Abstract
An important regulatory mechanism affecting mRNA translation involves various covalent modifications of RNA, which establish distinct epitranscriptomic signatures that actively influence various physiological processes. Dendritic translation in mammalian neurons is a potent target for RNA modification-based regulation. In this mini-review, we focus on the effect of potential RNA modifications on the spatiotemporal regulation of the dendritic translation of mRNAs, which are targeted by two important neuronal translational co-regulators, namely TDP-43 and Fragile X Mental Retardation Protein (FMRP).
1. Introduction
Genetic study in the last decade has extended beyond exploration of simple DNA coding. Highly regulated structural modifications of DNA and RNA are now believed to act as molecular bridges between genes and the environment and are responsible for several normal and pathogenic phenotypes. Epigenomics (‘epi-’ is derived from the Greek word meaning ‘over’ or ‘above’) defines the study of nucleic acid modifications that interfere with gene copy number, DNA transcription, translation and RNA editing processes. Epigenetic mechanisms include DNA methylation (5-methylcytosine and its oxidized products, namely, 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxycytosine), histone modifications and the actions of non-coding RNAs, e.g., miRNAs. Besides these, another regulatory epigenetic mechanism involves a plenitude of chemical modifications occurring in RNAs that collectively constitute the “epitranscriptome”. The advent of high throughput technologies specially, Next-Generation Sequencing platforms, so far allow us to identify 107 RNA modifications majority of which occur in tRNA and rRNAs [1]. Transcriptome-wide RNA modifications mainly include methyl-6-adenosine (m6A), methyl-5-cytosine (m5C) and conversion of adenosine-to-inosine (A-to-I). Both m6A and m5C occur in mRNAs with m6A being populated around stop codons [2,3,4], whereas m5C marks have been found in tRNAs and a variety of ncRNAs [5,6]. The m6A is a reversible RNA modification. Adenosine bases can be methylated by Methyltransferase Like protein (METTL)-14 and -3 or m6A marks on RNAs can be erased by demethylases like Fat mass and Obesity-associated protein (FTO) and Alkb homolog 5 (ALKBH5) [7]. Cytosine methylation of RNAs are catalyzed by m5C methyltransferases that transfer methyl groups from S-adenosylmethionine (SAM) [8]. The m5C methyltransferases were divided into four major families: NOP2/NOL1, YebU/Trm4, RsmB, and NSun6 [8,9]. RNA editing, mainly involving A-to-I conversion is also a part of epitranscriptome and catalyzed by adenosine deaminases (ADARs) [10,11]. Besides the above-mentioned conventional RNA modifications, recently other infrequent structural changes in RNAs have been reported, e.g., pseudo-uridylations (Ψ) in various ncRNAs and mRNAs by Ψ synthases [12,13]. It causes the isomerization of uridine residues [14] and is known to increase the translation efficiency of the modified mRNAs [15]. Another relatively less abundant RNA modification is N6, 2′-O-dimethyladenosine (m6Am) that involves methylation at the 2′-O as well as N6 positions of adenosine. The m6Am modification is generated by methylation of the N6 position of 2′-O-methyladenosine (Am) that follows the 7-methylguanosine caps of some mRNAs [16,17]. Different modifications in RNAs have been schematically represented in Figure 1A and described in Table 1. Also chemical structures of modified bases of RNAs and the enzymes catalyzing the reversible/irreversible modification reactions have been shown in Figure 1B. These modifications undoubtedly control many aspects of mRNA translation, and therefore likely control important biological processes such as regulation of circadian rhythms, embryonic stem cell differentiation, neuronal function, and early neuronal, as well as brain development. In this mini-review, we concentrate on m6A, m5C, and A-to-I modifications of RNAs in the context of neurological disorders and discuss potential influence of RNA methylation upon dendritic translation of mRNAs targeted by two translational regulators Fragile X Mental Retardation Protein (FMRP) and TDP-43.
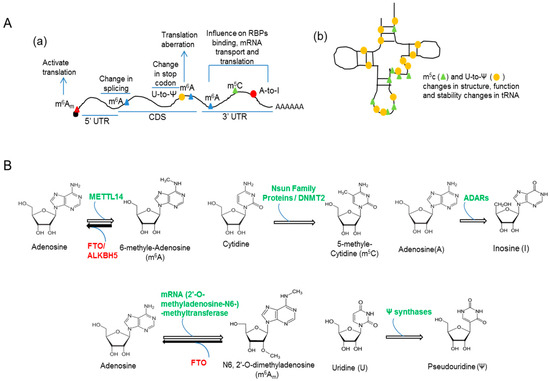
Figure 1.
RNA methylation and editing. (A) Schematic representation of the presence of m6A, m5C, m6Am methylation marks as well as deamination (A-to-I) and pseudouridylation (U-to-Ψ) modifications on different parts of mRNA (a) and tRNA (b); (B) Representation of chemical structures of unmodified and modified RNA bases as well as RNA modification enzymes (Table 1).

Table 1.
Chemical modifications of different RNAs and associated enzymes.
2. RNA Methylation and Neuronal Function
Adenosine methylation (m6A) has been found to appear copiously throughout transcriptome, and thus target many neuronal mRNAs with subsequent active modulation of the intracellular response to neuronal signaling events [2,18]. This is directly correlated with the occurrence of the highest levels of m6A in brain tissue compared to other tissues in mammals. The dynamic nature of m6A modification is revealed by the experience-dependent and locus specific accumulation of this modification near the stop codons of mRNAs of neural plasticity associated genes in mouse prefrontal cortex [19]. Recent studies have identified two m6A demethylases [20,21], one of which is FTO that has been reported to be associated with diseases such as cancer [22], obesity [23], attention-deficit hyperactivity disorder [24], and Alzheimer’s disease [25,26]. Impaired presynaptic dopamine receptor signaling in dopaminergic neuron was observed after targeted deletion of FTO indicating that it is necessary for the proper presynaptic response to extracellular dopamine levels [27]. Elucidation of mechanisms underlying m6A-mediated regulation of mRNA function in connection with neuronal signaling events will unfold various other roles of this epitranscriptomic mark in neuronal function.
