; v:v) was used (Control). On October 2016, the 40 seeds were soaked in 150 µL of either Control or 5azaC solutions and stored at 4 °C during 48 h. Imbibition of scarified seeds was fast, 27.5% of seeds had a visible radicle (at least twice as long as the seed coat) when they were transferred from vials to substrate (frequency was similar in control and treated seeds χ2 1 = 0.50, p = 0.48). After this period of 48 h of treatment, seeds were individually sown 2–5 mm deep in cells of seedling plug trays [31], that were placed in water vessels under greenhouse conditions (16 h light; 25–20 °C) until the end of the experiment.
For every seedling, leaf number was counted and the length of the longest leaf was measured (to the nearest mm) 11 and 17 d after sowing, respectively. Leaf and root material from all of the individuals were collected 39 d after sowing. For leaf collection, a sample of 2–3 fully grown leaves without signs of damage or senescence of each F2 seedling was placed in labeled paper bags and dried at ambient temperature in sealed containers with abundant silica gel. Roots were taken out of the soil, carefully washed in water, excess water was wiped away using absorbent paper, and a sample of fine roots (avoiding the primary thickest one) was collected. Each root sample was placed in labeled paper bags and dried at ambient temperature in sealed containers with abundant silica gel.
4.2. Laboratory Methods
Dried samples were homogenized to a fine powder using a Retsch MM 200 mill. Total genomic DNA was extracted using Bioline ISOLATE II Plant DNA Kit. A 100 ng aliquot of DNA extract was digested with 3 U of DNA Degradase PlusTM (Zymo Research, Irvine, CA, USA), a nuclease mix that degrades DNA to its individual nucleoside components. Digestion was carried out in a 40 µL volume at 37 °C for 3 h, and terminated by heat inactivation at 70 °C for 20 min. Two independent replicates of digested DNA per sample were processed to estimate global cytosine methylation more precisely. Selective derivatization of cytosine moieties with 2-bromoacetophenone under anhydrous conditions and subsequent reverse phase HPLC with spectrofluorimetric detection, were conducted. The 160 vials (40 seedlings × 2 tissues × 2 replicates) were processed in randomized order; two replicates failed to produce any signal and were discarded (N = 158). The percentage of total cytosine methylation on each replicate was estimated as 100 × 5mdC/(5mdC + dC), where 5mdC and dC are the integrated areas under the peaks for 5-methyl-2′-deoxycytidine and 2′-deoxycytidine, respectively (see [11,57] for details).
4.3. Statistical Analyses
All of the statistical analyses were carried out using the R environment [58]. Data were visually inspected and the absence of obvious outliers was confirmed. Linear models were used to analyze heterogeneity of the response variables studied [59]. The lm function was used to analyze the effect of treatment (Control vs. 5azaC), provenance (PT vs. CH) and its interaction, on leaf number and maximum leaf length. For the replicated cytosine methylation percentage data, the lme function from the nlme package [60] was used to fit mixed-effect models with Restricted Maximum Likelihood (REML) estimation, including plant as a random effect [59]. The full model included tissue (leaf vs. root), treatment (Control vs. 5azaC treated), provenance (PT vs. CH), and all of their interactions as fixed effects. Partial models were subsequently applied to leaf and root tissue independently to better interpret the sign of interactions. Inspection of residuals was conducted to assess goodness-of-fit. Normal distribution of residuals was not rejected in any of the fitted models presented. Significance of fixed effects and interactions were obtained by applying to fitted models the ANOVA function from the car package [61]. Model-adjusted marginal averages for the fixed main effects were obtained with the lsmeans function from the lsmeans package [62].
Supplementary Materials
The following are available online at www.mdpi.com/2075-4655/1/3/16/s1. Figure S1: Differences between Control (left) and 5azaC-treated sibling seedlings were evident at the time of tissue collection for global methylation analyses.
