DNA Methylation in Rice and Relevance for Breeding
Abstract
:1. Introduction
2. Epigenetic Regulations Are Involved in Agricultural/Adaptive Traits
3. Main Regulators of DNA Methylation in Rice
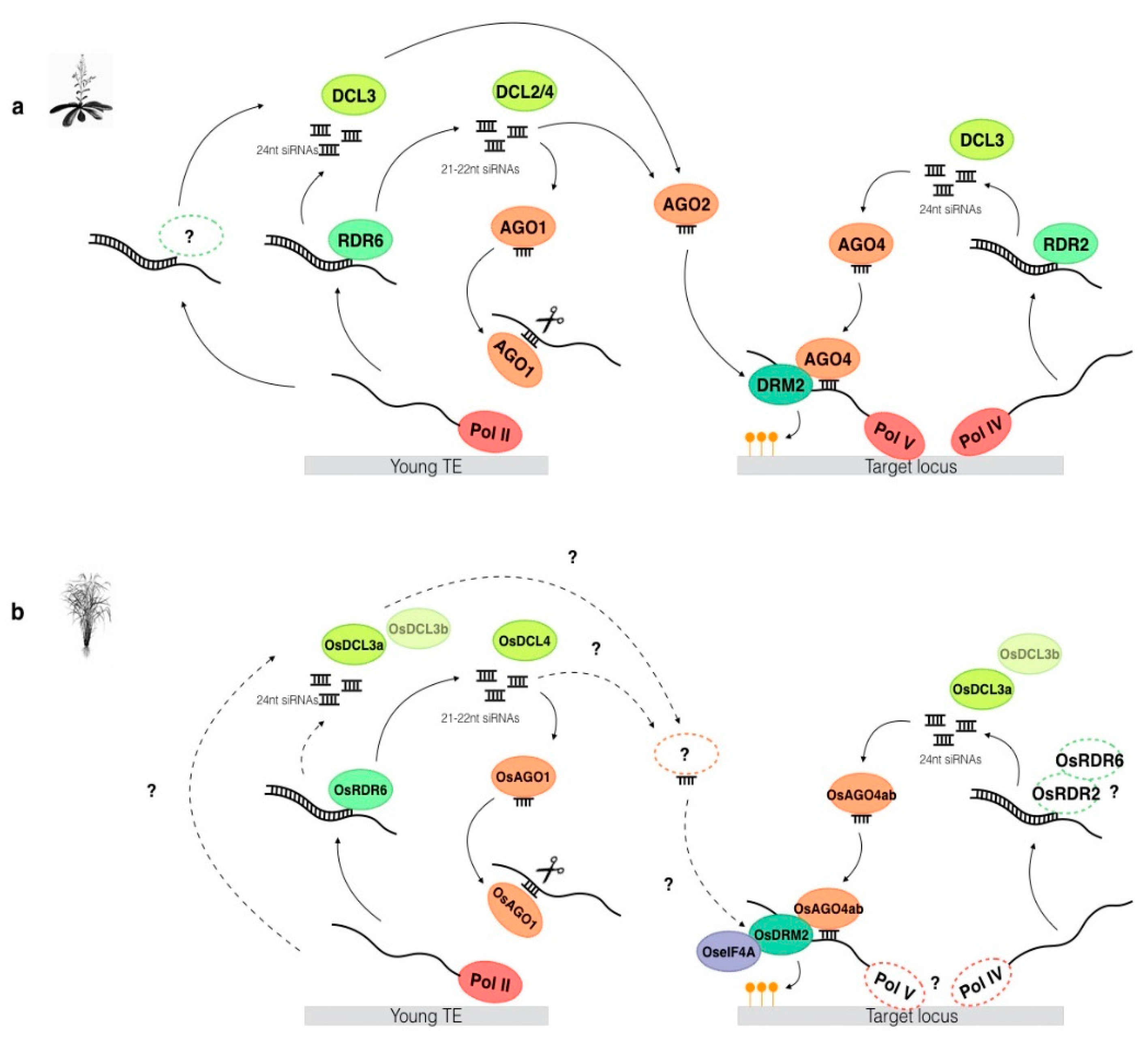
4. Establishment of De Novo DNA Methylation
5. Conclusions and Perspectives
Acknowledgments
Conflicts of Interest
References
- Du, J.; Johnson, L.M.; Jacobsen, S.E.; Patel, D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-Y.; Feng, S.; Joo, J.W.J.; Jacobsen, S.E.; Pellegrini, M. A comparative analysis of DNA methylation across human embryonic stem cell lines. Genome Biol. 2011, 12, R62. [Google Scholar] [CrossRef] [PubMed]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Springer, N.M. Epigenetics and crop improvement. Trends Genet. 2013, 29, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhou, D.-X. Rice epigenomics and epigenetics: Challenges and opportunities. Curr. Opin. Plant Biol. 2013, 16, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Song, X.; Wei, L.; Liu, C. Epigenetic regulation and epigenomic landscape in rice. Nat. Sci. Rev. 2016, 3, 309–327. [Google Scholar] [CrossRef]
- Gehring, M.; Bubb, K.L.; Henikoff, S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 2009, 324, 1447–1451. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Kim, M.Y.; Silva, P.; Rodrigues, J.A.; Dotson, B.; Brooks, M.D.; Zilberman, D. Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. USA 2010, 107, 18729–18734. [Google Scholar] [CrossRef] [PubMed]
- Saze, H.; Mittelsten Scheid, O.; Paszkowski, J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 2003, 34, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Gazzani, S.; Gendall, A.R.; Lister, C.; Dean, C. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 2003, 132, 1107–1114. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Dong, A.; Shen, W.-H. Epigenetic regulation of rice flowering and reproduction. Front. Plant Sci. 2014, 5, 803. [Google Scholar] [CrossRef] [PubMed]
- Xing, M.-Q.; Zhang, Y.-J.; Zhou, S.-R.; Hu, W.-Y.; Wu, X.-T.; Ye, Y.-J.; Wu, X.-X.; Xiao, Y.-P.; Li, X.; Xue, H.-W. Global analysis reveals the crucial roles of DNA methylation during rice seed development. Plant Physiol. 2015, 168, 1417–1432. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Colot, V. Epialleles in plant evolution. Genome Biol. 2012, 13, 249. [Google Scholar] [CrossRef] [PubMed]
- Quadrana, L.; Colot, V. Plant transgenerational epigenetics. Annu. Rev. Genet. 2016, 50, 467–491. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Huang, S.-S.C.; Jupe, F.; Sasaki, E.; Schmitz, R.J.; Urich, M.A.; Castanon, R.; Nery, J.R.; Barragan, C.; He, Y.; et al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 2016, 166, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Mirouze, M.; Paszkowski, J. Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 2011, 14, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Wang, C.; Shou, H.; Schultz, M.D.; Chiarenza, S. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 2015, 4, e09343. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Narayana Chevala, V.; Shankar, R.; Jain, M. Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci. Rep. 2015, 5, 14922. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Qin, Q.; Sun, F.; Wang, Y.; Xu, D.; Li, Z. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Front. Plant Sci. 2016, 39, 61–69. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, L.; Xia, H.; Wei, H.; Lou, Q.; Li, M.; Li, T.; Luo, L. Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant's adaptation to drought condition. Sci. Rep. 2017, 7, 39843. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.J.; Liu, X.S.; Tao, H.; Tan, S.K.; Chu, S.S.; Oono, Y.; Zhang, X.D.; Chen, J.; Yang, Z.M. Variation of DNA methylation patterns associated with gene expression in rice (Oryza sativa) exposed to cadmium. Plant Cell Environ. 2016, 39, 2629–2649. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.C.; Feng, S.J.; Zhang, J.J.; Luo, F.; Zhang, S. Genome-wide identification of DNA methylation provides insights into the association of gene expression in rice exposed to pesticide atrazine. Sci. Rep. 2016, 6, 18985. [Google Scholar] [CrossRef] [PubMed]
- Quadrana, L.; Bortolini Silveira, A.; Mayhew, G.F.; LeBlanc, C.; Martienssen, R.A.; Jeddeloh, J.A.; Colot, V. The Arabidopsis thaliana mobilome and its impact at the species level. eLife 2016, 5, e15176. [Google Scholar] [CrossRef] [PubMed]
- Lisch, D. How important are transposons for plant evolution? Nat. Rev. Genet. 2013, 14, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Troadec, C.; Boualem, A.; Rajab, M.; Fernandez, R.; Morin, H.; Pitrat, M.; Dogimont, C.; Bendahmane, A. A transposon-induced epigenetic change leads to sex determination in melon. Nature 2009, 461, 1135–1138. [Google Scholar] [CrossRef] [PubMed]
- Ong-Abdullah, M.; Ordway, J.M.; Jiang, N.; Ooi, S.-E.; Kok, S.-Y.; Sarpan, N.; Azimi, N.; Hashim, A.T.; Ishak, Z.; Rosli, S.K.; et al. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 2015, 525, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 2016, 18, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Rey, O.; Danchin, E.; Mirouze, M.; Loot, C.; Blanchet, S. Adaptation to global change: A transposable element-epigenetics perspective. Trends Ecol. Evol. 2016, 31, 514–526. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Cao, X. Transposon-mediated epigenetic regulation contributes to phenotypic diversity and environmental adaptation in rice. Curr. Opin. Plant Biol. 2017, 36, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Reinders, J.; Wulff, B.B.H.; Mirouze, M.; Marí-Ordóñez, A.; Dapp, M.; Rozhon, W.; Bucher, E.; Theiler, G.; Paszkowski, J. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009, 23, 939–950. [Google Scholar] [CrossRef] [PubMed]
- Johannes, F.; Porcher, E.; Teixeira, F.K.; Saliba-Colombani, V.; Simon, M.; Agier, N.; Bulski, A.; Albuisson, J.; Heredia, F.; Audigier, P.; et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009, 5, e1000530. [Google Scholar] [CrossRef] [PubMed]
- Stroud, H.; Ding, B.; Simon, S.A.; Feng, S.; Bellizzi, M.; Pellegrini, M.; Wang, G.-L.; Meyers, B.C.; Jacobsen, S.E. Plants regenerated from tissue culture contain stable epigenome changes in rice. eLife 2013, 2, e00354. [Google Scholar] [CrossRef] [PubMed]