Bisulfite sequencing and other transcriptome-wide approaches allowed us to identify another widespread RNA modification, m5C, in coding and non-coding RNAs, such as vault RNAs (vtRNAs) and tRNAs [28,29]. In the case of tRNAs, m5C modifications affect degradation and ribonuclease cleavage, thus can alter global protein translation [30,31,32,33,34]. Also it has been shown that deposition of m5C in the vtRNAs regulate correct processing of them to generate a specific set of small RNAs that functionally resemble microRNAs and act upon a specific set of mRNA targets [5]. In rRNAs and mRNAs, m5C also thought to affect translation [33,35]. Another important function of m5C deposition in the mRNA is to affect stability of target mRNA [36,37]. In higher eukaryotes deposition of m5C modification is carried out by RNA methyltransferases NSun2 and DNMT2 [38,39,40]. NSun2 is a nucleoler protein and encoded by highly conserved family of NOL1/NOP2/Sun domain (NSun) containing RNA methyltransferase genes, which comprises of six members including NSun2 or Misu [39,41]. High expression of NSun2 was found steadily from E7.5 to E10.5 during mouse embryogenesis [42] and its expression has been reported to enrich specifically in the brain [43]. Roles of NSun2 mediated m5C RNA modification has been reported in tissue development, differentiation, cancer, stem cell differentiation, and cellular signaling [34,39,41,43,44,45,46,47,48,49]. Interestingly, mutations in Nsun2 gene causing loss of mRNA leads to impairment of neurocognitive functions as observed in syndromic autosomal-recessive intellectual disability, Dubowitz-like syndrome and Noonan-like syndrome [44,50,51,52]. Also Loss of DNMT2 has been implicated in organ development in zebrafish [53,54]. Since m5C deposition on tRNA directly confers more stability, a loss of NSun2 or DNMT2-mediated m5C methylation induces, repression of protein translation in eukaryotes, which occurs via stress-induced tRNA cleavage [54,55,56,57,58,59,60,61,62]. How the loss of methylation causes symptoms of these neurological diseases is not yet fully understood. Loss of tRNA methylation could be the main defect causing these disorders as the vast majority of NSun2 targets are tRNAs [4,5,6]. Interestingly, increased tRNA cleavage has been implicated in neurodegenerative and neurodevelopmental disorders [63,64] that are commonly associated with oxidative stress [65,66].
3. RNA Editing
RNA editing is the mechanism, which acts upon RNAs post-transcriptionally, to modify specific bases thereby generates RNA and protein diversity. RNA editing includes two chemical modifications, namely, pseudouridylation, that is isomerization of uridine residues or deamination that involves removal of an amine group. The most prevalent form of RNA editing is the deamination reaction involving adenosine (A) to generate inosine (I) in double-stranded RNA. Adenosine-to-inosine conversion takes place at the pre-mRNA level and is catalyzed by the family of RNA-specific adenosine deaminases (ADARs) and subsequently the inosine is recognized as guanosine during translation [10,67,68,69,70]. ADARs-catalyzed A-to-I conversation has been implicated in many neurological disorders including depression, epilepsy, amyotrophic lateral sclerosis (ALS), and in several forms of cancer [71,72]. One prominent example citing requirement of RNA editing in brain function showed engineered RNA editing-impaired GluR-B allele synthesized calcium-permeable GluR-B subunit of glutamate receptor leading to excess calcium influx into neurons causing postnatal death in mice [73]. The most common RNA modification observed in patients with ALS is deamination of adenosine to inosine [74] and presence of Ca2+-permeable AMPA receptor-mediated pathogenic mechanism causing motor neuron death in ALS has been shown to operate due to reduction of ADAR2 leading to failure of GluA2 RNA editing [75].
4. Dendritic Local Translation and Neurological Disorders
Dendritic localization of mRNAs first hinted that RNA translation might occur locally within the dendrites [76]. Later, other translation related components e.g., initiation and elongation factors, ribosomal proteins, polyribosomes, and tRNAs were also discovered inside dendrites [77]. Further research revealed that dendritic local translation of synaptic mRNAs can be visualized in cultured neurons [78] and it can be stimulated by neuron activation [79,80,81,82]. It has also been shown that these mRNAs are transported to dendrites under neuron activity stimulation [83,84,85] and undergoes translation when they reach the destination [86,87,88]. Neuron activity-dependent dendritic translation can regulate synaptic plasticity, dendritic spinogenesis [89,90], as well as maintain long lasting changes in dendritic synapses such as Long-Term Potentiation (LTP) and Long-Term-Depression (LTD) [91,92,93].
In this context, FMRP is required to be mentioned because of its association with neurological diseases and its important role in stimulus-dependent translation and synaptic plasticity [94]. FMRP is a component of ribonucleoprotein complexes (RNPs) that transport mRNAs to dendrites, and it probably plays an active role in this process. This RNA binding protein (RBP) is a well-known translational repressor that inhibits either the initiation or elongation step [95]. Through its RNA-binding domains FMRP can bind mRNAs possessing the G-quadruplex structure and about 4% of total mouse brain mRNAs are known to interact with this protein [96,97]. In a mouse model for Fragile X mental retardation disease, loss of FMRP caused an increased number of dendritic spines [98], impaired brain development and deregulated synaptic plasticity [94] as a consequence of activation of global protein synthesis [99].