Acknowledgments
We are grateful to Laura Cabral, Esmeralda López, Daisy Johnson, Pablo Martín, Alejandro Mira and Noelia Zarza for assistance in greenhouse and laboratory; and Consejería de Medio Ambiente, Junta de Andalucía, for permission to work in Sierra de Cazorla. Partial financial support for this work was provided by grants CGL2013-43352-P and CGL2016-76605-P from the Spanish Ministerio de Economía y Competitividad.
Author Contributions
C.M.H., M.M. and C.A. conceived and designed the experiments; M.M. and C.A. performed the experiments; R.P. and P.B. contributed to sample analyses; C.A. analyzed the data and drafted the paper; all authors contributed to refine writing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Finnegan, E.J. DNA methylation: A dynamic regulator of genome organization and gene expression in plants. In Plant Developmental Biology—Biotechnological Perspectives: Volume 2; Pua, E.-C., Davey, M.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 2, pp. 295–323. [Google Scholar]
- Feil, R.; Fraga, M. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef]
- Seymour, D.K.; Koenig, D.; Hagmann, J.; Becker, C.; Weigel, D. Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet. 2014, 10, e1004785. [Google Scholar] [CrossRef]
- Burggren, W. Epigenetic inheritance and its role in evolutionary biology: Re-evaluation and new perspectives. Biology 2016, 5. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009, 12, 133–139. [Google Scholar] [CrossRef]
- Peng, H.; Zhang, J. Plant genomic DNA methylation in response to stresses: Potential applications and challenges in plant breeding. Prog. Nat. Sci. 2009, 19, 1037–1045. [Google Scholar] [CrossRef]
- Mirouze, M.; Paszkowski, J. Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 2011, 14, 267–274. [Google Scholar] [CrossRef]
- Herman, J.J.; Sultan, S.E. Adaptive transgenerational plasticity in plants: Case studies, mechanisms, and implications for natural populations. Front. Plant Sci. 2011, 2. [Google Scholar] [CrossRef]
- Walter, J.; Jentsch, A.; Beierkuhnlein, C.; Kreyling, J. Ecological stress memory and cross stress tolerance in plants in the face of climate extremes. Environ. Exp. Bot. 2013, 94, 3–8. [Google Scholar] [CrossRef]
- Pikaard, C.S.; Mittelsten Scheid, O. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 2014, 6, a019315. [Google Scholar] [CrossRef]
- Alonso, C.; Pérez, R.; Bazaga, P.; Medrano, M.; Herrera, C.M. MSAP markers and global cytosine methylation in plants: A literature survey and comparative analysis for a wild growing species. Mol. Ecol. Resour. 2016, 16, 80–90. [Google Scholar] [CrossRef]
- Pecinka, A.; Mittelsten Scheid, O. Stress-induced chromatin changes: A critical view on their heritability. Plant Cell Physiol. 2012, 53, 801–808. [Google Scholar] [CrossRef]
- Lopez, M.; Halby, L.; Arimondo, P.B. DNA methyltransferase inhibitors: Development and applications. In DNA Methyltransferases—Role and Function; Jeltsch, A., Jurkowska, R.Z., Eds.; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- Sano, H.; Kamada, I.; Youssefian, S.; Katsumi, M.; Wabiko, H. A single treatment of rice seedlings with 5-azacytidine induces heritable dwarfism and undermethylation of genomic DNA. Mol. Gen. Genet. 1990, 220, 441–447. [Google Scholar] [CrossRef]
