Abstract
Plant life-long organogenesis involves sequential, time and tissue specific expression of developmental genes. This requires activities of Polycomb Group (PcG) and trithorax Group complexes (trxG), respectively responsible for repressive Histone 3 trimethylation at lysine 27 (H3K27me3) and activation-related Histone 3 trimethylation at lysine 4 (H3K4me3). However, the genome-wide dynamics in histone modifications that occur during developmental processes have remained elusive. Here, we report the distributions of H3K27me3 and H3K4me3 along with expression changes, in a developmental series including Arabidopsis thaliana leaf and three stages of flower development. We found that chromatin mark levels are highly dynamic over the time series on nearly half of all Arabidopsis genes. Moreover, during early flower morphogenesis, changes in H3K4me3 prevail over changes in H3K27me3 and quantitatively correlate with expression changes, while H3K27me3 changes occur later. Notably, we found that H3K4me3 increase during the early activation of PcG target genes while H3K27me3 level remain relatively constant at the locus. Our results reveal that H3K4me3 predicts changes in gene expression better than H3K27me3, unveil unexpected chromatin mechanisms at gene activation and underline the relevance of tissue-specific temporal epigenomics.
1. Introduction
Morphogenesis in plants relies on continuous and iterative production of new organs with specific functions. In annual plants like Arabidopsis thaliana, upon transition to the reproductive phase, flower primordia are produced at the flanks of the inflorescence meristem. In these primordia, stem cell activity is re-established and the newly formed flower meristems produce founder cells for the different floral organs [1]. These cell-fate specifications coincide with changes in the expression pattern of developmental regulators [2]. The LEAFY (LFY), APETALA 1 (AP1) and CAULIFLOWER (CAL) transcription factors (TF) specify floral meristem identity via activation of downstream regulators, which include several genes encoding MADS domain TF. Spatially restricted expression of these genes then induces the differentiation of stem cells into specific cell types for formation of the different floral organs, as described in the ABC(D)E model [3]. Several reports on epigenetic mutants indicate that this regulation occurs through changes in the chromatin state of the corresponding loci, such as post-translational modifications of histones [4]. Histone 3 trimethylation at lysine 27 (H3K27me3) is a repressive modification targeting developmental regulators in all higher eukaryotes and is catalysed by Polycomb Group (PcG) proteins [5,6,7]. Genome-wide studies of H3K27me3 distribution in whole seedlings revealed that this mark targets more than one fourth of all Arabidopsis genes [8,9,10], comprising tissue- or stage-specific regulators of the vegetative to reproductive transition, floral meristem identity genes and all above-mentioned floral homeotic genes. Activation of PcG-repressed genes is conducted by the conserved trithorax Group (trxG) proteins, involved in three main activation processes: (i) removal of repressive H3K27me3 marks; (ii) deposition of activation-associated Histone 3 trimethylation at lysine 4 (H3K4me3) marks; and (iii) chromatin remodelling to allow access of the transcriptional machinery [4,11,12].
Thus, PcG and trxG antagonistically ensure the correct spatial and temporal expression of developmental regulators, such as those involved in flowering-time and flower development [13,14,15,16,17]. As a consequence, H3K27me3 and H3K4me3 chromatin marks should be highly dynamic throughout developmental stages and cell-types in plants. However, most genome-wide studies were so far performed for one developmental stage and often in a mixture of tissues during vegetative development, yielding only a static view of chromatin states. Some comparative studies used endosperm-specific nuclei [18], meristem-enriched tissue [19], roots [10], or mature versus senescing leaves [20,21]. An early study of comparing two developmental stages was conducted in 2009 by Charron et al. [22] comparing histone mark targeting in skotomorphogenic and photomorphogenic seedlings. All studies showed differential H3K27me3 marking between tissues or stages, at thousands of genes. Additionally, several hundreds of genes display quantitative changes in H3K27me3 intensity between undifferentiated and differentiated tissue, without losing the mark completely [19,23]. Several genome-wide analyses revealed an anti-correlation between changes in gene expression and changes in H3K27me3, and oppositely, a positive correlation for the H3K4me3 mark [19,20]. Studies at single gene level provided evidence that correlative changes in these histone marks are rather a cause than a consequence of the transcriptional state [24,25].
Nevertheless, many questions concerning the mechanistic relationship between H3K27me3 and H3K4me3 marks and gene expression remain open such as (i) the quantitative dynamics of histone mark between consecutive developmental stages; (ii) the correlation extent between changes in H3K27me3, H3K4me3 and gene expression; (iii) and the chronology of changes in H3K27me3, H3K4me3 and expression. To obtain answers to these questions, we used the Arabidopsis “cauliflower” inducible system, which allows collecting meristem tissue from synchronized floral stages, in sufficient amounts for genome-wide analyses of chromatin and expression [26,27,28,29]. Thus, the system enables both tissue and stage specific analysis of chromatin marks. We resolved the dynamics of the H3K4me3 and H3K27me3 marks and their correlation with expression changes in vegetative, inflorescence meristem, early developing flower and developed inflorescence tissues. This allowed establishing a comprehensive view of antagonistic histone modification dynamics during flower development in relation to gene activation or repression. We found that both chromatin marks are highly dynamic over development, with thousands of genes changing in their marking. H3K4me3 dynamics correlate quantitatively with expression changes, while pronounced H3K27me3 changes mostly occur later, with the exception of genes that are activated and required very early during flower morphogenesis, like SEPALLATA1-3 (SEP1-3), LFY and an organ boundary specifying gene (CUP-SHAPED COTYLEDON 1 (CUC1)). However, also for these genes, early changes in H3K27me3 are mild and less pronounced than H3K4me3 changes. Thus, we find that within the first two days of flower initiation, deposition of H3K4me3 is prevalent at gene activation, while strong reduction of H3K27me3 occurs at later stages. Together, our integrative analysis of histone marks and transcriptome provides a novel view on the mechanisms of gene regulation at the chromatin level.
2. Results
2.1. RNA-seq Reveals Specific Changes in Gene Expression during the Developmental Time Series
To assess the correlation between changes in expression patterns and dynamics of histone marks during early flower development, we employed the ap1cal 35S::AP1-GR floral induction system introduced in Wellmer et al. [26] (Figure 1A; Figure S1). This system allows harvesting synchronized flower buds along a developmental time course and insures (i) tissue homogeneity along the developmental time course; as well as (ii) sampling in sufficient amounts for chromatin immunoprecipitation (ChIP) [27,28,29].