- Finnegan, E.J.; Dennis, E.S. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 1993, 21, 2383–2388. [Google Scholar] [CrossRef] [PubMed]
- Pavlopoulou, A.; Kossida, S. Plant cytosine-5 DNA methyltransferases: Structure, function, and molecular evolution. Genomics 2007, 90, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Li, N.; Xu, C.; Zhong, S.; Lin, X.; Yang, J.; Zhou, T.; Yuliang, A.; Wu, Y.; Chen, Y.-R.; et al. Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc. Natl. Acad. Sci. USA 2014, 111, 10642–10647. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Moritoh, S.; Johzuka-Hisatomi, Y.; Ono, A.; Terada, R.; Nakamura, I.; Iida, S. Alternative splicing of the rice OsMET1 genes encoding maintenance DNA methyltransferase. Plant Physiol. 2008, 165, 1774–1782. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Kim, M.Y.; Hsieh, P.-H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Lindroth, A.M.; Cao, X.; Jackson, J.P.; Zilberman, D.; McCallum, C.M.; Henikoff, S.; Jacobsen, S.E. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 2001, 292, 2077–2080. [Google Scholar] [CrossRef] [PubMed]
- Stroud, H.; Do, T.; Du, J.; Zhong, X.; Feng, S.; Johnson, L.; Patel, D.J.; Jacobsen, S.E. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 2014, 21, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Ashapkin, V.V.; Kutueva, L.I.; Vanyushin, B.F. Plant DNA methyltransferase genes: Multiplicity, expression, methylation patterns. Biochemistry 2016, 81, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Tarutani, Y.; Miyao, A.; Ito, T.; Yamazaki, M.; Sakai, H.; Fukai, E.; Hirochika, H. Loss of function mutations in the rice chromomethylase OsCMT3a cause a burst of transposition. Plant J. 2015, 83, 1069–1081. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Singh, R.M.; Malik, G. Rice cytosine DNA methyltransferases-gene expression profiling during reproductive development and abiotic stress. FEBS J. 2009, 276, 6301–6311. [Google Scholar] [CrossRef] [PubMed]
- Moritoh, S.; Eun, C.-H.; Ono, A.; Asao, H.; Okano, Y.; Yamaguchi, K.; Shimatani, Z.; Koizumi, A.; Terada, R. Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation. Plant J. 2012, 71, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Tan, F.; Zhou, C.; Zhou, Q.; Zhou, S.; Yang, W.; Zhao, Y.; Li, G.; Zhou, D.-X. Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiol. 2016, 171, 2041–2054. [Google Scholar] [CrossRef] [PubMed]
- Higo, H.; Tahir, M.; Takashima, K.; Miura, A.; Watanabe, K.; Tagiri, A.; Ugaki, M.; Ishikawa, R.; Eiguchi, M.; Kurata, N.; et al. DDM1 (decrease in DNA methylation) genes in rice (Oryza sativa). Mol. Genet. Genom. 2012, 287, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Dong, M.; Li, N.; Zhao, Y.; Liu, B. Functional characterization of a rice de novo DNA methyltransferase, OsDRM2, expressed in Escherichia coli and yeast. Biochem. Biophys. Res. Commun. 2013, 432, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Dangwal, M.; Malik, G.; Kapoor, S.; Kapoor, M. De novo methyltransferase, OsDRM2, interacts with the ATP-dependent RNA helicase, OseIF4A, in rice. J. Mol. Biol. 2013, 425, 2853–2866. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Arora, R.; Lama, T.; Nijhawan, A.; Khurana, J.P.; Tyagi, A.K.; Kapoor, S. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genom. 2008, 9, 451. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Gu, L.; Song, X.; Cui, X.; Lu, Z.; Zhou, M.; Wang, L.; Hu, F.; Zhai, J.; Meyers, B.C.; et al. Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 3877–3882. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhou, H.; Zhang, Q.; Zhang, J.; Ni, F.; Liu, C.; Qi, Y. DNA methylation mediated by a microRNA pathway. Mol. Cell 2010, 38, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wang, D.; Ma, L.; Chen, Z.; Li, P.; Cui, X.; Liu, C.; Cao, S.; Chu, C.; Tao, Y.; et al. Rice RNA-dependent RNA polymerase 6 acts in small RNA biogenesis and spikelet development. Plant J. 2012, 71, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Chen, Z.; Song, X.; Liu, C.; Cui, X.; Zhao, X.; Fang, J.; Xu, W.; Zhang, H.; Wang, X.; et al. Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell 2007, 19, 2705–2718. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhang, D.; Wang, Z.; Xun, H.; Ma, J.; Wang, H.; Huang, W.; Liu, Y.; Lin, X.; Li, N.; et al. Mutation of the RDR1 gene caused genome-wide changes in gene expression, regional variation in small RNA clusters and localized alteration in DNA methylation in rice. BMC Plant Biol. 2014, 14, 177. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Qian, D.; Sun, R.; Jiang, L.; Wang, Y.; Wei, C.; Zhang, Z.; Li, Y. OsRDR6 plays role in host defense against double-stranded RNA virus, Rice Dwarf Phytoreovirus. Sci. Rep. 2015, 5, 11324. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Yoshikawa, T.; Nosaka, M.; Sakakibara, H.; Sato, Y.; Nagato, Y.; Itoh, J.-I. WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining microRNA and trans-acting small interfering RNA accumulation in rice. Plant Physiol. 2010, 154, 1335–1346. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Yamaguchi, K.; Fukada-Tanaka, S.; Terada, R.; Mitsui, T.; Iida, S. A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J. 2012, 71, 564–574. [Google Scholar] [CrossRef] [PubMed]
- La, H.; Ding, B.; Mishra, G.P.; Zhou, B.; Yang, H.; Bellizzi, M.D.R.; Chen, S.; Meyers, B.C.; Peng, Z.; Zhu, J.-K.; Wang, G.-L. A 5-methylcytosine DNA glycosylase/lyase demethylates the retrotransposon Tos17 and promotes its transposition in rice. Proc. Natl. Acad. Sci. USA 2011, 108, 15498–15503. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Jacobsen, S.E. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002, 12, 1138–1144. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Jacobsen, S.E. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 2002, 99, 16491–16498. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Springer, N.M.; Muszynski, M.G.; Phillips, R.L.; Kaeppler, S.; Jacobsen, S.E. Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc. Natl. Acad. Sci. USA 2000, 97, 4979–4984. [Google Scholar] [CrossRef] [PubMed]
