Abstract
Cell phenotype is influenced by the linear sequence of bases and by epigenetic changes. Despite the huge number of implants placed every year, epigenetic mechanisms controlling peri-implant processes remain unexplored. The purpose of this systematic qualitative review was to investigate the available articles dealing with the relationships between DNA methylations, histone modifications, or micro-RNA (miRNA) production and implant therapy. A large variety of different surfaces were evaluated based on their osteogenic stimulation of osteoblasts. Dental implant treatments like potassium hydroxide (KOH) alkali-etching, electrolytic etching, ionization, functionalization with miRNAs or anti-miRNAs, or osteogenic peptides enhanced osteoblast differentiation and gene activation by regulating miRNA production. Zirconia and anatase coating inhibited the activation of osteogenic genes. Epigenetic changes on peri-implant cells induced by smoking still remain unclear. Due to the heterogeneity of methodologies, a meta-analysis was not possible. Even if it is impossible to define which implant surface was best to genetically stimulate osteogenesis, there is evidence that implant surface features can upregulate or downregulate genes related to osseointegration.
1. Introduction
The rehabilitation of partial and total edentulism using dental implants has shown highly satisfactory clinical outcomes [1]. The success of implant therapies has been clinically and histologically documented [2] and is based on osseointegration, a direct structural and functional connection between native living bone and the implant surface [3].
Notwithstanding the wide use of osseointegrated implants, 80% of subjects and 50% of implants suffer from mucositis or other biological problems [4,5]. Biological complications around dental implants have been attributed to several factors, from the establishment of a pathogenic microflora [6] to the presence of inflammatory cells close to the implant-abutment interface [7,8].
Microbiological findings from sites of failing implants are similar to those in periodontally compromised teeth [9,10] with increased subgingival levels of Porphyromonas gingivalis, Prevotella intermedia, and Fusobacterium nucleatum.
Oral plaque is often considered the primary reason for implant failure after loading [11,12]. Furthermore, increased levels of prostaglandin E2 (PGE2), interleukins (IL), and platelet-derived growth factor (PDGF) at the sites of failed implants [13] indicate that the host’s response to environmental stress also plays a major role in peri-implant mechanisms. Despite the number of existing hypotheses, the genetic mechanisms controlling the peri-implant biological processes remain largely unexplored.
In 2008, epigenetics was defined as a “stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence” [14]. Epigenetic modifications alter the structure of chromatin and influence gene expression without alterations in the sequence of bases. Moreover, epigenetic inheritance influences phenotypes over multiple generations in the absence of any apparent genetic mutation [15,16] as if there is a “ghost in your genes”, as stated by Rothstein et al. [17].
The study of epigenetic mechanisms could also explain intergenerational disease susceptibility not directly hardwired into our genetic material [18]. Environmental stressors including toxins and microbial exposures [19], adrenaline and psychological stress [20], diet [21], and hormones [15] can change epigenetic patterns and thereby effect changes in gene activation and cell phenotype. Epigenetic modifications affect genetic expression by activating or repressing genes and preventing messenger RNA (mRNA) formation, or by affecting protein structure after translation from an mRNA template [18].
Epigenetic mechanisms mainly involve DNA methylation, histone modification, and mRNA regulation by non-coding RNAs (ncRNAs) called microRNAs (miRNAs) [22] (Table 1). DNA methylation is performed by DNA methyltransferases and causes a more condensed DNA structure, transcriptional repression, and gene silencing [23,24]. The DNA methylation state has an important role in bone metabolism as it regulates the expression of various genes involved in bone cell functions such as alkaline phosphatase, sclerosin, osterix, distal-less homeobox 5 protein (DLX5), oestrogen receptor alpha, osteoprotegerin, receptor activator of nuclear factor kappa-B ligand (RANKL), secreted frizzled-related protein 1 (SFRP1) , and leptin [25].

Table 1.
Overview of main epigenetic modifications. DNA methylation, histone modification, and micro RNA (miRNA) production influence DNA expression through chromatin modification and post transcriptional gene repression.
Histones, alkaline proteins that package the DNA into structural units, can be modified by various post-translational modifications. Acetylation of a core histone results in a more open chromatin structure that facilitates gene expression. Histone deacetylation causes the condensation of chromatin and inhibits gene transcription. Histone methylation can either result in an activated or repressed chromatin state [26]. Histone deacetylases (HDAC), highly sensitive to histone modification, strongly influence bone through the suppression of osteoblast differentiation [27,28], the promotion of RANKL-induced osteoclastogenesis [29], the maintenance of bone mass during development and aging [30], and many others.
miRNAs are brief sequences of ncRNAs composed of 18–22 nucleotides [31,32]. miRNA pathways regulate gene expression by inducing degradation and/or translational repression of target mRNAs. If miRNA production increases, levels of target mRNAs decrease and the gene expression is repressed. In the same way, if the miRNA levels decrease, gene expression is upregulated [31]. Numerous miRNAs have been demonstrated to directly regulate osteoblastogenesis and osteoclastogenesis, for example, miR-23a, miR-34c, miR-133a, miR-135a, miR-137, miR-204, miR-205, miR-211, miR-217, miR-218, miR-335, miR-338, miR-433, and miR-3077-5p target runt-related transcription factor-2 (RunX2), the master transcription factor in osteoblast differentiation [33,34,35].
Several authors have investigated the role of epigenetics in chronic or aggressive periodontitis [36,37,38], and have demonstrated that the expression levels of cytokines and chemokines [39], toll-like receptors [40], protease-activated receptors [41], interleukin-8 [42], and cyclooxygenase-2 [37,38] could be affected by oral bacteria. A recent study documented that the presence of different oral bacteria resulted in a differential methylation profile in gingival epithelia [43], while another article demonstrated hypomethylated oral epithelia in patients with generalized aggressive periodontitis [36].