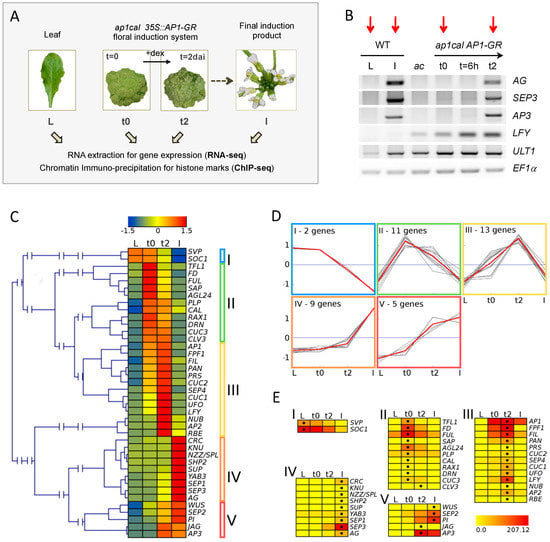
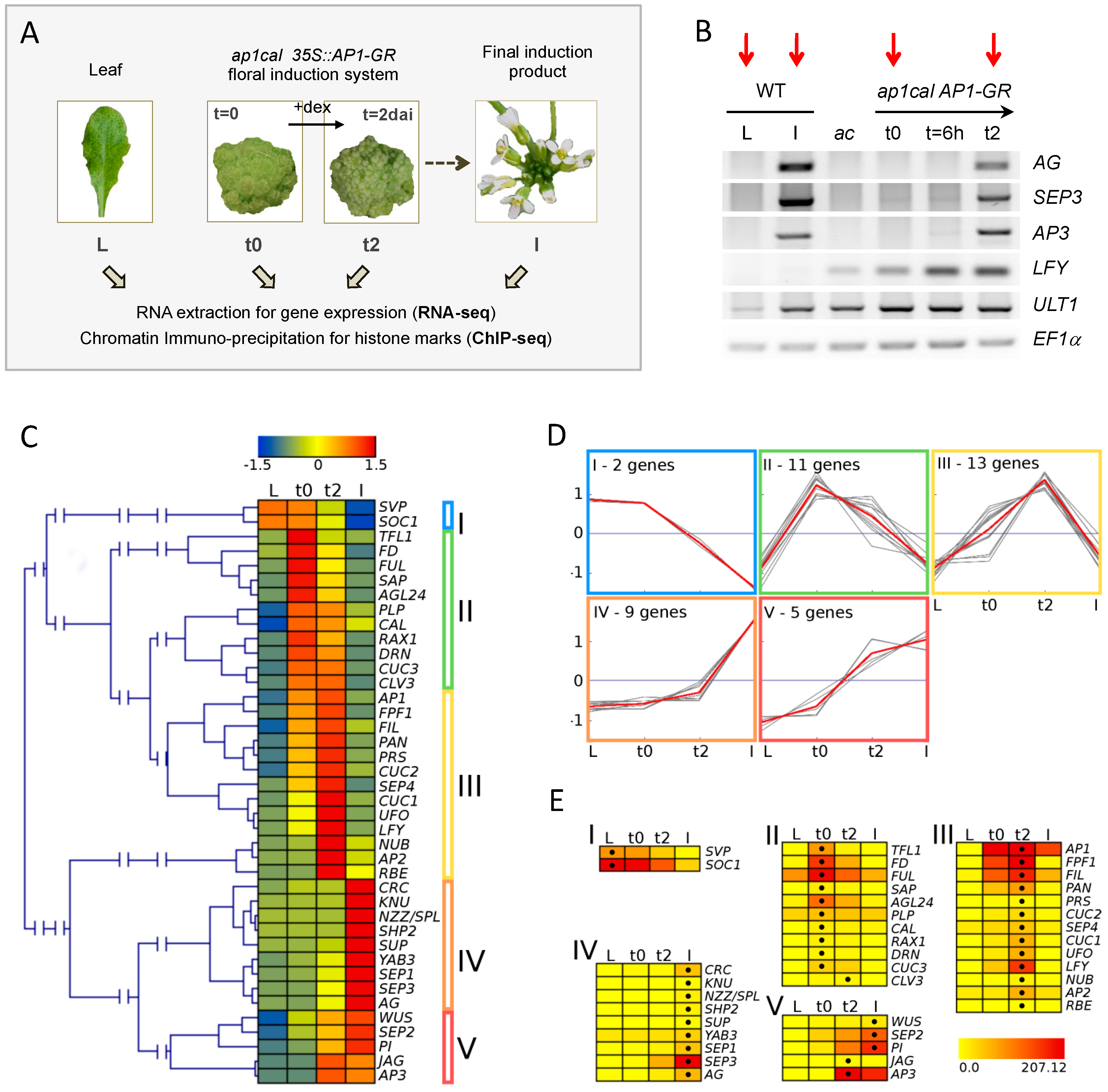
Figure 1.
Experimental setup and assessment of the floral induction system. (A) Tissue types and time-points employed in the study. dai: days after induction. Dex: dexamethasone; (B) RT-PCR analysis of expression of the MADS floral genes AG, SEP3, AP3, as well as the flower marker gene LFY and the inflorescence and flower marker gene ULTRAPETALA1 (ULT1), at t0, 6 h (t = 6 h) and 2 days (t2) after dex induction. EF1α is used as internal control. Wild-type (WT) leaf tissue serves as control state in which floral organ identity genes are not expressed (L). Cauliflower-like inflorescences of ap1cal mutant plants (ac) serve as control on the inherent leakage of the induction system. Dissected WT inflorescences contain flowers up to stage 12 (I). The expression analysis showed no or only very low expression of AP3, AG, and SEP3 homeotic genes prior to dex application, validating the experimental conditions. Strong up-regulation of MADS floral genes occurs at t2 and not at 6 h after induction. Red arrows point at the samples analysed by RNA-seq and ChIP-seq in this study; (C) Hierarchical clustering analysis for expression patterns of selected floral organ regulators, determined by RNA-seq. Relative expression values are expressed as z-scores to reveal similarities in expression patterns. Z-scores are represented by a heat map from blue (low expression in comparison to the other tissues) to red (high expression in comparison to the other tissues). Five major groups are visible; (D) Result of k-means clustering analysis with k = 5 to generate an overview of expression profiles in the five clusters marked in C. The grey lines represent the single genes in the cluster. Averages of z-scores in each cluster are depicted in red; (E) Detailed view of each cluster showing normalised absolute expression values in a heat map from yellow (no expression) to red (highest expression). The black dot indicates the highest expression value reached by a gene in the sample series. The highest displayed expression value corresponds to the highest observed value in the set (SOC1, L sample, with the exception of AP1, which is expressed from the 35S::AP1-GR transgene).
The early time points we chose cover the initial steps of flower development during which flower buds emerge from the inflorescence meristem and further initiate the flower patterning program. During this period, cells undergo two distinct fate transitions, first establishing flower stem cell characteristics, followed by differentiation into founder cells for specific floral organs. A suitable time point to study the early events of flower formation is one at which (i) changes in expression are already detectable for important regulators of flower development; and (ii) floral organ identity genes did not reach their full expression level, to enable observation of transition states from repression to activation. In our experimental setup, two days after induction (t2) stands as the appropriate time point for investigating activation of floral organ identity genes and at which floral meristem identity is still present (Figure 1B, [28]). Moreover, we previously showed using ChIP-PCR, that reduction of repressive H3K27me3 marks at the SEP3 locus is detectable at t2 [28] and thus decided to perform ChIP-seq for H3K27me3 and H3K4me3 and RNA-seq on a tissue series consisting of wild-type leaf (L) and inflorescence (I), and ap1cal 35S::AP1-GR t0 and t2 samples.
Clustering analysis of expression data for selected floral regulators (flowering time, flower development) detected five major groups of expression during the series (Figure 1C–E). The assignment of the genes to the clusters revealed that the observed expression patterns are in accordance with known functions of floral regulator genes. It also confirms the t2 time-point as an intermediate stage during the activation of floral organ and meristem identity genes (Appendix A.1).
Genome-wide analysis of differentially expressed genes (DEG) showed that most genes changed from L to I, emphasizing the different nature of the two tissues. 251 genes are up-regulated from t0 to t2, among them many developmental regulators like SEP2, SEP3, FLOWERING LOCUS T (FT), UNUSUAL FLORAL ORGANS (UFO), AP3, AG, PI, AGAMOUS-LIKE 15 (AGL15), LFY and CUC1, while 448 are down-regulated (Table 1, Table S1). K-means clustering and Gene Ontology (GO) analysis revealed that the gene expression is highly dynamic throughout the series and correlates with expected functionality of the tissues on a genome-wide scale (Appendices A.2 and A.3). Thus, RNA-seq analysis demonstrates transcriptome dynamics throughout the tested tissue series and validates the suitability of the selected tissue types and time-points to study the influence of chromatin marks on gene expression.

Table 1.
Numbers of differentially expressed genes (DEGs). Number of DEGs found in the different comparisons between tissues/time points and their overlap with each other. The grey diagonal displays the number of DEGs in one comparison, the white fields indicate the overlap with respective other comparisons.
2.2. Meristem and Differentiated Tissues Share Similar Over-All Distribution of H3K27me3 and H3K4me3 Marks
We performed quantitative ChIP-seq analysis for repression-associated H3K27me3 and activation-associated H3K4me3 marks in the same tissue samples as employed for the transcriptome analysis (L, I, t0 and t2) (Appendix A.4). The average expression values of H3K27me3 target genes are lower compared to all Arabidopsis genes, while H3K4me3 target genes display a higher average expression. This is true for all analysed tissue types and in accordance with H3K27me3 and H3K4me3 being associated with repressed and active expression states, respectively (Table S3).