- Janousek, B.; Siroky, J.; Vyskot, B. Epigenetic control of sexual phenotype in a dioecious plant, Melandrium album. Mol. Gen. Genet. 1996, 250, 483–490. [Google Scholar] [CrossRef]
- Fieldes, M.A.; Amyot, L.M. Evaluating the potential of using 5-azacytidine as an epimutagen. Can. J. Bot. 1999, 77, 1617–1622. [Google Scholar] [CrossRef]
- Kondo, H.; Shiraya, T.; Wada, K.C.; Takeno, K. Induction of flowering by DNA demethylation in Perilla frutescens and Silene armeria: Heritability of 5-azacytidine-induced effects and alteration of the DNA methylation state by photoperiodic conditions. Plant Sci. 2010, 178, 321–326. [Google Scholar] [CrossRef]
- Xu, J.; Tanino, K.K.; Robinson, S.J. Stable epigenetic variants selected from an induced hypomethylated Fragaria vesca population. Front. Plant Sci. 2016, 7, 1768. [Google Scholar] [CrossRef]
- Baubec, T.; Pecinka, A.; Rozhon, W.; Mittelsten Scheid, O. Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. Plant J. 2009, 57, 542–554. [Google Scholar] [CrossRef]
- Griffin, P.T.; Niederhuth, C.E.; Schmitz, R.J. A comparative analysis of 5-azacytidine- and zebularine-induced DNA demethylation. Genes Genomes Genet. 2016, 6, 2773–2780. [Google Scholar] [CrossRef]
- Bossdorf, O.; Arcuri, D.; Richards, C.L.; Pigliucci, M. Experimental alteration of DNA methylation affects the phenotypic plasticity of ecologically relevant traits in Arabidopsis thaliana. Evol. Ecol. 2010, 24, 541–553. [Google Scholar] [CrossRef]
- Verhoeven, K.J.F.; van Gurp, T.P. Transgenerational effects of stress exposure on offspring phenotypes in apomictic dandelion. PLoS ONE 2012, 7, e38605. [Google Scholar] [CrossRef]
- Herman, J.J.; Sultan, S.E. DNA methylation mediates genetic variation for adaptive transgenerational plasticity. Proc. R. Soc. B 2016, 283, 20160988. [Google Scholar] [CrossRef]
- Rendina González, A.P.; Chrtek, J.; Dobrev, P.I.; Dumalasová, V.; Fehrer, J.; Mráz, P.; Latzel, V. Stress-induced memory alters growth of clonal off spring of white clover (Trifolium repens). Am. J. Bot. 2016, 103, 1567–1574. [Google Scholar] [CrossRef]
- Cao, D.; Gao, X.; Liu, J.; Kimatu, J.N.; Geng, S.; Wang, X.; Zhao, J.; Shi, D. Methylation sensitive amplified polymorphism (MSAP) reveals that alkali stress triggers more DNA hypomethylation levels in cotton (Gossypium hirsutum L.) roots than salt stress. Afr. J. Biotechnol. 2011, 10, 18971–18980. [Google Scholar]
- Cao, D.; Gao, X.; Liu, J.; Wang, X.; Geng, S.; Yang, C.; Liu, B.; Shi, D. Root-specific DNA methylation in Chloris virgata, a natural alkaline-resistant halophyte, in response to salt and alkaline stresses. Plant Mol. Biol. Rep. 2012, 30, 1102–1109. [Google Scholar] [CrossRef]
- Greco, M.; Chiappetta, A.; Bruno, L.; Bitonti, M.B. Effects of light deficiency on genome methylation in Posidonia oceanica. Mar. Ecol. Prog. Ser. 2013, 473, 103–114. [Google Scholar] [CrossRef]
- Karan, R.; DeLeon, T.; Biradar, H.; Subudhi, P.K. Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PLoS ONE 2012, 7, e40203. [Google Scholar] [CrossRef]
- Fiz-Palacios, O.; Vargas, P.; Vila, R.; Papadopulos, A.S.T.; Aldasoro, J.J. The uneven phylogeny and biogeography of Erodium (Geraniaceae): Radiations in the Mediterranean and recent recurrent intercontinental colonization. Ann. Bot. 2010, 106, 871–884. [Google Scholar] [CrossRef]
- Francis, A.; Darbyshire, S.J.; Légère, A.; Simard, M.-J. The biology of Canadian weeds. 151. Erodium cicutarium (L.) L’Hér. Ex Aiton. Can. J. Plant Sci. 2012, 92, 1359–1380. [Google Scholar] [CrossRef]