Despite available data regarding epigenetics and periodontology, knowledge on epigenetics related to implant dentistry is weak. Therefore, the aim of this review was to evaluate the available evidence investigating the potential effects of DNA methylations, histone modifications, or micro-RNA production on dental implant therapy.
2. Materials and Methods
This review was conducted according to Cook’s principles with a pre-planned method and using explicit and reproducible criteria [44]. Potentially relevant articles were investigated in a comprehensive search. All presented data were appraised, synthesized, interpreted, and discussed [44].
2.1. Search Strategy
Systematic bibliographical electronic research was carried out selecting all potentially relevant publications dealing with the influences of implant surface features on gene activation and the influence of epigenetic changes on implant therapy outcomes.
The following terms and Boolean connectors were used: ((epigenetics) OR (DNA methylation) OR (DNA methyl transferase) OR (histone deacetylation) OR (histone deacetylase) OR (histone methyl transferase) OR (histone demethylase) OR (micro-rna)) AND ((dental implant) OR (dental implants) OR (implantology) OR (implant dentistry) OR (implant failure) OR (platform shifting) OR (platform switching) OR (implant-abutment connection) OR (osseointegration) OR (mucositis) OR (perimplantitis)).
The electronic outcome after query translation is listed in the Appendix A.
2.2. Selection of Studies
2.2.1. Inclusion Criteria
All scientific in vivo and in vitro publications investigating the impact of genetic expression levels on implant rehabilitations were included. According to Mulrow [45], studies with both direct and indirect evidence were also included. No filters like language or time limitation were applied. The last update for new published articles was done on 1 March 2017.
2.2.2. Exclusion Criteria
Published studies that did not meet the inclusion criteria and did not provide information concerning dental implant therapy, upregulation or downregulation of genes or their products were excluded. Scientific articles resulting from the use of confounding words, such as those investigated mucositis different from peri-implant mucositis, or orthopedic prosthetic joint, were excluded.
2.3. Development of the Review
This study was conducted in three phases. The first phase involved screening titles and abstracts. The second phase consisted of screening full-text articles. The final phase consisted of a review of included articles. For each included study, data regarding activated genes, influences of implant materials on gene expression, types of implant materials, types of surface modifications, and number of patients or types of cells studied were rewritten.
3. Results
The electronic search found 67 articles (Figure 1). During the first phase of screening, 18 studies were excluded as they did not address the existing connections between epigenetics and implant dentistry. Their main fields of interest were: oral mucositis during radiotherapy or chemotherapy, mucositis in patients with neoplastic diseases like cancer or leukemia, tolerability of temsirolimus, graft-versus-host disease, fluorouracil toxicity, hypomethylating agent therapy for neoplastic diseases, and radiation-induced tissue damages.
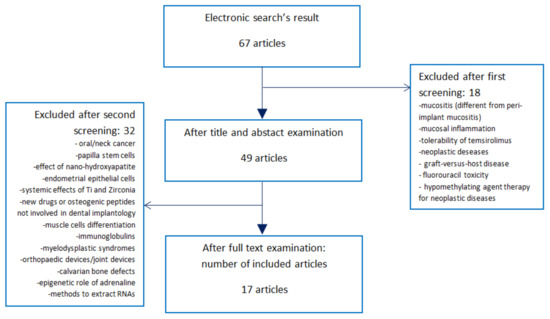
Figure 1.
Search strategy, screening for eligibility, and final number of included publications. The electronic search found 67 studies regarding changes in gene expression and implantology. After title and abstract screening, 18 of them were excluded as they focused mainly on mucositis after radio/chemotherapy, mucosal inflammation, neoplastic diseases, hypomethylating agent therapy for neoplastic diseases, and radiation-induced tissue damages. Forty-nine articles were downloaded and studied. Thirty-two of them were excluded as they did not investigate the effects of DNA methylations, histone modifications or micro-RNA (miRNA) production on implant survival, osseointegration, peri-implant mucositis, perimplantitis, or implant-abutment leakage. Thus, this systematic review was finalized with 17 articles.
After the first screening, 49 articles were downloaded and studied (Figure 1). During the second phase of screening, 32 articles were excluded as they did not investigate the effects of gene expression changes on implant therapy, but evaluated data regarding oral or neck cancer (seven articles); papilla stem cells (one article); nano-hydroxyapatite (one article); endometrial epithelial cells (one article); systemic influence of titanium and zirconia (one article); chemical drugs for systemic diseases (one article); osteogenic peptides, but not associated with implant surfaces (two articles); muscle cells (one article); immunoglobulins (one article); myelodysplastic syndromes (one article); orthopedic defects (five articles); critical limb ischemia (one article); cementoblasts (one article); bone-ligament cells (one article); calvarial bone defect (one article); genetic effects of adrenaline (one article); evaluation of systemic miRNAs (one article); or publications without information regarding gene expression (one article), or aimed to discuss different methods to detect RNAs from cells (three articles).
Thus, this review was based on 17 scientific articles (Figure 2). All 17 included articles were evaluated during phase three. Publication types of the included articles are expressed in Figure 2. Fifteen of 17 articles were in vitro or in vivo studies [46,47,48,49,50,51,52,53,54,55,56,57,58,59,60]. One was a randomized clinical trial with immune-histochemical analysis [61], and the other was a narrative review [62].
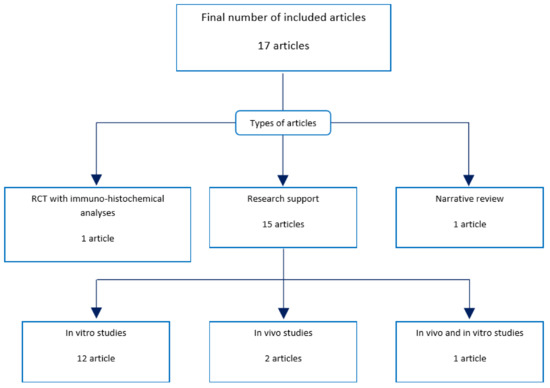
Figure 2.