Recently, it has been established that FMRP and Frontotemporal Lobar Dementia (FTLD)/ALS pathology related protein TDP-43 physically interact and associate with same mRNPs [100]. A functional link between these two RBPs in dendritic translation regulation has also been elucidated [100,101]. In another study, overexpression of TDP-43 inhibited translation of Futsch (Drosophila homolog of Map1b) mRNA in motor neurons [102], which was re-activated by overexpression of FMRP [103]. Moreover, 1140 common mRNA targets of FMRP and TDP-43 have been identified. Among them 160 targets are related to neuron structure, function, and neuron development. A significant portion of these mRNAs might be co-regulated by FMRP and TDP-43 at the translational level. Interestingly, the above mentioned 160 common targets of FMRP and TDP-43 also include candidate genes for Autism Spectrum Disorder (ASD; e.g., Rac1, Mapk1, Reln, and Shank3), Alzheimer’s disease (AD; e.g., Ank1, App, and Apoe) and Schizophrenia (e.g., Grin2b and Gsk3b) indicating that loss-of-function of either of these two RBPs might develop similar phenotypes [100].
5. Potential Roles of RNA Modifications in TDP-43/FMRP-Associated Neurological Diseases
As described above, FMRP and TDP-43 probably play important roles in modulating both neurodevelopmental (e.g., ASD, Schizophrenia) and neurodegenerative (e.g., AD) disorders by co-regulating dendritic local translation. It has been established that TDP-43 recruits the FMRP-CYFIP1-eIF4E inhibitory complex to the target mRNAs present at dendrites, thereby repressing initiation of translation [100]. Thus TDP-43 acts as an adaptor protein between target mRNAs and FMRP. Upon relevant synaptic stimulation, FMRP is dephosphorylated [104] and either FMRP-CYFIP1 is dissociated from eIF4E [105] or FMRP is dissociated from mRNAs, leading to re-activation of dendritic local translation [106]. However, it remains elusive how distinct sets of mRNAs are translationally regulated by FMRP/TDP-43 in the context of different neurological disorders. Also, underlying mechanisms that elicit differential responses in different regions of the brain under same pre-synaptic signals are not fully conceived. Here, it is intriguing to speculate that alike epigenetic code mediated regulation of gene transcription, the epitranscriptomic code drives evolution of dynamic modes of translational regulation in post-mitotic neurons via installation of potentially reversible RNA modifications such as methylation/demethylation of mRNAs, tRNAs and rRNAs [107,108,109,110].
In this context, the m5C methylation of the RNAs by NSun2 is of great importance for fine-tuning in dendritic local translation as well as for neuronal function. This methylase is associated with human intellectual disability syndromes, ASD, and epilepsy [44,50,51,52,111]. Drosophila and mouse models with down regulated NSun2 showed significant problems with learning and memory [44,112]. Interestingly, the hippocampus region of the mouse brain showed the highest expression of NSun2 [112,113]. NSun2-deficient mice also showed fragmentation of tRNAs followed by stress responses and apoptosis of neurons [112], a significant decrease in global translation [34], and mis-regulation of miRNAs that are known important regulators of translation along with RBPs [114]. Many indirect evidences could be put forward in support of the notion that NSun2-mediated methylation may regulate dendritic local translation and neurodevelopmental diseases. For example, NSun2 partially co-localizes with FMRP in dendrites but, in axons, no appreciable co-localization of this RNA methylase with FMRP has been detected [115]. Also a significant overlap between mRNA targets of FMRP and NSun2 has been reported [115]. Among different methylation targets of NSun2, mTOR mRNA is of particular interest, because of its activity-dependent translational up regulation in dendrites that facilitates phosphorylation of 4E-BP (eIF4E binding protein, e.g., CYFIP1) and release of CYFIP1-FMRP-TDP-43 mediated translational repression [116,117]. The potential role of mTOR mRNA methylation on synaptic mRNA translation co-regulated by TDP-43 and FMRP is schematically represented in Figure 2 (right schemes).
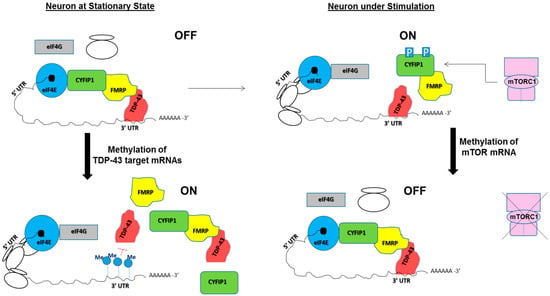
Figure 2.
A model showing potential roles of RNA methylation on spatiotemporal translational control of mRNAs, co-regulated by TDP-43 and FMRP. (Left) In the stationary state of immature neurons FMRP (yellow) or the FMRP-CYFIP1 complex is recruited to the vicinity of the mRNA(s) by mRNA-bound TDP-43 (red). CYFIP1 (green) interacts with eIF4E (blue) and blocks eIF4G (grey) to bind with eIF4E and to form the translation initiation complex eIF4F (not shown). Thus, translation of synaptic mRNAs like Rac1 mRNA remain in “OFF” state (top left scheme). Methylation marks at the 3′UTR region of mRNAs targeted by TDP-43 may inhibit its interaction with TDP-43 and thus potentially block the recruitment of CYFIP1-FMRP complex to these mRNAs. This leads to initiation of translation marked as “ON” state (lower left scheme); (Right) in neurons under stimulation, mTOR pathway is activated and mTORC1 (purple) phosphorylates CYFIP1 that inhibits eIF4E/CYFIP1-FMRP-TDP-43 interaction. Thus translation machinery becomes “ON” (top right scheme). The mTOR mRNA methylation-induced inhibition of mTOR pathway may influence its downstream signaling events at neuronal synapses (e.g., phosphorylation of CYFIP1 protein), to inhibit translation initiation resulting in the “OFF” state even under stimulation (lower right scheme).