- Blackshaw, R.E. Soil, temperature, soil moisture, and seed burial depth effects on redstem filaree (Erodium cicutarium) emergence. Weed Sci. 1992, 40, 204–207. [Google Scholar]
- Cox, J.A.; Conran, J.G. The effect of water stress on the life cycles of Erodium crinitum Carolin and Erodium cicutarium (L.) L’Herit. ex Aiton (Geraniaceae). Aust. J. Ecol. 1996, 21, 235–240. [Google Scholar] [CrossRef]
- Heger, T.; Jacobs, B.S.; Latimerb, A.M.; Johannes, K.; Kevin, J.R. Does experience with competition matter? Effects of source competitive environment on mean and plastic trait expression in Erodium cicutarium. Perspect. Plant Ecol. Evol. Syst. 2014, 16, 236–246. [Google Scholar] [CrossRef]
- Kimball, S.; Gremer, J.R.; Barron-Gafford, G.A.; Angert, A.L.; Huxman, T.E.; Venable, D.L. High water-use efficiency and growth contribute to success of non-native Erodium cicutarium in a Sonoran desert winter annual community. Conserv. Physiol. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Paparella, S.; Araújo, S.S.; Rossi, G.; Wijayasinghe, M.; Carbonera, D.; Balestrazzi, A. Seed priming: State of the art and new perspectives. Plant Cell Rep. 2015, 34, 1281–1293. [Google Scholar] [CrossRef] [PubMed]
- Leck, M.A.; Parker, V.T.; Simpson, R.L. Seedling Ecology and Evolution; Cambridge University Press: Cambridge, UK, 2008. [Google Scholar]
- Grant-Downton, R.T.; Dickinson, H.G. Epigenetics and its implications for plant biology 2. The ’Epigenetic Epiphany’: Epigenetics, evolution and beyond. Ann. Bot. 2006, 97, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Jablonka, E.; Raz, G. Transgenerational epigenetic inheritance: Prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 2009, 84, 131–176. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, S.; Baumberger, R.; Grossniklaus, U. Epigenetic variation, inheritance, and selection in plant populations. Cold Spring Harb. Symp. Quant. Biol. 2013, 77, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Richards, C.L.; Verhoeven, K.J.F.; Bossdorf, O. Evolutionary significance of epigenetic variation. In Plant Genome Diversity; Wendel, J., Greilhuber, J., Dolezel, J., Leitch, I., Eds.; Springer: Vienna, Austria, 2012; Volume 1, pp. 257–274. [Google Scholar]
- Fieldes, M.A. Heritable effects of 5-azacytidine treatments on the growth and development of flax (Linum usitatissimum) genotrophs and genotypes. Genome 1994, 37, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tanino, K.K.; Horner, K.N.; Robinson, S.J. Quantitative trait variation is revealed in a novel hypomethylated population of woodland strawberry (Fragaria vesca). BMC Plant Biol. 2016, 16, 240–257. [Google Scholar] [CrossRef] [PubMed]
- Amoah, S.; Kurup, S.; Rodriguez Lopez, C.M.; Welham, S.J.; Powers, S.J.; Hopkins, C.J.; Wilkinson, M.J.; King, G.J. A hypomethylated population of Brassica rapa for forward and reverse epi-genetics. BMC Plant Biol. 2012, 12, 193–210. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Fei, Z.; Chen, Y.-R.; Zheng, Y.; Huang, M.; Vrebalov, J.; McQuinn, R.; Gapper, N.; Liu, B.; Xiang, J.; et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013, 31, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Pérez, J. Breeding system, flower visitors and seedling survival of two endangered species of Helianthemum (Cistaceae). Ann. Bot. 2005, 95, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Messeguer, R.; Ganal, M.; Steffens, J.; Tanksley, S. Characterization of the level, target sites and inheritance of cytosine methylation in tomato nuclear DNA. Plant Mol. Biol. 1991, 16, 753–770. [Google Scholar] [CrossRef] [PubMed]