Types of included articles. Out of the 17 included articles, 12 were in vitro studies; two were in vivo studies; one was both an in vivo and in vitro study; one was a randomized controlled trial (RCT) with immune-histochemical and RNA analyses; and one was a narrative review. All 17 articles were reviewed.
3.1. In Vivo and In Vitro Studies
Fifteen of the 17 articles included publications aimed to compare changes in the gene expression profiles of cells cultured on different implant surfaces [46,47,48,49,50,51,52,53,54,55,56,57,58,59,60] (Table 2).

Table 2.
Effects of different surfaces, effects of two different clinical treatments, and effects of smoking and diabetes on genetic expression.
Eight articles documented miRNA production [47,51,52,53,55,56,57,58]. Seven articles did not provide information on miRNA production [46,48,49,50,54,59,60]. No articles reported information on chromatin alteration due to DNA methylation or histone modification.
3.2. Cell Population
A large variability of cell selection was tested. Regarding in vitro studies, four studies used human osteoblast like cells (MG-63) for cell culture [48,53,56,57] while the other four used human alveolar stem cells from human donors [51,52,55,59]. Human mesenchymal stem cells [60], dental pulp stem cells [47], rat bone marrow cells [58], and marrow stromal cells obtained from the iliac crest [46] were used in one study each.
Two were in vivo studies aimed at investigating genetic expression levels of implant adherent cells taken from the tibias of rats [54] and humans [49].
Finally, one study was both an in vitro and in vivo study using MG-63 cells and implant adherent cells from the tibias of beagle dogs [50].
3.3. Implant Surfaces
A huge variety of surfaces and their effects on gene activation were tested. With reference to in vitro studies, KOH alkali-etched surfaces enhanced bone sialo protein and matrix metalloprotease 2 levels [46]. Ionized surfaces induced higher levels of Runx-2, Smad5, and osteocalcin [47]. Electrolytic etched surfaces were associated with higher levels of alkaline phosphatase, osteocalcin, Runx-2, osteopontin, and collagen type Iα1 [48]. Sandblasted and acid-etched (SLA) and modSLA surfaces downregulated miRNAs influencing osteoprogenitor cells [51]. CS/HA/miR-21 surfaces increased the production levels of collagen type IIIα1, osteocalcin, Runx-2, osteopontin, and collagen type Iα1 [52]. Zirconia surfaces inhibited the production of bone morphogenic protein-4 and -7 [53]. Nanotextured surfaces showed the highest alkaline phosphatase production, while the microtextured surface group had the greatest amount of calcium and mineralized nodules [55]. Zirconia surfaces upregulated 18 miRNAs involved in the repression of osteogenic genes [56]. Anatase surfaces upregulated nine miRNAs with repression of collagen 9α2, disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS4), and alkaline phosphatase [57]. The anti-miR-138 functionalized surface induced higher expression of bone morphogenic protein, osteocalcin, osterix, and Runx2. The miR-29b functionalized surface induced a higher expression of alkaline phosphatase and collagen type Iα1 [58]. Macro-sandblasted surfaces had higher levels of TGFβ2, osteopontin, Runx-2, and bone sialoprotein [59]. The Ti6A14V#9 surface showed greater alkaline phosphatase (ALP), osteocalcin, vascular endothelial growth factor-A (VEGF-A), fibroblast growth factor-2 (FGF-2), bone morphogenic protein, and osteoprotegerin production [60]. SLA surfaces with 80 nm of nanotubes were shown to induce higher formation of filopodia, lamellipodia, cellular extensions, and gene expression [50].
With reference to in vivo studies, in one study, TiOblast and osseospeed implants were placed in human smoking and nonsmoking patients. Implants were harvested after two and four days of submerged healing, and implant adherent cells were studied [49]. Interestingly, the authors found that the time variable influenced gene expression more that the effect of different surfaces or nicotine. Similar trends in gene expression were noted in implant-adherent cells regardless of implant surface and smoking status. In another study, nano- or microroughened implants were placed in the tibias of rats and harvested after two and four days of submerged healing [54]. Significant differences at the gene level were not noted when the two implant surfaces were compared at each timepoint. However, genes were differentially regulated at different days for both implant surfaces.
3.4. Comparative Analysis
Nine studies found that specific surfaces induced osteoblast differentiation, expression of osteogenic genes or repression of the miRNAs that downregulate osteogenic genes [46,47,48,50,52,53,58,59,60], while six articles did not indicate a better surface [49,51,54,55,56,57]. Two articles documented similar trends in gene expression regardless of implant surface and found that the effect of time influenced gene expression more than the surfaces [49,54]. Only two publications evaluated the genetic effect of zirconia surfaces. In the first study, zirconia surfaces were associated with worse genetic activation than machined titanium ones [53]. In the second, the number of miRNAs upregulated was much higher than the number of miRNAs downregulated [56].
3.5. Randomized Clinical Trial
One article out of the 17 was a randomized clinical trial [61] (Table 2). Twenty-six patients with one buccal implant dehiscence defect were each randomly treated with particulate allograft bone (control) or particulate allograft bone and pericardium membrane (test). After six months of healing, an analysis of bone volume and gene expression was performed. Greater volume levels were found in the test group than in the control group. Bone biopsies were harvested and processed. Genetic expression of osteogenic genes was evaluated with an immune-histochemical analysis. Positive periostin, sclerostin, and Runx2 immunoreactivities were detected in both the control and test groups without statistical differences. Tartrate-resistant acid phosphatase (TRAP) positives were mostly noted in the control group. Analysis of DNA methylation, histone modifications, and miRNA production was not provided.