Interestingly, NSun2-mediated m5C methylation of ncRNAs regulates cellular levels of the transmembrane AMPA receptor regulatory protein, CACNG8 [5]. The point to be noted here is that the transmembrane AMPA receptor subunit is important in TDP-43-mediated spinogenesis [101]. Lastly, NSun2 mRNA forms an in vivo complex with the TDP-43-associated protein, FUS, indicating the presence of a complex mechanism involving different RNA metabolism pathways [118]. The potential role of m6A residues in dendritic localization and stability of ASD associated mRNAs have been recently reviewed [119]. Although there is no direct evidence of m6A methylation affecting TDP-43/FMRP-regulated neuronal translation or neurological disorders, it should be noted that in human cells more than 12,000 m6A sites in about 7000 mRNAs have been identified [119] and the most of m6A RNA methylation found in the last exons. This might influence 3′-UTR-mediated translational regulation of mRNAs [120,121]. Therefore, m6A methylation potentially exerts fine-tuning on the dendritic translation mechanisms by inhibiting mRNA binding of TDP-43 at the 3′UTR region. This probably would affect TDP-43 mediated mRNA recruitment to a translation inhibitory complex. A recent study, examining the consensus RNA-binding sites of FMRP protein, speculated the role of m6A as the negative determinant of FMRP-binding to target mRNAs [122]. Schematic representation of potential role of RNA methylation on spatiotemporal translational control of mRNAs co-regulated by TDP-43 and FMRP has been shown in Figure 2 (left schemes).
In addition to RNA methylation, aberrant A-to-I RNA editing by adenosine deaminase enzyme in ADAR2-KO mice caused mis-localization and aggregation of TDP-43 [123] and delayed death of motor neurons. It is also reported that motor neurons from ALS patients are more susceptible to RNA-editing deficiencies [124]. Finally, an example of translational regulation at synapses due to 2′-O-methylation of RNA altering the BC1-FMRP interaction with target mRNA [125] indicates the importance of studying non-conventional RNA modifications and their roles in regulating the dendritic translation mechanism in neurons.
As we have described above, RNA modification may play a very important role in dendritic mRNA translation. Several RNA-modifying enzymes exist, and they probably regulate different types of RNA modifications. Systematic study of these enzymes may reveal a complexity long postulated for RNA and its involvement in neurological diseases.
Acknowledgments
We want to acknowledge the quick and effective editing effort of John O’Brien, English Editor, IMB, Academia Sinica, Taipei, Taiwan.
Author Contributions
P.M. and C.-K.J.S. conceived the theme of the review. P.M. and B.C. consulted the literature and wrote the review.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Agris, P.C.P.; Rozenski, J.; Fabris, D.; Vendeix, F. The RNA Modification Database. Available online: http://mods.rna.albany.edu (accessed on 27 June 2017).
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Sajini, A.A.; Blanco, S.; Dietmann, S.; Lombard, P.; Sugimoto, Y.; Paramor, M.; Gleeson, J.G.; Odom, D.T.; Ule, J.; et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013, 4, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Khoddami, V.; Cairns, B.R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 2013, 31, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Liu, J.; He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015, 29, 1343–1355. [Google Scholar] [CrossRef] [PubMed]
- Bujnicki, J.M.; Feder, M.; Ayres, C.L.; Redman, K.L. Sequence-structure-function studies of tRNA:M5c methyltransferase trm4p and its relationship to DNA:M5c and RNA:M5u methyltransferases. Nucleic Acids Res. 2004, 32, 2453–2463. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jia, G. Methylation modifications in eukaryotic messenger RNA. J. Genet. Genom. 2014, 41, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Li, J.B.; Church, G.M. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat. Neurosci. 2013, 16, 1518–1522. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, J.J.; Seeburg, P.H. A-to-I RNA editing: Effects on proteins key to neural excitability. Neuron 2012, 74, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; Leon-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Yu, Y.T. RNA pseudouridylation: New insights into an old modification. Trends Biochem. Sci. 2013, 38, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Harcourt, E.M.; Kietrys, A.M.; Kool, E.T. Chemical and structural effects of base modifications in messenger RNA. Nature 2017, 541, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiong, X.; Yi, C. Epitranscriptome sequencing technologies: Decoding RNA modifications. Nat. Methods 2016, 14, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Agarwala, S.D.; Mumbach, M.R.; Jovanovic, M.; Mertins, P.; Shishkin, A.; Tabach, Y.; Mikkelsen, T.S.; Satija, R.; Ruvkun, G.; et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 2013, 155, 1409–1421. [Google Scholar] [CrossRef] [PubMed]