Mark abundance distributions over loci for both marks is comparable to previous reports [8,9,10,30]: H3K27me3 is spread over the whole transcribed region, into the promoter and the 3′ UTR, while H3K4me3 is more concentrated to the transcription start site (TSS), covering the proximal promoter and the 5′ region of the gene (Figure 2A).
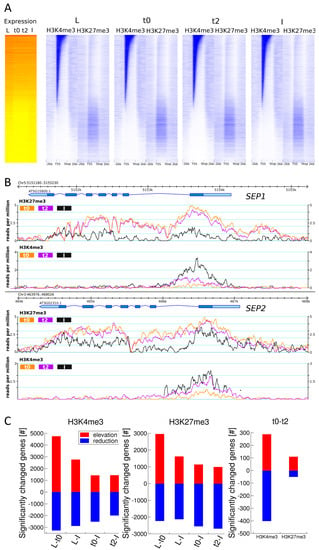
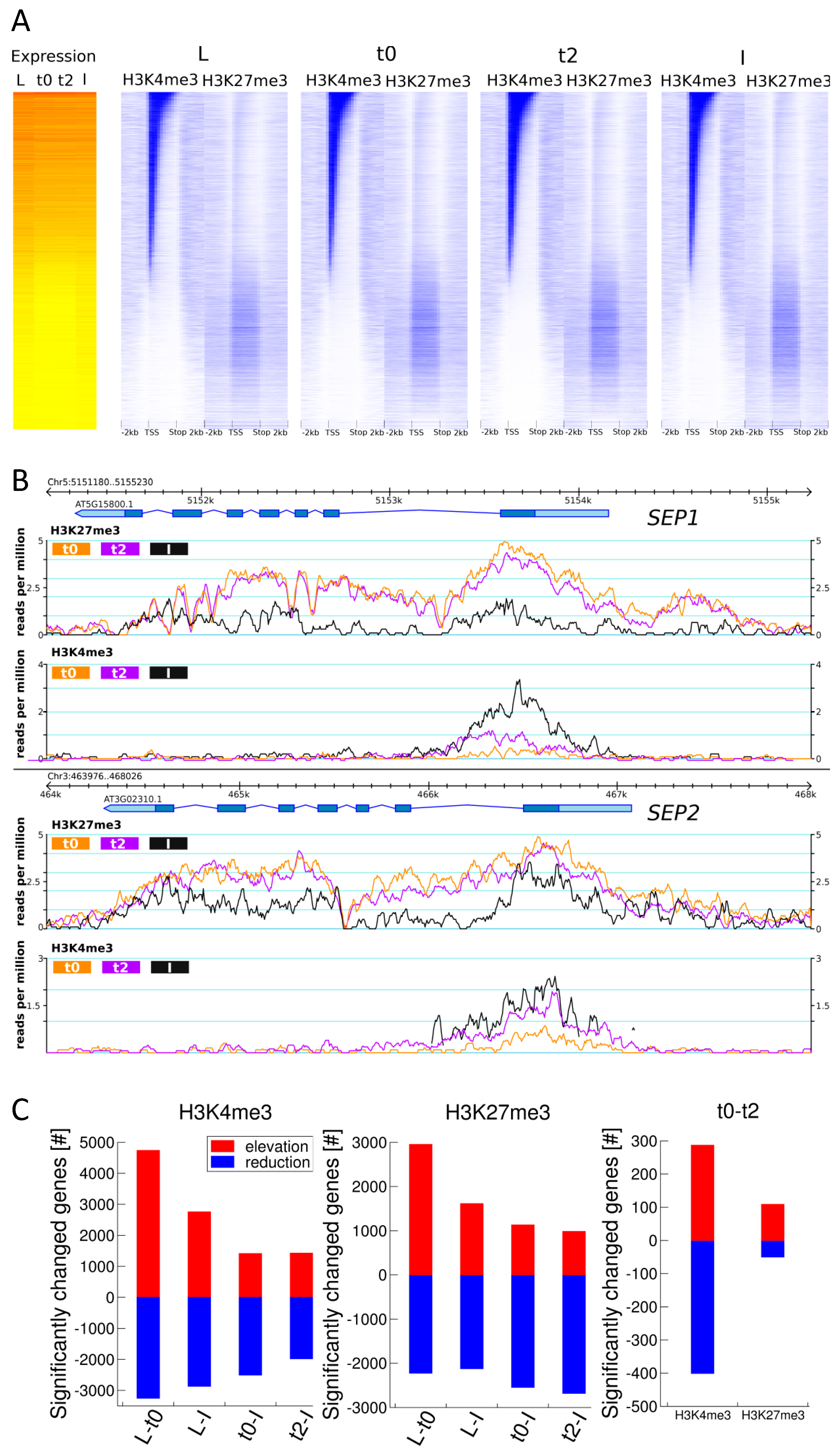
Figure 2.
Genome-wide H3K27me3 and H3K4me3 distribution over loci and quantitative changes in H3K27me3 or/and H3K4me3 marks in the tissue series. (A) Heat maps showing expression values and H3K4me3 and H3K27me3 abundance for all Arabidopsis genes in all four tissues of the developmental series. Genes are ordered by the average H3K4me3 signal over the gene (transcription start site (TSS) to transcription stop) at t0. Expression data is displayed as an average of three biological replicates, histone mark data is derived from one replicate. We saw the same tendencies in the other replicate; (B) Genome browser view of H3K27me3 and H3K4me3 distribution on the SEP1 and SEP2 loci in ap1cal 35S::AP1-GR at t0, t2 and in fully expanded inflorescences. Pictures are generated from one replicate; (C) Numbers of significantly differentially marked genes (DMG) for H3K27me3 and H3K4me3. FPKM: fragments per kilobase of exon per million fragments mapped.
When sorted by H3K4me3 abundance and assessed for expression in the four tissues (Figure 2A), three main categories of genes emerge: First, highly expressed H3K4me3-marked genes with a strong correlation between expression levels and both intensity and spread of the H3K4me3 signal. Second, lowly expressed genes that carry low H3K4me3 levels and high H3K27me3 levels. Many genes in this group display higher expression in the differentiated L and I samples than in the meristematic tissue. This is in line with the previous observation that expression patterns of H3K27me3 target genes are more tissue specific than other genes [8]. Third, genes with low marking in both marks. Interestingly, this is the group of genes with the lowest expression, probably representing genes carrying other repressive marks like DNA methylation or H3K9me2. These distributions are observed for all tissues including the meristematic stages t0 and t2, indicating that plant inflorescence meristems share similar overall H3K27me3 and H3K4me3 marking with differentiated cells but differ in the quantitative abundance of the marks.
2.3. H3K27me3 and H3K4me3 Levels are Highly Dynamic over the Time Series, in Accordance with Tissue Functionality
The examples of histone mark distribution over two important floral organ identity gene loci (SEP1 and SEP2) for t0, t2 and I (Figure 2B) indicate the presence of quantitative changes during flower morphogenesis. Particularly for H3K27me3, these changes are subtle but span a wide range of the marked region.
Genome-wide determination of differentially marked genes (DMGs) for the L-t0, L-I, t0-t2, t0-I and t2-I comparisons revealed that thousands of genes display quantitative change in H3K27me3 and H3K4me3 marks (Figure 2C, Table S4). In the case of very different tissue types (L-t0 or L-I), more than 60% or 45% of the corresponding target genes, show changes in H3K27me3 or H3K4me3 respectively. Noticeably, the vast majority of genes showing an elevation of any of the two marks from L to t0 shows a reduction of the same mark from t0 or t2 to I, and vice versa (Table S4, Figure S7), indicating that these changes are meristem specific.
For several hundred genes we detected changes in both antagonistic marks in opposite directions (H3K27me3 elevated and H3K4me3 reduced or vice-versa), while only few genes (none for the t0-t2 comparison) changed in the same direction for both marks (Table S4).
Our quantitative analysis allowed to specifically identify genes with marks changing in the two first days of flower initiation, from t0 to t2 (early flower morphogenesis). Among these, 13 DMGs change in both marks towards activation (H3K27me3 reduced, H3K4me3 elevated) and 15 towards repression (H3K27me3 elevated, H3K4me3 reduced) (Table 2). These changes relate to coordinated activation or repression of key genes for flower development (Appendix A.5).