3.6. Narrative Review
A narrative review investigating the epigenetic effect of smoking and diabetes on osseointegration [62] was also included in this study. Findings indicated that global DNA methylation is influenced by smoking behavior (Table 2). Smoking had an impact on bone metabolism and estrogen production, leading to a phenotype of low-bone mineral density. In fact, gene expression of bone matrix proteins, including osteopontin, Type II collagen, bone morphogenetic protein-2, and osteoprotegerin resulted in significant downregulation by smoking components. Diabetes was associated with the decreased gene expression of bone matrix proteins like osteocalcin and parathyroid hormone-related protein (PTHrP), downregulation of transcription factors involved in osteoblast differentiation like Runx2 and osterix, and repression of osteoprotegerin through the activation of RANKL [63]. Histone lysine methylation and other post transcriptional changes were implicated in aberrant gene regulation associated with the pathology of diabetes and its complications. Moreover, serum osteocalcin levels were found to be significantly increased in patients with Type II diabetes [64]. As a result, alteration of bone healing in diabetes and epigenetic modifications are deeply connected.
4. Discussion
Genetic information is encoded not only by the linear sequence of DNA, but by epigenetic modifications [12] such as DNA methylation, histone modification, and miRNA production. The purpose of this review was to investigate the available literature dealing with the role of epigenetics in implant treatment.
Even though the 17 included articles documented changes in gene expression due to different implant surfaces or different clinical treatments, only eight studies documented how these changes were affected by miRNAs [47,51,52,53,55,56,57,58]. The narrative review investigated the effects of global DNA methylation and histone modification [62]. More articles performed miRNA analysis than other epigenetic analyses because of the more precise results due to the functional link between specific miRNAs and target mRNAs. Some included articles showed an alteration in protein production without explaining the genetic processes behind them. Factors other than epigenetic modifications could also contribute to up- or downregulating protein production. The presence of 3D scaffolds might play a role in the induction of cells differentiation [65]. Mechanical perturbation of osteoblasts may enhance bone sialoprotein (BSP) levels [66]. Cell differentiation and protein production could also be influenced by additives and factors used in the medium [47]. However, cells cultured on titanium disks showed an early differentiation processes, influenced by the macro/micro/nano structure, regardless of the presence of dexamethasone, glycerol phosphate, and ascorbic acid in the medium [47,60].
Zirconia and anatase surfaces increased the production of the miRNAs that downregulate osteogenic genes [53,56,57]. Thus, zirconia and anatase surfaces are not eligible to genetically stimulate new deposition of mineralized bone. Alkali-etching treatments [46], titanium-alloy [60], ionization [47], electrolytic etching [48], surfaces with nanotubes [50], N2 treatments [51], and surfaces functionalized with miRNAs [52,58] stimulated the differentiation of mesenchymal cells in osteoblasts. Moreover, they enhanced the activation of osteogenic genes. Micro-sandblasted, macro-sandblasted [59], SLA, and electrolytic etched [48] surfaces upregulated osteogenic genes more than machined ones. As zirconia showed worse results than the machined surfaces [53], it seems that micro-sandblasted, macro-sandblasted, SLA, and electrolytic etched surfaces also upregulate osteogenic genes more than zirconia.
Interestingly, the results from in vitro studies are different from those resulting from clinical trials. A systematic review investigating zirconia clinical outcomes reported high survival rates [67] after one year of follow-up. On the other hand, results of in vitro studies did not necessarily agree with clinical outcomes that can be influenced by other factors. For example, the passage of time could influence the cells’ phenotype much more than implant surfaces [49]. However, a recent review on zirconia implant outcomes concluded that early failure rate of zirconia implants was higher when compared with titanium ones [68].
Regarding the types of cells used, most of the reported data derived from studies were performed on osteoblast-like cells (MG-63) or animal cells, which are not normal human osteoblasts. Notwithstanding this, the advantages of using MG-63 cells are related to the fact that the reproducibility of data is higher. Primary human cell cultures provide a source of human cells, but also contain contaminating cells of different types and cells in variable differentiation states. Furthermore, most of the reviewed studies were in vitro studies that were not influenced by interindividual variability of human patients; indeed, adrenaline released after psychological stress may inhibit osteogenic differentiation through histone acetylation and downregulation of miR-2 [20]. On the other hand, in vivo human studies require a large sample and long follow-up for a better comprehension of epigenetic influences.
Regarding the influence of time, similar trends in gene expression were noted in implant-adherent cells regardless of implant surface if they were evaluated at the same early time point [49,54]. However, when the time-course was evaluated, statistical differences in genetic patterns were identified. Therefore, the influence of time affected gene expression more than different surfaces. Regarding smoking, one in vivo study noted that the impact of smoking did not occur at early time points when all implants were submerged [49]. As the absorption of nicotine through the oral mucosal tissues is pH dependent, given that the pH of tobacco smoke in most cigarettes is acidic, nicotine is primarily ionized, resulting in minimal absorption of nicotine from cigarette smoke. These data indicated that the gene expression profiles of submerged implant adherent cells were similar among smokers and non-smokers.
Articles investigating the epigenetic effects of orthopedic implant surfaces showed similar results to those investigating the effects of dental implants surfaces. Some miRNAs induced the reduction of the expression of VEGF (a key regulator of inflammatory osteoclastogenesis correlated to the aseptic loosening of orthopedic implants), and the use of miRNAs could be used to safely lower inflammation [69]. In two of the included articles, miRNAs were loaded to dental implant surfaces to selectively activate or repress genes. Surfaces functionalized with miRNAs were shown to upregulate osteogenic genes more than non-functionalized ones [52,58]. Furthermore, five of the included articles tested surfaces used for dental and orthopedic rehabilitations [46,50,51,55,60].
According to Cook’s criteria, the present review is a systematic qualitative review [44] because of the expressed methodology and reproducible results. The large heterogeneity of types of cells, time point, surfaces used, and genes tested did not allow for meta-analysis. Only indirect evidence was possible to evaluate [45] based on Mulrow indications.