- Widagdo, J.; Zhao, Q.Y.; Kempen, M.J.; Tan, M.C.; Ratnu, V.S.; Wei, W.; Leighton, L.; Spadaro, P.A.; Edson, J.; Anggono, V.; et al. Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 2016, 36, 6771–6777. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Iles, M.M.; Law, M.H.; Stacey, S.N.; Han, J.; Fang, S.; Pfeiffer, R.; Harland, M.; Macgregor, S.; Taylor, J.C.; Aben, K.K.; et al. A variant in FTO shows association with melanoma risk not due to BMI. Nat. Genet. 2013, 45, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.C.; Yeo, G.S. From GWAS to biology: Lessons from FTO. Ann. N. Y. Acad. Sci. 2011, 1220, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Choudhry, Z.; Sengupta, S.M.; Grizenko, N.; Thakur, G.A.; Fortier, M.E.; Schmitz, N.; Joober, R. Association between obesity-related gene FTO and ADHD. Obesity (Silver Spring) 2013, 21, E738–E744. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Xu, W.; Wang, H.X.; Winblad, B.; Fratiglioni, L.; Graff, C. The obesity related gene, FTO, interacts with APOE, and is associated with alzheimer’s disease risk: A prospective cohort study. J. Alzheimer’s Dis. 2011, 23, 461–469. [Google Scholar]
- Reitz, C.; Tosto, G.; Mayeux, R.; Luchsinger, J.A. Genetic variants in the fat and obesity associated (FTO) gene and risk of alzheimer’s disease. PLoS ONE 2012, 7, e50354. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.E.; Hess, S.; Meyer, K.D.; Verhagen, L.A.; Koch, L.; Bronneke, H.S.; Dietrich, M.O.; Jordan, S.D.; Saletore, Y.; Elemento, O.; et al. The fat mass and obesity associated gene (FTO) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 2013, 16, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Amort, T.; Souliere, M.F.; Wille, A.; Jia, X.Y.; Fiegl, H.; Worle, H.; Micura, R.; Lusser, A. Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 2013, 10, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Edelheit, S.; Schwartz, S.; Mumbach, M.R.; Wurtzel, O.; Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in Bacteria, Archaea, and yeast reveals m5C within Archaeal mRNAs. PLoS Genet. 2013, 9, e1003602. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.; Pang, Y.L.; Deng, W.; Babu, I.R.; Dyavaiah, M.; Begley, T.J.; Dedon, P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012, 3, 937. [Google Scholar] [CrossRef] [PubMed]
- Chernyakov, I.; Whipple, J.M.; Kotelawala, L.; Grayhack, E.J.; Phizicky, E.M. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 2008, 22, 1369–1380. [Google Scholar] [CrossRef]
- Chow, C.S.; Lamichhane, T.N.; Mahto, S.K. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2007, 2, 610–619. [Google Scholar] [CrossRef]
- Tuorto, F.; Liebers, R.; Musch, T.; Schaefer, M.; Hofmann, S.; Kellner, S.; Frye, M.; Helm, M.; Stoecklin, G.; Lyko, F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012, 19, 900–905. [Google Scholar] [CrossRef]
- Metodiev, M.D.; Spahr, H.; Loguercio Polosa, P.; Meharg, C.; Becker, C.; Altmueller, J.; Habermann, B.; Larsson, N.G.; Ruzzenente, B. NSun4 is a dual function mitochondrial protein required for both methylation of 12s rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014, 10, e1004110. [Google Scholar] [CrossRef]
- Hussain, S.; Aleksic, J.; Blanco, S.; Dietmann, S.; Frye, M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 2013, 14, 215. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Z.; Yi, J.; Tang, H.; Xing, J.; Yu, M.; Tong, T.; Shang, Y.; Gorospe, M.; Wang, W. The tRNA methyltransferase Nsun2 stabilizes p16INK(4) mRNA by methylating the 3′-untranslated region of p16. Nat. Commun. 2012, 3, 712. [Google Scholar] [CrossRef]
- Brzezicha, B.; Schmidt, M.; Makalowska, I.; Jarmolowski, A.; Pienkowska, J.; Szweykowska-Kulinska, Z. Identification of human tRNA:M5c methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res. 2006, 34, 6034–6043. [Google Scholar] [CrossRef]
- Frye, M.; Watt, F.M. The RNA methyltransferase MIsu (NSun2) mediates myc-induced proliferation and is upregulated in tumors. Curr. Biol. 2006, 16, 971–981. [Google Scholar] [CrossRef]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of trnaasp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef]
- Sakita-Suto, S.; Kanda, A.; Suzuki, F.; Sato, S.; Takata, T.; Tatsuka, M. Aurora-b regulates RNA methyltransferase NSun2. Mol. Biol. Cell 2007, 18, 1107–1117. [Google Scholar] [CrossRef]
- Chi, L.; Delgado-Olguin, P. Expression of NOL1/NOP2/sun domain (NSun) RNA methyltransferase family genes in early mouse embryogenesis. Gene Expr. Patterns 2013, 13, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Kurowski, A.; Nichols, J.; Watt, F.M.; Benitah, S.A.; Frye, M. The RNA-methyltransferase MIsu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011, 7, e1002403. [Google Scholar] [CrossRef] [PubMed]
- Abbasi-Moheb, L.; Mertel, S.; Gonsior, M.; Nouri-Vahid, L.; Kahrizi, K.; Cirak, S.; Wieczorek, D.; Motazacker, M.M.; Esmaeeli-Nieh, S.; Cremer, K.; et al. Mutations in NSun2 cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012, 90, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Bandiera, R.; Popis, M.; Hussain, S.; Lombard, P.; Aleksic, J.; Sajini, A.; Tanna, H.; Cortes-Garrido, R.; Gkatza, N.; et al. Stem cell function and stress response are controlled by protein synthesis. Nature 2016, 534, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Dragoni, I.; Chin, S.F.; Spiteri, I.; Kurowski, A.; Provenzano, E.; Green, A.; Ellis, I.O.; Grimmer, D.; Teschendorff, A.; et al. Genomic gain of 5p15 leads to over-expression of MIsu (NSun2) in breast cancer. Cancer Lett. 2010, 289, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Benavente, S.B.; Nascimento, E.; Dragoni, I.; Kurowski, A.; Gillich, A.; Humphreys, P.; Frye, M. The nucleolar RNA methyltransferase MIsu (NSun2) is required for mitotic spindle stability. J. Cell Biol. 2009, 186, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Tuorto, F.; Menon, S.; Blanco, S.; Cox, C.; Flores, J.V.; Watt, S.; Kudo, N.R.; Lyko, F.; Frye, M. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol. Cell Biol. 2013, 33, 1561–1570. [Google Scholar] [CrossRef] [PubMed]