Table 2.
Function of differentially marked genes (DMGs) where marks change towards activation or repression.
A GO analysis further revealed that the functions of the H3K27me3 and H3K4me3 DMGs in the series correlate with the functionality of the tissues at the analysed time-points (Appendix A.6).
2.4. DMGs Largely Overlap with DEGs and Histone Mark Changes Are Stronger among DEGs
To evaluate if changes in repressive or active histone marks correlate with changes in expression of the corresponding loci, we determined the fractions of differentially up- and down-regulated genes that display changes in histone mark abundance (Figure 3).
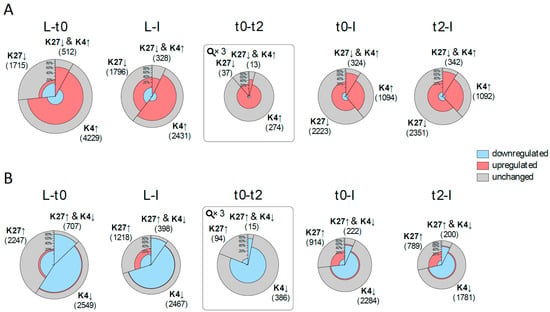
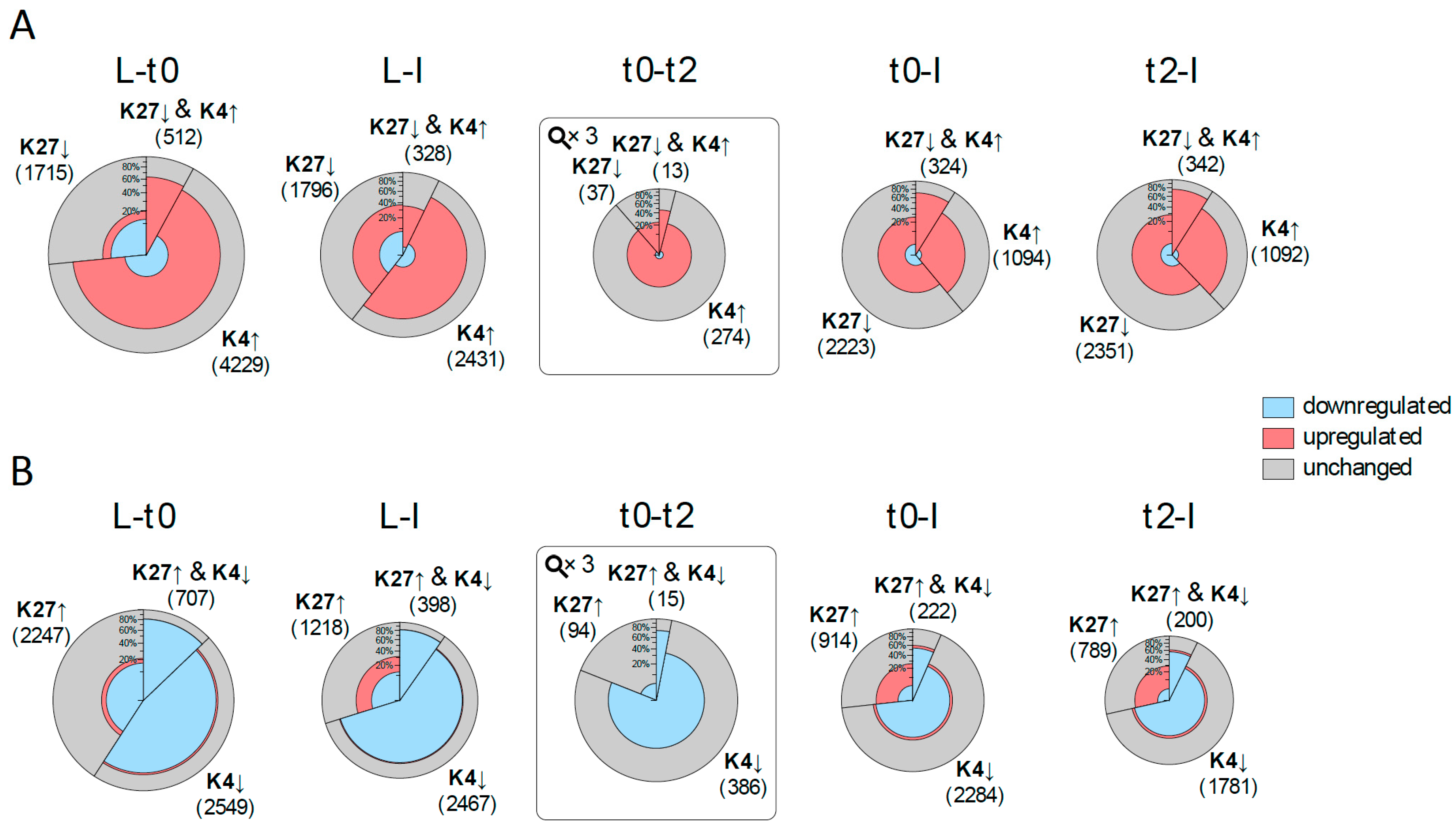
Figure 3.
Correlation of changes in histone marks and expression changes prior to and during flower development. For each comparison between tissues/time points, two pies are drawn: (A) one for changes in histone marks which are expected to be correlated with activation (elevation (↑) of H3K4me3, reduction (↓) of H3K27me3); and (B) one for repression. The size of the pie circle represents the number of genes (for t0-t2 the size is zoomed three times) and is partitioned into three sectors, which sizes correspond to the number of genes changing in H3K4me3 (K4), H3K27me3 (K27) or both. Each sector is coloured according to the number of differentially expressed genes (DEGs) (from inside to outside: down-regulated genes (blue), up-regulated genes (red) and genes with no significant change in expression (grey).
For most comparisons, including all for H3K4me3, expression changes occur more frequently in the expected direction (up-regulation for elevated H3K4me3 and reduced H3K27me3 and vice-versa). This is especially true when comparing t0 to t2. In all cases, the number of DEG in the expected direction is significantly higher among DMGs than for all targets (based on a hypergeometric distribution with probability cut-off of 0.01). With the exception of the L to I comparison, DMGs in both marks display a higher percentage of DEGs compared to DMGs in one mark. These findings are significant for all cases in the expected direction with the exception of t0-t2 in the activation direction (based on a hyper-geometric distribution with probability cut-off of 0.01), indicating that combined changes in both marks are more prone to coincide with changes in expression.
Since transcription is initiated by the assembly of the pre-initiation complex (PIC) near the TSS of a gene, histone marks might primarily change at this region to allow activation or repression of transcription. We thus analysed the marks average distribution along the locus for genes displaying changes during induction of flower development (DMG from t0 to t2 or I) (Figure 4). The t0-t2 and t0-I differences are clearly visible among the DMGs and occur rather uniformly over the covered regions than locally, except for a stronger change at the peak close to the TSS for H3K4me3 in the t0-t2 activation comparison. Changes in H3K4me3 are stronger when DEG subsets are considered, while for H3K27me3 this is only true for the t0-I activated genes and the t0-t2 repressed genes.