Despite current difficulty in defining which implant surface is best to genetically stimulate osteogenesis, there is evidence that implant surface features can upregulate or downregulate genes related to osseointegration. The importance of epigenetic mechanisms will largely influence the industry and the clinical application of implant treatment. As reported by Williams et al. [18], epigenetics is a new frontier in dentistry.
5. Conclusions
This systematic qualitative review showed that genetic stimulation of osteogenic genes was performed by surface treatments like alkali-etching, ionization, electrolytic etching, surfaces with nanotubes, isotonic solution, and N2 treatments. Osteogenic inhibition was associated with zirconia surfaces and anatase coatings. Micro and nanoporous surfaces may provide a larger area for loading miRNAs, anti-miRNAs, peptides, or other osteogenic drugs.
Author Contributions
Riccardo Di Gianfilippo worked on the research of studies, selection of studies, critical analysis, and writing. Carmine Di Gianfilippo worked on the research and selection of studies. Giovan Paolo Pini Prato supervised the phase of research and selection, worked on the critical analysis of included studies, and contributed to the writing. The systematic bibliographical electronic research was carried out independently by two authors (R.D.G. and C.D.G.). Two authors (R.D.G. and C.D.G.) worked independently and compared their results about research and selection of studies. One author (G.P.P.) supervised each phase in the role of quality control. Two authors (R.D.G. and G.P.P.) worked on the critical analysis of the included studies.
Conflicts of Interest
The authors declared no conflict of interest.
Abbreviations
List of Abbreviated Surfaces
| SLA | sandblasted acid-etched titanium surface |
| modSLA | SLA surface with an N2 protection and stored in an isotonic saline solution |
| MAO | microarch-oxidated titanium |
| CH/HA/mR-21 | chitosan/hyaluronic acid surface with miRNA-21 |
| Ti6A14V | micron-scale rough titanium alloy |
| EE | micro/nanostructured surface electrolytic etched |
| M | machined surface |
| TiO | TiOBlast; surface blasted with TiO2 |
| OS | osseospeed; surface blasted with TiO2 then treated with hydrofluoric acid |
| SMO | smooth polished surface |
| AT-1 | oxalic acid and hydrofluoric acid treated surface |
| AT-2 | oxalic acid treated surface |
| Ti6A14V | micron-scale rough titanium alloy (# indicates different dimension of roughness parameters) |
| TCPS | polystyrene surface |
List of Abbreviated Genes and Proteins
| ALP | alkaline phosphatase |
| BMP | bone morphogenic protein |
| BSP | bone sialoprotein |
| Runx | runt-related transcription factor |
| OCN | Osteocalcin |
| PTHrp | parathyroid hormone-related protein |
| PTH | parathyroid hormone |
| BIC | bone-implant contact |
| POSTN | periostin related factor |
| VEGF | vascular endothelial growth factor |
| COL1 | collagen type Iα1 |
| COL3 | collagen type IIIα1 |
| OPN | Osteopontin |
| FGF | fibroblast growth factor |
| ITGA | integrin subunit |
| SHOX | short stature homeobox-containing gene |
| IGF | insulin-like grow factor |
| NOG | noggin gene |
| PRDX | Peroxiredoxin |
| ADAMTS | gene encoding for disintegrin and metalloproteinase with thrombospondin motifs |
| AMBN | Ameloblastin |
| PHEX | phosphate-regulating neutral endopeptidase |
| FBN | Fibrillin |
| CALCA | calcitonin related polypeptide alpha |
| TFIP | tissue factor pathway inhibitor |
| OSX | osteoblast-specific transcription factor, osterix |
Appendix A
Query translation of the typed formula was automatically calculated by PubMed’s electronic program as:
(("epigenomics"[MeSH Terms] OR "epigenomics"[All Fields] OR "epigenetics"[All Fields]) OR ("dna methylation"[MeSH Terms] OR ("dna"[All Fields] AND "methylation"[All Fields]) OR "dna methylation"[All Fields]) OR (("dna"[MeSH Terms] OR "dna"[All Fields]) AND methyl[All Fields] AND ("transferases"[MeSH Terms] OR "transferases"[All Fields] OR "transferase"[All Fields])) OR (("histones"[MeSH Terms] OR "histones"[All Fields] OR "histone"[All Fields]) AND deacetylation[All Fields]) OR ("histone deacetylases"[MeSH Terms] OR ("histone"[All Fields] AND "deacetylases"[All Fields]) OR "histone deacetylases"[All Fields] OR ("histone"[All Fields] AND "deacetylase"[All Fields]) OR "histone deacetylase"[All Fields]) OR (("histones"[MeSH Terms] OR "histones"[All Fields] OR "histone"[All Fields]) AND methyl[All Fields] AND ("transferases"[MeSH Terms] OR "transferases"[All Fields] OR "transferase"[All Fields])) OR (("histones"[MeSH Terms] OR "histones"[All Fields] OR "histone"[All Fields]) AND demethylase[All Fields]) OR ("micrornas"[MeSH Terms] OR "micrornas"[All Fields] OR ("micro"[All Fields] AND "rna"[All Fields]) OR "micro rna"[All Fields])) AND (("dental implants"[MeSH Terms] OR ("dental"[All Fields] AND "implants"[All Fields]) OR "dental implants"[All Fields] OR ("dental"[All Fields] AND "implant"[All Fields]) OR "dental implant"[All Fields]) OR ("dental implants"[MeSH Terms] OR ("dental"[All Fields] AND "implants"[All Fields]) OR "dental implants"[All Fields]) OR implantology[All Fields] OR ("Implant Dent"[Journal] OR ("implant"[All Fields] AND "dentistry"[All Fields]) OR "implant dentistry"[All Fields]) OR (implant[All Fields] AND failure[All Fields]) OR (platform[All Fields] AND shifting[All Fields]) OR (platform[All Fields] AND switching[All Fields]) OR (implant-abutment[All Fields] AND connection[All Fields]) OR ("osseointegration"[MeSH Terms] OR "osseointegration"[All Fields]) OR ("mucositis"[MeSH Terms] OR "mucositis"[All Fields]) OR perimplantitis[All Fields]).