- Satterlee, J.S.; Basanta-Sanchez, M.; Blanco, S.; Li, J.B.; Meyer, K.; Pollock, J.; Sadri-Vakili, G.; Rybak-Wolf, A. Novel RNA modifications in the nervous system: Form and function. J. Neurosci. 2014, 34, 15170–15177. [Google Scholar] [CrossRef] [PubMed]
- Fahiminiya, S.; Almuriekhi, M.; Nawaz, Z.; Staffa, A.; Lepage, P.; Ali, R.; Hashim, L.; Schwartzentruber, J.; Abu Khadija, K.; Zaineddin, S.; et al. Whole exome sequencing unravels disease-causing genes in consanguineous families in Qatar. Clin. Genet. 2014, 86, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Rafiq, M.A.; Noor, A.; Hussain, S.; Flores, J.V.; Rupp, V.; Vincent, A.K.; Malli, R.; Ali, G.; Khan, F.S.; et al. Mutation in NSun2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012, 90, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.J.; Lee, J.H.; Lee, J.E.; Blanco, S.; Nickerson, E.; Gabriel, S.; Frye, M.; Al-Gazali, L.; Gleeson, J.G. Whole exome sequencing identifies a splicing mutation in NSun2 as a cause of a dubowitz-like syndrome. J. Med. Genet. 2012, 49, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.; Chidester, S.; Zavala, C.V.; Manos, E.J.; James, S.R.; Karpf, A.R.; Jones, D.A.; Cairns, B.R. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007, 21, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Emara, M.M.; Ivanov, P.; Hickman, T.; Dawra, N.; Tisdale, S.; Kedersha, N.; Hu, G.F.; Anderson, P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010, 285, 10959–10968. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Feng, J.; Liu, Q.; Sun, F.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009, 583, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Gebetsberger, J.; Zywicki, M.; Kunzi, A.; Polacek, N. Trna-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012, 2012, 260909. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 2011, 43, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Sobala, A.; Hutvagner, G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013, 10, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Spriggs, K.A.; Bushell, M.; Willis, A.E. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010, 40, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Lu, C.; Green, P.J.; Parker, R. Trna cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008, 14, 2095–2103. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Ivanov, P.; Hu, G.F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Karaca, E.; Weitzer, S.; Pehlivan, D.; Shiraishi, H.; Gogakos, T.; Hanada, T.; Jhangiani, S.N.; Wiszniewski, W.; Withers, M.; Campbell, I.M.; et al. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell 2014, 157, 636–650. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, A.E.; Eggens, V.R.; Caglayan, A.O.; Reuter, M.S.; Scott, E.; Coufal, N.G.; Silhavy, J.L.; Xue, Y.; Kayserili, H.; Yasuno, K.; et al. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 2014, 157, 651–663. [Google Scholar] [CrossRef] [PubMed]
- De Felice, C.; Signorini, C.; Leoncini, S.; Pecorelli, A.; Durand, T.; Valacchi, G.; Ciccoli, L.; Hayek, J. The role of oxidative stress in rett syndrome: An overview. Ann. N. Y. Acad. Sci. 2012, 1259, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Lintas, C.; Sacco, R.; Persico, A.M. Genome-wide expression studies in autism spectrum disorder, rett syndrome, and down syndrome. Neurobiol. Dis. 2012, 45, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K. Functions and regulation of RNA editing by adar deaminases. Annu. Rev. Biochem. 2010, 79, 321–349. [Google Scholar] [CrossRef] [PubMed]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002, 71, 817–846. [Google Scholar] [CrossRef] [PubMed]
- Gerber, A.P.; Keller, W. RNA editing by base deamination: More enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001, 26, 376–384. [Google Scholar] [CrossRef]
- Valente, L.; Nishikura, K. ADAR gene family and A-to-I RNA editing: Diverse roles in posttranscriptional gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 2005, 79, 299–338. [Google Scholar] [PubMed]
- Slotkin, W.; Nishikura, K. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013, 5, 105. [Google Scholar] [CrossRef] [PubMed]
- Tariq, A.; Jantsch, M.F. Transcript diversification in the nervous system: A to I RNA editing in CNS function and disease development. Front. Neurosci. 2012, 6, 99. [Google Scholar] [CrossRef] [PubMed]
- Brusa, R.; Zimmermann, F.; Koh, D.S.; Feldmeyer, D.; Gass, P.; Seeburg, P.H.; Sprengel, R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 1995, 270, 1677–1680. [Google Scholar] [CrossRef] [PubMed]