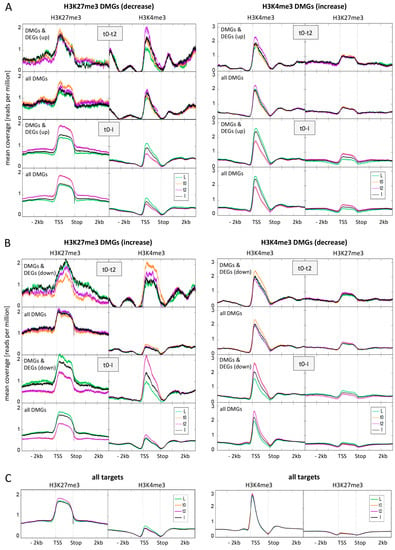
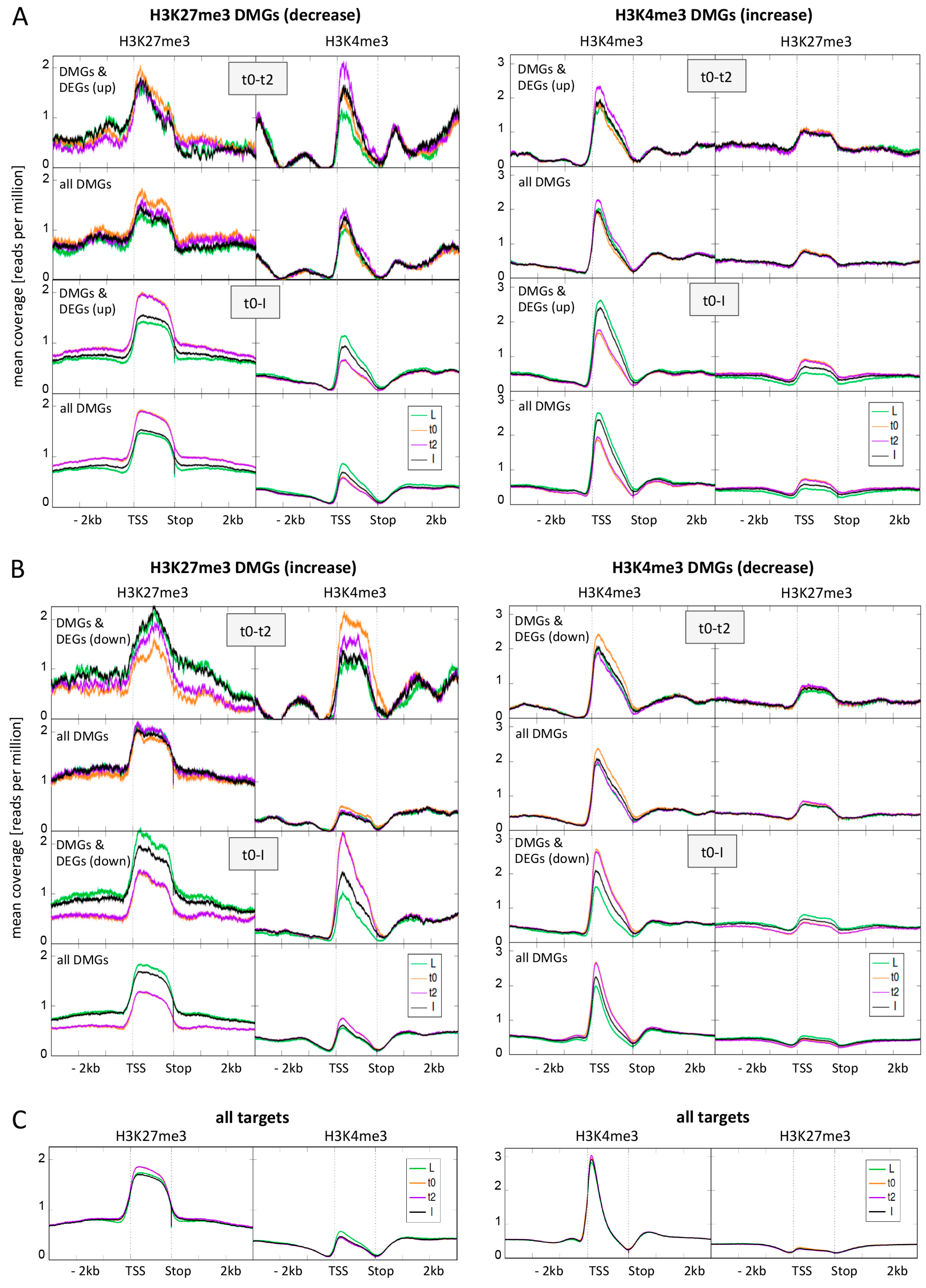
Figure 4.
Average distribution of H3K27me3 and H3K4me3 over differentially marked genes (DMGs) during flower morphogenesis and at all target genes. (A) Average distribution of H3K27me3 and H3K4me3 over H3K27me3 DMGs (left panel) and H3K4me3 DMGs (right panel) changing in activation direction (reduced H3K27me3, elevated H3K4me3). For both early (t0-t2) and later (t0-I) DMGs we first depicted the distribution for those DMGs that are also DEGs (up-regulated) and below the distribution for all DMGs at this time point; (B) Average distribution of H3K27me3 and H3K4me3 over H3K27me3 DMGs (left panel) and H3K4me3 DMGs (right panel) changing in repression direction (elevated H3K27me3, reduced H3K4me3). For both early (t0-t2) and later (t0-I) DMGs we first depicted the distribution for those DMGs that are also DEGs (down-regulated) and below the distribution for all DMGs at this time point; (C) Average distribution of H3K27me3 and H3K4me3 over all H3K27me3 target genes (left panel) and all H3K4me3 target genes (right panel). Genes are scaled to the same length (1500 bins), only the second replicate is displayed here. We saw the same tendencies in the first replicate.
Notably, DMGs in one mark (regardless if H3K27me3 or H3K4me3) show a higher abundance of the other mark compared to all target genes, and especially DMGs changing in expression. This indicates that these DMGs are in a transition state. For H3K27me3 DMGs genes, a change in H3K4me3 abundance is visible in the opposite direction for all comparisons, and this change is stronger for genes changing in expression. This is in accordance with our observation that genes changing in both marks more frequently display changes in expression than genes changing in one mark.
Taken together, our analysis of DMGs and DEGs established a relation between H3K27me3/H3K4me3 dynamics and changes in expression during flower development. Furthermore, it shows that during early stages of flower morphogenesis, H3K4me3 dynamics affects more genes and is in general more strongly correlated with expression changes than H3K27me3 dynamics.
2.5. Expression Changes during Early Flower Morphogenesis Quantitatively Correlate with Changes in H3K4me3 While Changes in H3K27me3 Mostly Occur after Prolonged Expression Changes
Next, we aimed to test for a quantitative correlation between histone mark changes and expression changes. For this, we mainly focused our analysis on the genes changing during early flower morphogenesis (t0 to t2) as these constitute the most homogenous tissue samples, and because genes changing in this time frame are highly dynamic and comprise many important developmental regulators. A clear correlation was observed between H3K4me3 changes and expression changes: mark changes are of higher intensity, of wider spread, and greater towards the TSS for genes with stronger expression changes (Figure 5A).
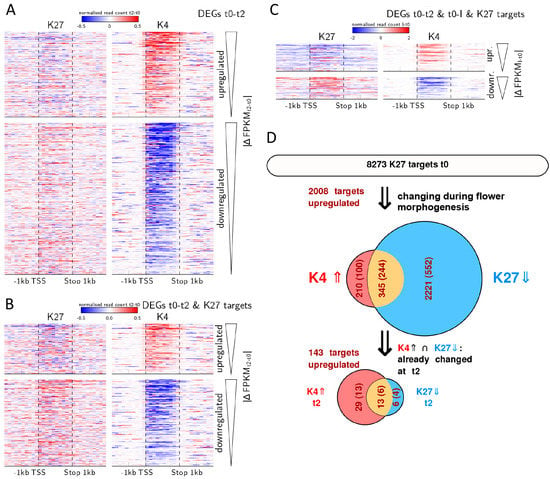
Figure 5.