References
- Berglundh, T.; Persson, L.; Klinge, B. A systematic review of the incidence of biological and technical complications in implant dentistry reported in prospective longitudinal studies of at least 5 years. J. Clin. Periodontol. 2002, 29 (Suppl. S3), 197–212. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, A.; Pohler, O.; Sutter, F. Tissue reaction to an implant of a titanium hollow cylinder with a titanium surface spray layer. SSO Schweiz. Monatsschr. Zahnheilkd. 1976, 86, 713–727. [Google Scholar] [PubMed]
- Adell, R.; Lekholm, U.; Rockler, B.; Brånemark, P.I. A 15-year study of osseointegrated implants in the treatment of the edentulous jaw. Int. J. Oral Surg. 1981, 10, 387–416. [Google Scholar] [CrossRef]
- Roos-Jansåker, A.M.; Lindahl, C.; Renvert, H.; Renvert, S. Nine to fourteen-year follow-up of implant treatment. Part I: Implant loss and associations to various factors. J. Clin. Periodontol. 2006, 33, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Roos-Jansåker, A.M.; Lindahl, C.; Renvert, H.; Renvert, S. Nine to fourteen-year follow-up of implant treatment. Part II: Presence of peri-implant lesions. J. Clin. Periodontol. 2006, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Lindhe, J.; Meyle, J. Peri-implant diseases: Consensus Report of the Sixth European Workshop on Periodontology. J. Clin. Periodontol. 2008, 35 (Suppl. S8), 282–285. [Google Scholar] [CrossRef] [PubMed]
- Broggini, N.; McManus, L.M.; Hermann, J.S.; Medina, R.; Schenk, R.K.; Buser, D.; Cochran, D.L. Peri-implant inflammation defined by the implant-abutment interface. J. Dent. Res. 2006, 85, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Piattelli, A.; Vrespa, G.; Petrone, G.; Iezzi, G.; Annibali, S.; Scarano, A. Role of the microgap between implant and abutment: A retrospective histologic evaluation in monkeys. J. Periodontol. 2003, 74, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Mombelli, A.; Buser, D.; Lang, N.P. Colonization of osseointegrated titanium implants in edentulous patients. Early results. Oral Microbiol. Immunol. 1988, 3, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Mombelli, A. Microbiology and antimicrobial therapy of peri-implantitis. Periodontology 2000 2002, 28, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Esposito, M.; Thomsen, P.; Ericson, L.E.; Lekholm, U. Histopathologic observations on early oral implant failures. Int. J. Oral Maxillofac. Implant. 1999, 14, 798–810. [Google Scholar]
- Esposito, M.; Hirsch, J.; Lekholm, U.; Thomsen, P. Differential diagnosis and treatment strategies for biologic complications and failing oral implants: A review of the literature. Int. J. Oral Maxillofac. Implant. 1999, 14, 473–490. [Google Scholar]
- Salcetti, J.M.; Moriarty, J.D.; Cooper, L.F.; Smith, F.W.; Collins, J.G.; Socransky, S.S.; Offenbacher, S. The clinical, microbial, and host response characteristics of the failing implant. Int. J. Oral Maxillofac. Implant. 1997, 12, 32–42. [Google Scholar]
- Berger, S.L.; Kouzarides, T.; Shiekhattar, R.; Shilatifard, A. An operational definition of epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar] [CrossRef] [PubMed]
- Martos, S.N.; Tang, W.Y.; Wang, Z. Elusive inheritance: Transgenerational effects and epigenetic inheritance in human environmental disease. Prog. Biophys. Mol. Biol. 2015, 118, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Fazzari, M.J.; Greally, J.M. Epigenomics: Beyond CpG islands. Nat. Rev. Genet. 2004, 5, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Rothstein, M.A.; Cai, Y.; Marchant, G.E. The ghost in our genes: Legal and ethical implications of epigenetics. Health Matrix Clevel. 2009, 19, 1–62. [Google Scholar] [PubMed]
- Williams, S.D.; Hughes, T.E.; Adler, C.J.; Brook, A.H.; Townsend, G.C. Epigenetics: A new frontier in dentistry. Aust. Dent. J. 2014, 59 (Suppl. S1), 23–33. [Google Scholar] [CrossRef] [PubMed]
- Barros, S.P.; Offenbacher, S. Epigenetics: Connecting environment and genotype to phenotype and disease. J. Dent. Res. 2009, 88, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wang, Z. Adrenaline inhibits osteogenesis via repressing miR-21 expression. Cell Biol. Int. 2017, 41, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Bishop, K.S.; Ferguson, L.R. The interaction between epigenetics, nutrition and the development of cancer. Nutrients 2015, 7, 922–947. [Google Scholar] [CrossRef] [PubMed]
- Egger, G.; Liang, G.; Aparicio, A.; Jones, P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004, 429, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Liang, G. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 2009, 10, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Hark, A.T.; Schoenherr, C.J.; Katz, D.J.; Ingram, R.S.; Levorse, J.M.; Tilghman, S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 2000, 405, 486–489. [Google Scholar] [PubMed]
- Vrtačnik, P.; Marc, J.; Ostanek, B. Epigenetic mechanisms in bone. Clin. Chem. Lab. Med. 2014, 52, 589–608. [Google Scholar] [CrossRef] [PubMed]
- Bayarsaihan, D. Epigenetic mechanisms in inflammation. J. Dent. Res. 2011, 90, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Razidlo, D.F.; Whitney, T.J.; Casper, M.E.; McGee-Lawrence, M.E.; Stensgard, B.A.; Li, X.; Secreto, F.J.; Knutson, S.K.; Hiebert, S.W.; Westendorf, J.J. Histone deacetylase 3 depletion in osteo/chondroprogenitor cells decreases bone density and increases marrow fat. PLoS ONE 2010, 5, e11492. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Alliston, T.; Delston, R.; Derynck, R. Repression of Runx2 function by TGF-β through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005, 24, 2543–2555. [Google Scholar] [CrossRef] [PubMed]