- Paez-Colasante, X.; Figueroa-Romero, C.; Sakowski, S.A.; Goutman, S.A.; Feldman, E.L. Amyotrophic lateral sclerosis: Mechanisms and therapeutics in the epigenomic era. Nat. Rev. Neurol. 2015, 11, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Chai, H.L.; Teramoto, S.; Tsuji, S.; Shimazaki, K.; Muramatsu, S.; Kwak, S. Rescue of amyotrophic lateral sclerosis phenotype in a mouse model by intravenous AAV9-ADAR2 delivery to motor neurons. EMBO Mol. Med. 2013, 5, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- Bodian, D. A suggestive relationship of nerve cell RNA with specific synaptic sites. Proc. Natl. Acad. Sci. USA 1965, 53, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Steward, O.; Levy, W.B. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J. Neurosci. 1982, 2, 284–291. [Google Scholar] [PubMed]
- Torre, E.R.; Steward, O. Demonstration of local protein synthesis within dendrites using a new cell culture system that permits the isolation of living axons and dendrites from their cell bodies. J. Neurosci. 1992, 12, 762–772. [Google Scholar] [PubMed]
- Aakalu, G.; Smith, W.B.; Nguyen, N.; Jiang, C.; Schuman, E.M. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 2001, 30, 489–502. [Google Scholar] [CrossRef]
- Feig, S.; Lipton, P. Pairing the cholinergic agonist carbachol with patterned schaffer collateral stimulation initiates protein synthesis in hippocampal CA1 pyramidal cell dendrites via a muscarinic, NMDA-dependent mechanism. J. Neurosci. 1993, 13, 1010–1021. [Google Scholar] [PubMed]
- Weiler, I.J.; Greenough, W.T. Potassium ion stimulation triggers protein translation in synaptoneurosomal polyribosomes. Mol. Cell Neurosci. 1991, 2, 305–314. [Google Scholar] [CrossRef]
- Weiler, I.J.; Greenough, W.T. Metabotropic glutamate receptors trigger postsynaptic protein synthesis. Proc. Natl. Acad. Sci. USA 1993, 90, 7168–7171. [Google Scholar] [CrossRef] [PubMed]
- Antar, L.N.; Afroz, R.; Dictenberg, J.B.; Carroll, R.C.; Bassell, G.J. Metabotropic glutamate receptor activation regulates fragile X mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J. Neurosci. 2004, 24, 2648–2655. [Google Scholar] [CrossRef] [PubMed]
- Tiruchinapalli, D.M.; Oleynikov, Y.; Kelic, S.; Shenoy, S.M.; Hartley, A.; Stanton, P.K.; Singer, R.H.; Bassell, G.J. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J. Neurosci. 2003, 23, 3251–3261. [Google Scholar] [PubMed]
- Tongiorgi, E.; Righi, M.; Cattaneo, A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J. Neurosci. 1997, 17, 9492–9505. [Google Scholar] [PubMed]
- Gong, R.; Park, C.S.; Abbassi, N.R.; Tang, S.J. Roles of glutamate receptors and the mammalian target of rapamycin (mTOR) signaling pathway in activity-dependent dendritic protein synthesis in hippocampal neurons. J. Biol. Chem. 2006, 281, 18802–18815. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.N.; Smith, L.T.; Logan, S.M.; Simpkins, J.W. Estrogen-induced activation of extracellular signal-regulated kinase signaling triggers dendritic resident mRNA translation. Neuroscience 2010, 170, 1080–1085. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.O.; Kim, S.M.; Zhao, Y.; Hwang, H.; Miura, S.K.; Sossin, W.S.; Martin, K.C. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science 2009, 324, 1536–1540. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Martin, A.; Crespo, M.; Portera-Cailliau, C. Delayed stabilization of dendritic spines in fragile X mice. J. Neurosci. 2010, 30, 7793–7803. [Google Scholar] [CrossRef] [PubMed]
- Engert, F.; Bonhoeffer, T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature 1999, 399, 66–70. [Google Scholar] [PubMed]
- Huber, K.M.; Kayser, M.S.; Bear, M.F. Role for rapid dendritic protein synthesis in hippocampal mglur-dependent long-term depression. Science 2000, 288, 1254–1257. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Schuman, E.M. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science 1996, 273, 1402–1406. [Google Scholar] [CrossRef] [PubMed]
- Raymond, C.R.; Thompson, V.L.; Tate, W.P.; Abraham, W.C. Metabotropic glutamate receptors trigger homosynaptic protein synthesis to prolong long-term potentiation. J. Neurosci. 2000, 20, 969–976. [Google Scholar] [PubMed]
- Sidorov, M.S.; Auerbach, B.D.; Bear, M.F. Fragile X mental retardation protein and synaptic plasticity. Mol. Brain 2013, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Joseph, S. Fragile X mental retardation protein: A paradigm for translational control by RNA-binding proteins. Biochimie 2015, 114, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Menon, L.; Mader, S.A.; Mihailescu, M.R. Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA. RNA 2008, 14, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Vasilyev, N.; Polonskaia, A.; Darnell, J.C.; Darnell, R.B.; Patel, D.J.; Serganov, A. Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP. Proc. Natl. Acad. Sci. USA 2015, 112, E5391–E5400. [Google Scholar] [CrossRef] [PubMed]
- Buffington, S.A.; Huang, W.; Costa-Mattioli, M. Translational control in synaptic plasticity and cognitive dysfunction. Annu. Rev. Neurosci. 2014, 37, 17–38. [Google Scholar] [CrossRef] [PubMed]
- Osterweil, E.K.; Krueger, D.D.; Reinhold, K.; Bear, M.F. Hypersensitivity to mGLUR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J. Neurosci. 2010, 30, 15616–15627. [Google Scholar] [CrossRef] [PubMed]
- Majumder, P.; Chu, J.F.; Chatterjee, B.; Swamy, K.B.; Shen, C.J. Co-regulation of mRNA translation by TDP-43 and fragile X syndrome protein fmrp. Acta Neuropathol. 2016, 132, 721–738. [Google Scholar] [CrossRef] [PubMed]