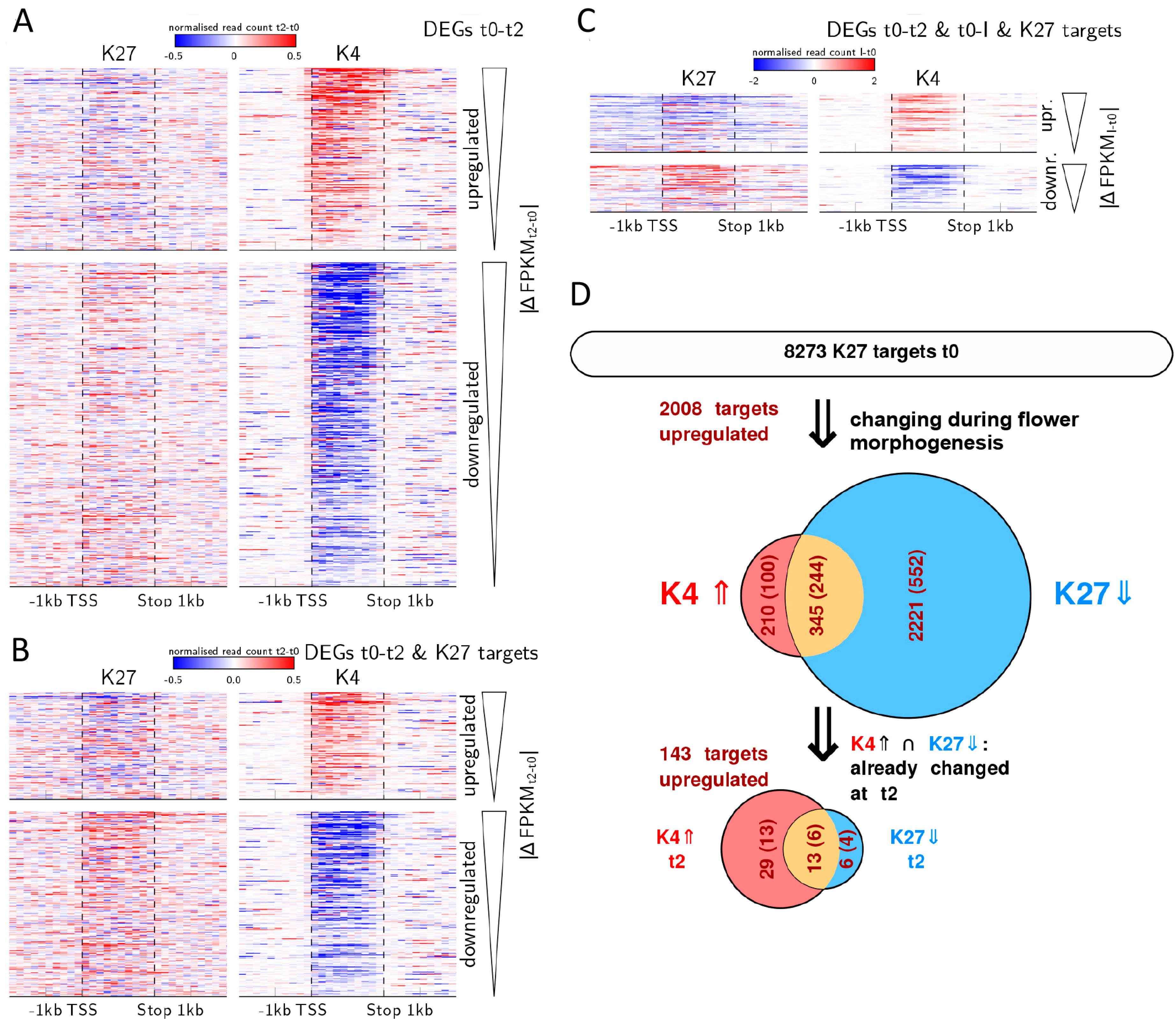
Quantitative changes in H3K4me3 correlate with gene expression dynamics and are prevalent during early flower morphogenesis. (A–C) Heat maps displaying the average change in histone mark abundance (in reads per million) calculated as the difference between the earlier and the later compared time point in 200 bp windows/10% bins over the gene (i.e., t2 minus t0 or I minus t0). Each line in the heat map represents a DEG, genes are sorted by their difference in expression value (t2 minus t0 or I minus t0) starting with the strongest up- or down regulated gene, respectively; (D) Flow chart of histone marks and expression changes during activation of H3K27me3 targets in the process of flower morphogenesis. Only genes marked with repressive H3K27me3 marks at t0 are considered. The upper Venn-diagram shows all genes changing during flower morphogenesis (that is genes that change either from t0 to t2, t0 to I or both) with fraction of those genes changes in expression (dark red numbers in ()), in H3K27me3 (blue fraction), in H3K4me3 (red fraction) or both (orange). The lower Venn-diagram shows all genes that change in both marks (345 genes) which already show activation related changes in marks and expression from t0 to t2.
As H3K4me3 changes are on average slightly stronger at the TSS, we further considered all DEGs that change early but remain DEGs in the same direction for the t0 to I comparison, thus allowing to follow activation at early and later stages. For those genes, we analysed the average distribution of changes from t0 to t2 and from t2 to I, over their locus (Figure S8). A clear shift from the TSS towards further downstream region is visible from early to late changes for up-regulated genes, indicating that H3K4me3 changes occur mainly at the TSS during early activation but then spread more into the locus. For down-regulation, the opposite effect is observed but less pronounced.
For H3K27me3, very little correlation between expression changes and mark changes is observed for all DEGs from t0 to t2. This could be due to only some of the DEGs being H3K27me3 targets, while H3K4me3 marks target more genes. Therefore, we considered only those among the early DEGs that are H3K27me3 targets (147 of 251 up-regulated DEGs and 255 of 448 down-regulated DEGs). We again, could observe only a slight or no preference for changes in the expected direction for down-regulated and up-regulated DEGs, respectively. Furthermore, the correlation with the intensity in expression change rank is also much lower than for H3K4me3 (Figure 5B, Table 3). Only when considering H3K27me3 target genes that had been up- or down-regulated for a longer time (i.e., DEGs which expression changed early but remained in the same direction for the t0 to I comparison, 80 up-regulated DEGs and 65 down-regulated DEGs), we could observe a clear prevalence of changes in the expected direction (Figure 5C, Table 3). Especially for activated genes of this category, the H3K27me3 signal changes over the whole locus including the promoter region for most genes and the change is more pronounced for genes with higher expression changes. This indicates that changes in H3K27me3 rather occur after prolonged changes in expression and is in accordance with a low number of H3K27me3 DMGs during early flower morphogenesis. However, there are some changes in H3K27me3 already observed for the t0 to t2 comparison (e.g., the MADS developmental regulators SEP1-3). We hypothesised that these genes might be very early activated/repressed genes. Indeed, we found that most DEGs with high average H3K27me3 change in the expected direction displayed already an increase/decrease in expression during the first day of flower development as determined by Wellmer et al. [26] (Figure S9).

Table 3.
Overall correlation between quantitative changes in histone mark abundance with rank of gene expression changes in the three gene sets described in Figure 4A–C. DEGs were ranked according to their change in expression and the Pearson product-moment correlation coefficient (PCC) between this rank and the change in histone mark abundance was calculated. Only average changes over the gene body were taken into account. To provide a measure for the overall changes in histone marks among DEGs, average changes in normalised read counts over gene bodies for all the DEGs in the same categories were calculated (Δ normalised read count).
Taken together, during flower morphogenesis, we observe that H3K4me3 dynamics are strongly predictive of expression changes, while H3K27me3 changes are rather predicted by already on-going or recently happened expression changes (Appendix A.7).
2.6. Early Activation of PcG Target Genes is Mainly Accomponied by H3K4me3 Changes
Many regulators of flower development are known to be H3K27me3 targets and regulated by antagonising trxG and PcG factors [8,31]. Therefore, we were interested to see whether changes in genes that will eventually significantly loose H3K27me3 and gain H3K4me3 are also characterised by H3K4me3 dynamics during early flower morphogenesis. We analysed DMGs that are targets of H3K27me3 at t0 in more detail (Figure 5D). Of these 8273 H3K27me3 target genes, 2776 become differentially methylated in one of the marks or both during flower morphogenesis i.e., from t0 to t2 and/or from t0 to I. The majority of them show only reduction in H3K27me3 (2221 genes), while 210 show only elevation in H3K4me3. The 345 genes that show a change in both marks are suitable to address the question of which mark is changed first. We thus further considered the sub-population of these 345 genes, which show changes in marks during early activation, i.e., from t0 to t2 (48 genes). Among these, the proportions of changes in marks are inverted compared to all genes changing during flower morphogenesis: 6 genes change only in H3K27me3, 13 change in both marks (corresponding to the 13 genes discussed above and mentioned in Table 2) and the majority (29 genes) changes only in H3K4me3. This again indicates that during initiation of flower development (t0-t2) genes carrying H3K27me3 can display an increase in H3K4me3 without significantly changing in their H3K27me3 level within the observed tissue. Such effects have been shown also for stress-induced transcription at single gene level [32].