- Jeddeloh, J.A.; Stokes, T.L.; Richards, E.J. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 1999, 22, 94–97. [Google Scholar] [PubMed]
- Brzeski, J.; Jerzmanowski, A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J. Biol. Chem. 2003, 278, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Lippman, Z.; Gendrel, A.-V.; Black, M.; Vaughn, M.W.; Dedhia, N.; McCombie, W.R.; Lavine, K.; Mittal, V.; May, B.; Kasschau, K.D.; et al. Role of transposable elements in heterochromatin and epigenetic control. Nature 2004, 430, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Agius, F.; Kapoor, A.; Zhu, J.-K. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl. Acad. Sci. USA 2006, 103, 11796–11801. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Gehring, M.; Johnson, L.; Hannon, M.; Harada, J.J.; Goldberg, R.B.; Jacobsen, S.E.; Fischer, R.L. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell 2002, 110, 33–42. [Google Scholar] [CrossRef]
- Penterman, J.; Zilberman, D.; Huh, J.H.; Ballinger, T.; Henikoff, S.; Fischer, R.L. DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. USA 2007, 104, 6752–6757. [Google Scholar] [CrossRef] [PubMed]
- Mirouze, M.; Vitte, C. Transposable elements, a treasure trove to decipher epigenetic variation: Insights from Arabidopsis and crop epigenomes. J. Exp. Bot. 2014, 65, 2801–2812. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Eichten, S.R.; Hermanson, P.J.; Zaunbrecher, V.M.; Song, J.; Wendt, J.; Rosenbaum, H.; Madzima, T.F.; Sloan, A.E.; Huang, J.; et al. Genetic perturbation of the maize methylome. Plant Cell 2014, 26, 4602–4616. [Google Scholar] [CrossRef] [PubMed]
- Bucher, E.; Reinders, J.; Mirouze, M. Epigenetic control of transposon transcription and mobility in Arabidopsis. Curr. Opin. Plant Biol. 2012, 15, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Cuerda-Gil, D.; Slotkin, R.K. Non-canonical RNA-directed DNA methylation. Nat. Plants 2016, 2, 16163. [Google Scholar] [CrossRef] [PubMed]
- Lahmy, S.; Pontier, D.; Bies-Etheve, N.; Laudié, M.; Feng, S.; Jobet, E.; Hale, C.J.; Cooke, R.; Hakimi, M.-A.; Angelov, D.; et al. Evidence for ARGONAUTE4–DNA interactions in RNA-directed DNA methylation in plants. Genes Dev. 2016, 30, 2565–2570. [Google Scholar] [CrossRef] [PubMed]
- Panda, K.; Ji, L.; Neumann, D.A.; Daron, J.; Schmitz, R.J.; Slotkin, R.K. Full-length autonomous transposable elements are preferentially targeted by expression-dependent forms of RNA-directed DNA methylation. Genome Biol. 2016, 17, 170. [Google Scholar] [CrossRef] [PubMed]
- Arikit, S.; Zhai, J.; Meyers, B.C. Biogenesis and function of rice small RNAs from non-coding RNA precursors. Curr. Opin. Plant Biol. 2013, 16, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Ream, T.S.; Haag, J.R.; Wierzbicki, A.T.; Nicora, C.D.; Norbeck, A.D.; Zhu, J.-K.; Hagen, G.; Guilfoyle, T.J.; Pasa-Tolić, L.; Pikaard, C.S. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol. Cell 2009, 33, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Tucker, S.L.; Reece, J.; Ream, T.S.; Pikaard, C.S. Evolutionary history of plant multisubunit RNA polymerases IV and V: Subunit origins via genome-wide and segmental gene duplications, retrotransposition, and lineage-specific subfunctionalization. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Kendall, T.; Forsythe, E.S.; Dorantes-Acosta, A.; Li, S.; Caballero-Pérez, J.; Chen, X.; Arteaga-Vázquez, M.; Beilstein, M.A.; Mosher, R.A. Ancient origin and recent innovations of RNA polymerase IV and V. Mol. Biol. Evol. 2015, 32, 1788–1799. [Google Scholar] [CrossRef] [PubMed]