- Majumder, P.; Chen, Y.T.; Bose, J.K.; Wu, C.C.; Cheng, W.C.; Cheng, S.J.; Fang, Y.H.; Chen, Y.L.; Tsai, K.J.; Lien, C.C.; et al. TDP-43 regulates the mammalian spinogenesis through translational repression of Rac1. Acta Neuropathol. 2012, 124, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Coyne, A.N.; Siddegowda, B.B.; Estes, P.S.; Johannesmeyer, J.; Kovalik, T.; Daniel, S.G.; Pearson, A.; Bowser, R.; Zarnescu, D.C. Futsch/MAP1B mRNA is a translational target of TDP-43 and is neuroprotective in a drosophila model of amyotrophic lateral sclerosis. J. Neurosci. 2014, 34, 15962–15974. [Google Scholar] [CrossRef] [PubMed]
- Coyne, A.N.; Yamada, S.B.; Siddegowda, B.B.; Estes, P.S.; Zaepfel, B.L.; Johannesmeyer, J.S.; Lockwood, D.B.; Pham, L.T.; Hart, M.P.; Cassel, J.A.; et al. Fragile X protein mitigates TDP-43 toxicity by remodeling RNA granules and restoring translation. Hum. Mol. Genet. 2015, 24, 6886–6898. [Google Scholar] [CrossRef] [PubMed]
- Bernard, P.B.; Castano, A.M.; O’Leary, H.; Simpson, K.; Browning, M.D.; Benke, T.A. Phosphorylation of fmrp and alterations of fmrp complex underlie enhanced mltd in adult rats triggered by early life seizures. Neurobiol. Dis. 2013, 59, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Napoli, I.; Mercaldo, V.; Boyl, P.P.; Eleuteri, B.; Zalfa, F.; De Rubeis, S.; Di Marino, D.; Mohr, E.; Massimi, M.; Falconi, M.; et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 2008, 134, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.R.; Bray, S.M.; Warren, S.T. Molecular mechanisms of fragile X syndrome: A twenty-year perspective. Annu. Rev. Pathol. 2012, 7, 219–245. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Dai, Q.; Zhang, W.; Ren, J.; Pan, T.; He, C. The ALKB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew. Chem. Int. Ed. Engl. 2010, 49, 8885–8888. [Google Scholar] [CrossRef] [PubMed]
- Saikia, M.; Fu, Y.; Pavon-Eternod, M.; He, C.; Pan, T. Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA 2010, 16, 1317–1327. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, C. Dynamic RNA modifications in posttranscriptional regulation. Mol. Cell 2014, 56, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Komara, M.; Al-Shamsi, A.M.; Ben-Salem, S.; Ali, B.R.; Al-Gazali, L. A novel single-nucleotide deletion (c.1020delA) in NSun2 causes intellectual disability in an emirati child. J. Mol. Neurosci. 2015, 57, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Frye, M. Role of RNA methyltransferases in tissue renewal and pathology. Curr. Opin. Cell Biol. 2014, 31, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Flores, J.V.; Cordero-Espinoza, L.; Oeztuerk-Winder, F.; Andersson-Rolf, A.; Selmi, T.; Blanco, S.; Tailor, J.; Dietmann, S.; Frye, M. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Rep. 2017, 8, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Schratt, G. Micrornas at the synapse. Nat. Rev. Neurosci. 2009, 10, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Bashir, Z.I. The epitranscriptome in modulating spatiotemporal RNA translation in neuronal post-synaptic function. Front. Cell Neurosci. 2015, 9, 420. [Google Scholar] [CrossRef] [PubMed]
- Ceman, S.; O’Donnell, W.T.; Reed, M.; Patton, S.; Pohl, J.; Warren, S.T. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum. Mol. Genet. 2003, 12, 3295–3305. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Pepio, A.M.; Sossin, W.S. Serotonin activates s6 kinase in a rapamycin-sensitive manner in aplysia synaptosomes. J. Neurosci. 2001, 21, 382–391. [Google Scholar] [PubMed]
- Colombrita, C.; Onesto, E.; Megiorni, F.; Pizzuti, A.; Baralle, F.E.; Buratti, E.; Silani, V.; Ratti, A. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J. Biol. Chem. 2012, 287, 15635–15647. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Zang, L.Q.; Li, X.K.; Shu, Q. An epigenetic view of developmental diseases: New targets, new therapies. World J. Pediatr. 2016, 12, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.; Alemu, E.A.; Mertens, C.; Gantman, E.C.; Fak, J.J.; Mele, A.; Haripal, B.; Zucker-Scharff, I.; Moore, M.J.; Park, C.Y.; et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015, 29, 2037–2053. [Google Scholar] [CrossRef] [PubMed]
- Rowles, J.; Wong, M.; Powers, R.; Olsen, M. FTO, RNA epigenetics and epilepsy. Epigenetics 2012, 7, 1094–1097. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.R.; Chopra, P.; Suhl, J.A.; Warren, S.T.; Bassell, G.J. Identification of consensus binding sites clarifies FMRP binding determinants. Nucleic Acids Res. 2016, 44, 6649–6659. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Kwak, S. The molecular link between inefficient GluA2 Q/R site-RNA editing and TDP-43 pathology in motor neurons of sporadic amyotrophic lateral sclerosis patients. Brain Res. 2014, 1584, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; Ito, K.; Sun, H.; Aizawa, H.; Kanazawa, I.; Kwak, S. Glutamate receptors: RNA editing and death of motor neurons. Nature 2004, 427, 801. [Google Scholar] [CrossRef] [PubMed]
- Lacoux, C.; Di Marino, D.; Boyl, P.P.; Zalfa, F.; Yan, B.; Ciotti, M.T.; Falconi, M.; Urlaub, H.; Achsel, T.; Mougin, A.; et al. BC1-FMRP interaction is modulated by 2′-O-methylation: RNA-binding activity of the tudor domain and translational regulation at synapses. Nucleic Acids Res. 2012, 40, 4086–4096. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).