- Harakalova, M.; Van den Boogaard, M.J.; Sinke, R.; Van Lieshout, S.; Van Tuil, M.C.; Duran, K.; Renkens, I.; Terhal, P.A.; De Kovel, C.; Nijman, I.J.; et al. X-exome sequencing identifies a HDAC8 variant in a large pedigree with X-linked intellectual disability, truncal obesity, gynaecomastia, hypogonadism and unusual face. J. Med. Genet. 2012, 49, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Suh, J.H.; Kim, A.Y.; Lee, Y.S.; Park, S.Y.; Kim, J.B. Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Mol. Endocrinol. 2006, 20, 2432–2443. [Google Scholar] [CrossRef] [PubMed]
- Shookhoff, J.M.; Gallicano, G.I. The emerging role of microRNAs in adult stem cells. In Stem Cell Biology and Regenerative Medicine; Turksen, K., Ed.; Humana Press: New York, NY, USA; Springer: Berlin, Germany, 2011; Volume 1, pp. 57–94. [Google Scholar]
- Fakhry, M.; Hamade, E.; Badran, B.; Buchet, R.; Magne, D. Molecular mechanisms of mesenchymal stem cell differentiation towards osteoblasts. World J. Stem Cells 2013, 5, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xie, R.L.; Croce, C.M.; Stein, J.L.; Lian, J.B.; Van Wijnen, A.J.; Stein, G.S. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc. Natl. Acad. Sci. USA 2011, 108, 9863–9868. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, L.; Xing, L.; Chen, D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells 2010, 28, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Kang, I.H.; Lee, J.W.; Jang, W.G.; Koh, J.T. MiR-433 mediates ERRγ-suppressed osteoblast differentiation via direct targeting to Runx2 mRNA in C3H10T1/2 cells. Life Sci. 2013, 92, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Andia, D.C.; De Oliveira, N.F.; Casarin, R.C.; Casati, M.Z.; Line, S.R.; De Souza, A.P. DNA methylation status of the IL-8 gene promoter in aggressive periodontitis. J. Periodontol. 2010, 81, 1336–1341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Barros, S.P.; Niculescu, M.D.; Moretti, A.J.; Preisser, J.S.; Offenbacher, S. Alteration of PTGS2 promoter methylation in chronic periodontitis. J. Dent. Res. 2010, 89, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Crivello, A.; Offenbacher, S.; Moretti, A.; Paquette, D.W.; Barros, S.P. Interferon-gamma promoter hypomethylation and increased expression in chronic periodontitis. J. Clin. Periodontol. 2010, 37, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.O.; An, J.Y.; Yin, L.; Hacker, B.M.; Rohani, M.G.; Dommisch, H.; DiJulio, D.H. Interplay of protease activated receptors and NOD pattern recognition receptors in epithelial innate immune responses to bacteria. Immunol. Lett. 2010, 131, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Darveau, R.P.; Pham, T.T.; Lemley, K.; Reife, R.A.; Bainbridge, B.W.; Coats, S.R.; Howald, W.N.; Way, S.S.; Hajjar, A.M. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect. Immun. 2004, 72, 5041–5051. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.O.; Hansen, S.R.; Rao, D.; Dale, B.A. Protease-activated receptor signaling increases epithelial antimicrobial peptide expression. J. Immunol. 2004, 173, 5165–5170. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, N.F.; Damm, G.R.; Andia, D.C.; Salmon, C.; Nociti, F.H., Jr.; Line, S.R.; De Souza, A.P. DNA methylation status of the IL8 gene promoter in oral cells of smokers and non-smokers with chronic periodontitis. J. Clin. Periodontol. 2009, 36, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Chung, W.O. Epigenetic regulation of human ß-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria. Mucosal Immunol. 2011, 4, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.J.; Mulrow, C.D.; Haynes, R.B. Systematic reviews: Synthesis of best evidence for clinical decisions. Ann. Intern. Med. 1997, 126, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Mulrow, C.; Langhorne, P.; Grimshaw, J. Integrating heterogeneous pieces of evidence in systematic reviews. Ann. Intern. Med. 1997, 127, 989–995. [Google Scholar] [CrossRef] [PubMed]
- Giannoni, P.; Muraglia, A.; Giordano, C.; Narcisi, R.; Cancedda, R.; Quarto, R.; Chiesa, R. Osteogenic differentiation of human mesenchymal stromal cells on surface-modified titanium alloys for orthopedic and dental implants. Int. J. Artif. Organs 2009, 32, 811–820. [Google Scholar] [PubMed]
- Iaculli, F.; Di Filippo, E.S.; Piattelli, A.; Mancinelli, R.; Fulle, S. Dental pulp stem cells grown on dental implant titanium surfaces: An in vitro evaluation of differentiation and microRNAs expression. J. Biomed. Mater. Res. B Appl. Biomater. 2017, 105, 953–965. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.; Zhou, Y.; Zhang, Y.; Cai, Q.; Yang, L.; Wang, B. Effects of hierarchical micro/nano-textured titanium surface features on osteoblast-specific gene expression. Implant Dent. 2013, 22, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Thalji, G.; Cooper, L.F.; Nares, S. Gene Expression Profiles of Early Implant Adherent Cells in Smokers and Nonsmokers. J. Oral Implantol. 2015, 41, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Zhou, L.; Wang, J.; Zhao, Q.; Lin, X.; Gao, Y.; Li, S.; Wu, J.; Rong, M.; Guo, Z.; et al. The effects of hierarchical micro/nanosurfaces decorated with TiO2 nanotubes on the bioactivity of titanium implants in vitro and in vivo. Int. J. Nanomed. 2015, 10, 6955–6973. [Google Scholar]