- Demeulemeester, M.A.C.; Van Stallen, N.; De Proft, M.P. Degree of DNA methylation in chicory (Cichorium intybus l.): Influence of plant age and vernalization. Plant Sci. 1999, 142, 101–108. [Google Scholar] [CrossRef]
- Ruiz-García, L.; Cervera, M.T.; Martínez-Zapater, J.M. DNA methylation increases throughout Arabidopsis development. Planta 2005, 222, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.C.L.; De Decker, M.M.; Fieldes, M.A. A comparative analysis of developmental profiles for DNA methylation in 5-azacytidine-induced early-flowering flax lines and their control. Plant Sci. 2008, 175, 217–225. [Google Scholar] [CrossRef]
- Osabe, K.; Clement, J.D.; Bedon, F.; Pettolino, F.A.; Ziolkowski, L.; Llewellyn, D.J.; Finnegan, E.J.; Wilson, I.W. Genetic and DNA methylation changes in cotton (Gossypium) genotypes and tissues. PLoS ONE 2014, 9, e86049. [Google Scholar] [CrossRef] [PubMed]
- Eichten, S.R.; Vaughn, M.W.; Hermanson, P.J.; Springer, N.M. Variation in DNA methylation patterns is more common among maize inbreds than among tissues. Plant Genome 2013, 6. [Google Scholar] [CrossRef]
- Roessler, K.; Takuno, S.; Gaut, B.S. CG methylation covaries with differential gene expression between leaf and floral bud tissues of Brachypodium distachyon. PLoS ONE 2016, 11, e0150002. [Google Scholar] [CrossRef] [PubMed]
- Chwialkowska, K.; Nowakowska, U.; Mroziewicz, A.; Szarejko, I.; Kwasniewski, M. Water-deficiency conditions differently modulate the methylome of roots and leaves in barley (Hordeum vulgare L.). J. Exp. Bot. 2016, 67, 1109–1121. [Google Scholar] [CrossRef] [PubMed]
- Fieldes, M.A.; Schaeffer, S.M.; Krech, M.J.; Brown, J.C.L. DNA hypomethylation in 5-azacytidine-induced early-flowering lines of flax. Theor. Appl. Genet. 2005, 111, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Sano, H.; Kamada, I.; Youssefian, S.; Wabiko, H. Correlation between DNA undermethylation and dwarfism in maize. Biochim. Biophys. Acta 1989, 1009, 35–38. [Google Scholar] [CrossRef]
- Latzel, V. Pitfalls in ecological research—Transgenerational effects. Folia Geobot. 2015, 50, 75–85. [Google Scholar] [CrossRef]
- Lopez-Torres, A.; Yanez-Barrientos, E.; Wrobel, K.; Wrobel, K. Selective derivatization of cytosine and methylcytosine moieties with 2-bromoacetophenone for submicrogram DNA methylation analysis by reversed phase HPLC with spectrofluorimetric detection. Anal. Chem. 2011, 83, 7999–8005. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing, 3.3.3; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Crawley, M.J. The R Book; John Wiley & Sons Ltd.: Chichester, UK, 2007. [Google Scholar]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; R Core Team. Nlme: Linear and nonlinear mixed effects models. R Found. Stat. Comput. 2017, 3, 107–156. [Google Scholar]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 2nd ed.; Sage: Thousand Oaks, CA, USA, 2011. [Google Scholar]
- Lenth, R.V. Least-squares means: The R package lsmeans. J. Stat. Softw. 2016, 69, 1–33. [Google Scholar]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).