- Bousios, A.; Gaut, B.S. Mechanistic and evolutionary questions about epigenetic conflicts between transposable elements and their plant hosts. Curr. Opin. Plant Biol. 2016, 30, 123–133. [Google Scholar] [CrossRef] [PubMed]
| Proteins | Locus ID | Mutation | Expression | Description/Phenotype | Functions | References | |
|---|---|---|---|---|---|---|---|
| Maintenance of DNA methylation | OsMET1-2 (DNA METHYLTRANSFERASE 1) | LOC_Os 07g08500 | T-DNA insertion (Tos17) | KO | All germinated seedlings undergo quick necrotic death | Maintain DNA methylation at CG sites during DNA replication. Two copies MET1-1 and MET1-2 | [37,44] |
| OsDRM1a | LOC_Os 11g01810 | / | / | Downregulated by jasmonic acid | Not expressed, lack of methyltransferase motifs | [45] | |
| OsDRM1b | LOC_Os 12g01800 | / | / | Downregulated by jasmonic acid | |||
| OsCMT3 (CHROMOMETHYLTRANSFERASE) | LOC_Os 10g01570 | T-DNA insertion (Tos17) | KO | No difference in the vegetative phase. Early reproductive stage, 15% shorter stature and decreased fertility | Maintain DNA methylation at CHG sites during DNA replication | [43,44] | |
| Chromatin remodeler | OsDDM1a (DECREASE in DNA METHYLATION) | LOC_Os 09g27060 | RNAi mutants | KD | 93% identity between both DDM1 homologs; dwarf phenotype; hypomethylation in later generations of selfed progenies | Remodeling histones ATPases. Maintenance of cytosine methylation; Required for maintenance of TE silencing | [46,47] |
| OsDDM1b (DECREASED in DNA METHYLATION) | LOC_Os 03g51230 | RNAi mutants | KD | / | Maintenance of cytosine methylation | [46,47] | |
| RdDM | OsDRM2 (DOMAINS REARRANGED METHYLTRANSFERASE) | LOC_Os 03g02010 | Gene targeting through homologous recombinaison | KO | Reduction of vegetative growth and semi-dwarf phenotype. Reduction in the de novo methylation at transposons and 5S repeat sequences | De novo DNA methylation at CHH sites directed by siRNAs. Major DRM1/2-type methyltransferase gene in rice | [44,45,48,49] |
| OsDCL3a (DICER LIKE PROTEIN 3) | LOC_Os 01g68120 | RNAi mutant | KD | Pleiotropic phenotypes affecting agricultural traits: plant height, angle of flag leaf, smaller panicles. Similar phenotypes as RNAi mutants of AGO4ab-1 and RDR2-2 | Biogenesis of 24-nt long miRNAs (lmiRNAs) which can direct DNA methylation (cis and trans); 24 nt siRNA biogenesis | [50,51,52] | |
| OsDCL3b (DICER LIKE PROTEIN 3) | LOC_Os 10g34430 | RNAi mutant | KD | / | Panicle and early seed-specific and require for 24 nt phased small RNAs. DCL3a is expressed at a much higher level than DCL3b | [50,53] | |
| OsDCL4 (DICER LIKE PROTEIN 4) | LOC_Os 04g43050 | / | KO | Severe spikelet defects including thread-like lemma and male sterility | Biogenesis of 21 nt siRNA in panicles and seedlings | [53,54] | |