- Chakravorty, N.; Ivanovski, S.; Prasadam, I.; Crawford, R.; Oloyede, A.; Xiao, Y. The microRNA expression signature on modified titanium implant surfaces influences genetic mechanisms leading to osteogenic differentiation. Acta Biomater. 2012, 8, 3516–3523. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, G.; Feng, Z.; Bai, S.; Dong, Y.; Wu, G.; Zhao, Y. Microarc-oxidized titanium surfaces functionalized with microRNA-21-loaded chitosan/hyaluronic acid nanoparticles promote the osteogenic differentiation of human bone marrow mesenchymal stem cells. Int. J. Nanomed. 2015, 10, 6675–6687. [Google Scholar]
- Palmieri, A.; Pezzetti, F.; Brunelli, G.; Lo Muzio, L.; Scarano, A.; Scapoli, L.; Martinelli, M.; Arlotti, M.; Guerzoni, L.; Rubini, C.; et al. Short-period effects of zirconia and titanium on osteoblast microRNAs. Clin. Implant Dent. Relat. Res. 2008, 10, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Thalji, G.; Gretzer, C.; Cooper, L.F. Comparative molecular assessment of early osseointegration in implant-adherent cells. Bone 2013, 52, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Wimmers Ferreira, M.R.; Rodrigo Fernandes, R.; Freire Assis, A.; Dernowsek, J.A.; Passos, G.A.; Variola, F.; Fittipaldi Bombonato-Prado, K. Oxidative nanopatterning of titanium surface influences mRNA and MicroRNA expression in human alveolar bone osteoblastic cells. Int. J. Biomater. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, A.; Pezzetti, F.; Brunelli, G.; Zollino, I.; Lo Muzio, L.; Martinelli, M.; Scapoli, L.; Arlotti, M.; Masiero, E.; Carinci, F. Zirconium oxide regulates RNA interfering of osteoblast-like cells. J. Mater. Sci. Mater. Med. 2008, 19, 2471–2476. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, A.; Pezzetti, F.; Brunelli, G.; Arlotti, M.; Lo Muzio, L.; Scarano, A.; Rubini, C.; Sollazzo, V.; Massari, L.; Carinci, F. Anatase nanosurface regulates microRNAs. J. Craniofac. Surg. 2008, 19, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Song, W.; Zhao, L.; Liu, M.; Yan, J.; Andersen, M.Ø.; Kjems, J.; Gao, S.; Zhang, Y. MicroRNA functionalized microporous titanium oxide surface by lyophilization with enhanced osteogenic activity. ACS Appl. Mater. Interfaces 2013, 5, 2733–2744. [Google Scholar] [CrossRef] [PubMed]
- Marinucci, L.; Balloni, S.; Becchetti, E.; Belcastro, S.; Guerra, M.; Calvitti, M.; Lilli, C.; Calvi, E.M.; Locci, P. Effect of titanium surface roughness on human osteoblast proliferation and gene expression in vitro. Int. J. Oral Maxillofac. Implant. 2006, 21, 719–725. [Google Scholar]
- Olivares-Navarrete, R.; Hyzy, S.L.; Berg, M.E.; Schneider, J.M.; Hotchkiss, K.; Schwartz, Z.; Boyan, B.D. Osteoblast lineage cells can discriminate microscale topographic features on titanium-aluminum-vanadium surfaces. Ann. Biomed. Eng. 2014, 42, 2551–2561. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.H.; Rios, H.; Al-Hezaimi, K.; Oh, T.J.; Benavides, E.; Wang, H.L. A randomized clinical trial evaluating the efficacy of the sandwich bone augmentation technique in increasing buccal bone thickness during implant placement. II. Tomographic, histologic, immunohistochemical, and RNA analyses. Clin. Oral Implant. Res. 2015, 26, 1150–1157. [Google Scholar] [CrossRef] [PubMed]
- Razzouk, S.; Sarkis, R. Smoking and Diabetes Epigenetics Involvement in Osseointegration. N. Y. State Dent. J. 2013, 79, 27–30. [Google Scholar] [PubMed]
- Lozano, D.; De Castro, L.F.; Dapia, S.; Andrade-Zapata, L.; Manzarbeitia, F.; Alvarez-Arroyo, M.V.; Gómez-Barrena, E.; Esbrit, P. Role of parathyroid hormone-related protein in the decreased osteoblast function in diabetes-related osteopenia. Endocrinology 2009, 150, 2027–2035. [Google Scholar] [CrossRef] [PubMed]
- Ferron, M.; Wei, J.; Yoshizawa, T.; Del Fattore, A.; DePinho, R.A.; Teti, A.; Ducy, P.; Karsenty, G. Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism. Cell 2010, 142, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Mangano, C.; Paino, F.; d’Aquino, R.; De Rosa, A.; Iezzi, G.; Piattelli, A.; Laino, L.; Mitsiadis, T.; Desiderio, V.; Mangano, F.; et al. Human dental pulp stem cells hook into biocoral scaffold forming an engineered biocomplex. PLoS ONE 2011, 6, e18721. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.S.; Bumann, A.; Schaffer, J.L.; Gerstenfeld, L.C. Pre-dominant integrin ligands expressed by osteoblasts show preferential regulation in response to both cell adhesion and mechanical perturbation. J. Cell. Biochem. 2002, 84, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Pieralli, S.; Kohal, R.J.; Jung, R.E.; Vach, K.; Spies, B.C. Clinical Outcomes of Zirconia Dental Implants. J. Dent. Res. 2017, 96, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Cionca, N.; Hashim, D.; Mombelli, A. Zirconia dental implants: Where are we now, and where are we heading? Periodontology 2000 2017, 73, 241–258. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, Y.; Cheng, L. miR-21 expression is related to particle-induced osteolysis pathogenesis. J. Orthop. Res. 2012, 30, 1837–1842. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).