2.7. Relative Dynamics of Histone Marks, MADS TF Binding and Chromatin Opening during Early Stages of Gene Activation
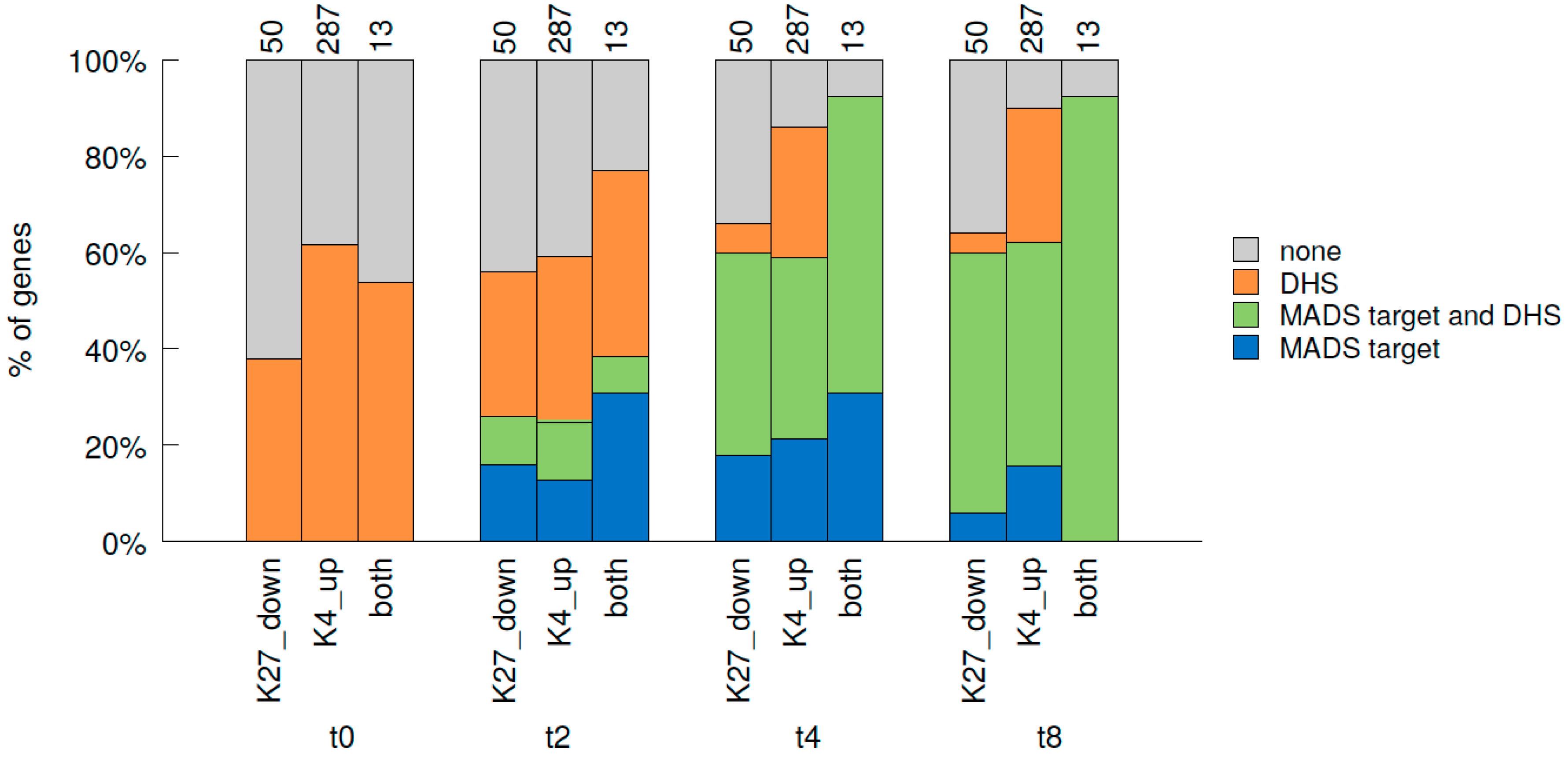
Since flower morphogenesis is mainly regulated by activation of ABC(D)E genes, and especially MADS domain TFs, we were interested in the correlation between MADS binding to chromatin [29] and histone mark changes (this study). We therefore analysed which fractions of t0-t2 DMGs in our data set are targets of the MADS TFs SEP3 and AP1 in the floral induction system as determined by Pajoro et al. at two, four and eight days (t2, t4, t8) after induction [29] (Figure 6, Figure S11A). At t2, slightly more than 20% of genes with reduced H3K27me3 or elevated H3K4me3 are targeted by the MADS TFs, while almost 40% of the genes changing in both marks are. At t4, the percentage of genes being MADS targets raises to around 60% for those changing in a single mark and to more than 90% for those changing in both marks (12 of 13 genes are MADS targets at t4). Towards t8 only minute changes occur in this distribution. These percentages of DMGs bound by the MADS TFs are higher than that observed for all H3K27me3 or H3K4me3 target genes: only 6.7% of H3K27me3 (8.9% for H3K4me3) target genes are MADS targets at t2, increasing to 23.4% (33% H3K4me) at t4 and 26.9% (35.7% H3K4me3) at t8.
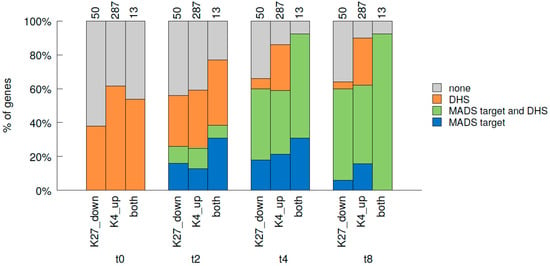
Figure 6.
Correlation of genome-wide changes in histone marks from t0 to t2, binding of MADS TFs and DNaseI hypersensitivity during early flower development. Fractions of DMGs for H3K27me3, H3K4me3 and both marks from t0 to t2 (K27 down: H3K27me3 reduced t0-t2, K4 up: H3K4me3 elevated t0-t2, both: H3K27me3 reduced and H3K4me3 elevated t0-t2) that are bound by MADS TFs (AP1 and/or SEP3) and/or overlap with DNase I hypersensitive sites (DHS) during early flower development. TF binding at three time points after dex-induction was considered: t2, t4 and t8, while DHS was additionally considered at t0 [29]. “none” identifies genes that were not DHS nor MADS target.
As MADS TFs were proposed to assist in opening chromatin, we analysed if the genes changing in histone marks from t0 to t2 would also change in their chromatin conformation as determined in Pajoro et al. [29] by assessing DNase I hypersensitivity (DHS, Figure 6, Figure S11B). Interestingly, about 40% or 60% of those DMGs are already located in DNase I hypersensitive (accessible) regions at t0. This is quite unchanged in t2 and the percentage of genes in open regions only increases slightly towards t4 and more towards t8. MADS binding seems to mainly affect other, still closed genes at t2, while a larger overlap between MADS binding and DHS is observed at t4 and even more at t8 (Figure 6).
This dataset comparison indicates that the majority of genes changing in H3K27me3 and H3K4me3 histone marks during early steps of flower morphogenesis are targets of the floral organ identity regulators SEP3 and AP1, and that binding of these MADS TFs occurs at the same time as histone mark changes or slightly later. Furthermore, it appears that opening of previously closed chromatin may occur after histone mark changes and MADS binding (Figure 6).
3. Discussion
3.1. Chromatin Marks are Highly Dynamic in Association with Tissue Fate during Flower Morphogenesis
Several mutants in PcG and trxG components display defects in initiation of flowering and in floral architecture [3]. Moreover, almost all floral organ identity genes and many flowering time regulators are targeted by H3K27me3, and detailed studies at single gene level (e.g., FT and FLOWERING LOCUS C) have shown that activation of these genes is accompanied by histone mark dynamics. These facts led to the assumption that PcG-associated H3K27me3 and trxG-associated H3K4me3 dynamics operate at the onset of flowering and flower development, affecting a wide range of target genes. However, the extent of the chromatin marks remodelling remained unknown. Through the floral induction system we could generate a genome-wide map of dynamics in histone mark abundance and correlate it with expression dynamics, during early steps of flower development. Our study demonstrates that histone marks change between vegetative and reproductive structures, and during flower initiation and morphogenesis affect a total of 14,826 genes (changed at least in one mark in one comparison, Tab S4) in A. thaliana.