| OsRDR1 (RNA DEPENDENT RNA POLYMERASE 1) | LOC_Os 02g50330 | T-DNA insertion (Tos17) | KO | Ephemeral phenotypic fluctuations occurred only under some abiotic stress conditions | Role in the production and amplification of exogenous, virus-derived siRNAs (vsiRNAs) in infected plants and in some abiotic stress responses. Role in maintaining the intrinsic locus-specific CHH methylation patterns | [55] | |
| OsRDR2 (RNA DEPENDENT RNA POLYMERASE 2) | LOC_Os 04g39160 | RNAi mutant | KD | Similar phenotypes as RNAi mutants of AGO4ab-1 and OsDCL3a. | Role not studied yet but could be similar to AtRDR2 (according to its expression pattern) | [50] | |
| OsRDR6 (RNA DEPENDENT RNA POLYMERASE 6) | LOC_Os 01g34350 | SNP (G -> T) | Temperature dependent | Spikelet defects | Biogenesis of 21 nt and 24 nt siRNAs (different from Arabidopsis) and resistance against virus | [53,56] | |
| AGO4a/AGO4b | LOC_Os 01g16870/LOC_Os 04g06770 | RNAi mutants | KD | Similar phenotypes as RNAi mutants of OsDCL3a and RDR2-2 | High similarity with Arabidopsis AGO4 | [50] | |
| OsAGO1s (4 OsAGO1 homologs OsAGO1a, OsAGO1b, OsAGO1c, OsAGO1d) | LOC_Os 02g45070/LOC_Os 04g47870/LOC_Os 02g58490/LOC_Os 06g51310 | RNAi mutants | KD | Various developmental defects | miRNA mediated gene regulation | [50] | |
| WAF1 (WAVY LEAF1) | LOC_Os 07g06970 | NMU mutagenesis | KO | Seedling lethality due to defects of SAM maintenance or pleiotropic phenotypes in leaf morphology and floral development. Phenotypes similar to sho1 and sho2 mutants deficient in DCL4 and AGO7, respectively | Methylates 3’ terminal nucleotide of siRNAs; HEN1 (HUA ENHANCER 1) homolog | [57] | |
| 5-meC DNA glycosylase/lyases | OsROS1a | LOC_Os 01g11900 | knock-in targeting | KO | Severe underdeveloped endosperm phenotype | There are 4 ROS1 orthologs (ROS1a-d). ROS1a is the most expressed gene compared to ROS1b-d. ROS1a and Arabidopsis DME gene could have analogous functions in the endosperm | [58] |
| DNG701 (OsROS1c) | LOC_Os 05g37350 | T-DNA insertion, RNAi | KO; KD; OE | The progeny of ros1c mutant present two seed phenotypes, normal seeds and wrinkled seeds | ROS1a and ROS1c could play different roles in seed development. Could be involved in the control of transposition. | [58,59] | |
| DML3a (DEMETER LIKE 3) and DML3b | LOC_Os 04g28860/LOC_Os 02g29380 | / | / | / | / | [10,58] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lanciano, S.; Mirouze, M. DNA Methylation in Rice and Relevance for Breeding. Epigenomes 2017, 1, 10. https://doi.org/10.3390/epigenomes1020010
Lanciano S, Mirouze M. DNA Methylation in Rice and Relevance for Breeding. Epigenomes. 2017; 1(2):10. https://doi.org/10.3390/epigenomes1020010
Chicago/Turabian StyleLanciano, Sophie, and Marie Mirouze. 2017. "DNA Methylation in Rice and Relevance for Breeding" Epigenomes 1, no. 2: 10. https://doi.org/10.3390/epigenomes1020010
APA StyleLanciano, S., & Mirouze, M. (2017). DNA Methylation in Rice and Relevance for Breeding. Epigenomes, 1(2), 10. https://doi.org/10.3390/epigenomes1020010