Several thousands of genes changed in histone marks from leaf towards inflorescence meristems and then changed in the opposite direction from inflorescence meristems towards fully expanded inflorescences. This indicates that histone marks antagonistically change in the different tissue types according to the underlying meristematic activities. These reverted patterns of histone mark changes throughout the time series thus likely reflect the chromatin features of differentiated versus meristem-like tissues and highlight the continuous and iterative mode of plant development.
3.2. Integrative Analysis of Chromatin and Expression Datasets Provides Insights on the Mechanistic of Gene Activation, Revealing Unexpected Order of Events
Combination of the transcriptomics and chromatin dynamics datasets revealed that changes in histone marks are widely correlated with changes in expression in the anticipated direction. Especially, changes in both marks are more prone to coincide with expression changes.
The temporal resolution brought by our dataset revealed that activation of a locus does rather not follow the canonically assumed order of events, with repressive marks being removed and active marks being added during transcriptional activation. Some studies had challenged this view in the past, at single gene level in plants. They report that transcription can take place while repressive marks are still present, and reduction in these marks was proposed to rather be a consequence of expression of the target gene [24,25,66]. Our genome-wide study shows that H3K4me3 is both quantitatively and temporally closely correlated with expression changes, while changes in H3K27me3 most likely require prior transcriptional changes to occur (Figure 7). This is in line with the recent report in mouse that re-establishment of H3K4me3 occurs more rapidly than that of H3K27me3 during activation of the zygote genome [67].
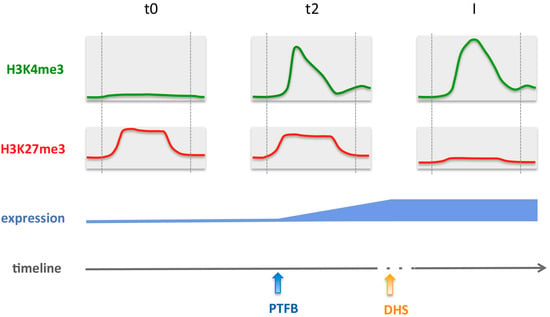
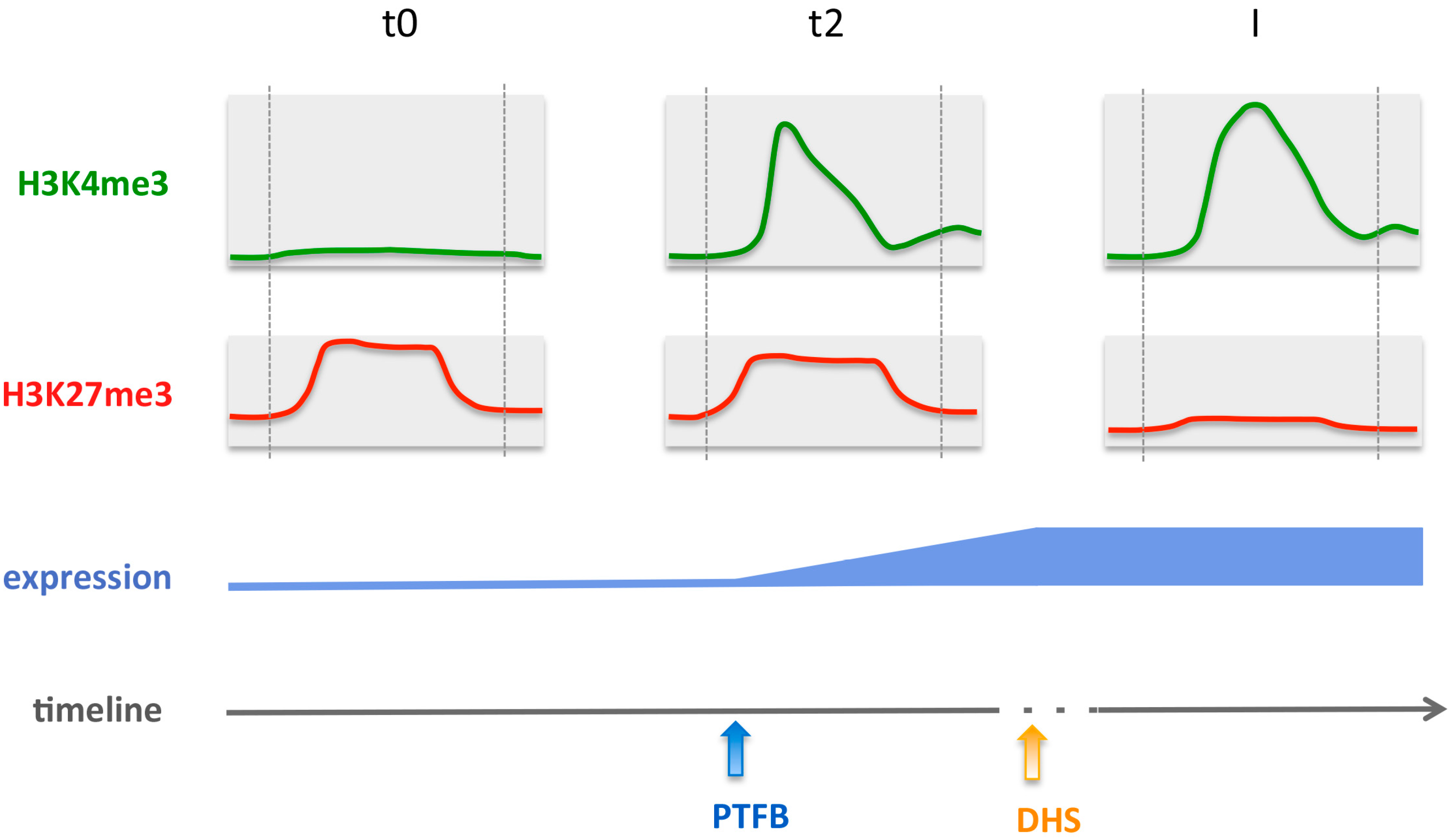
Figure 7.
Model for gene activation during flower morphogenesis, showing changes in H3K4me3 and H3K27me3 chromatin marks, pioneer transcription factor binding (PTFB) and DNA accessibility (DHS). The dotted vertical grey lines mark the TSS (left) and the Stop (right) positions. The majority of genes changing in H3K27me3 and H3K4me3 during early steps of flower morphogenesis are targets of the SEP3 and AP1 pioneer transcription factors. PTBF occurs at the same time as histone mark changes or slightly later at these genes, while opening of previously closed chromatin occurs rather after histone mark changes and PTFB. H3K4me3 deposition displays temporal correlation with expression changes while H3K27me3 reduction occurs later.
Moreover, we found that during flower formation, activation of expression of PcG target genes is predominantly accompanied by changes in H3K4me3 while H3K27me3 marks remain at the loci, and only decline later during flower morphogenesis. This indicates that initiation of floral morphogenesis involves more activation than de-repression events at the chromatin.
Decrease in H3K27me3 would require either active demethylation or dilution of the mark over cell divisions. In a tissue that undergoes cell division, in order to maintain a H3K27 methylated state at a specific locus, new chromatids would need to be methylated at the same locus as their template, after each cell cycle. Oppositely, in absence of de novo methylation, the H3K27me3 mark would be diluted at every cell division. Thus, a cell division-dependent dilution of marks, rather than an active demethylation process could explain the temporal delay we observe in H3K27me3 decrease after gene activation.
Because flower development in the induced ap1cal AP1-GR system occurs at a similar speed as that of the wild type in our conditions (Figure S1 and [68]), one can expect a maximum of two to three cell divisions to occur during the t0-t2 transition, as reported by 4-D imaging and cell lineage analysis during early stages of A. thaliana flower development [69]. In this quite slow dividing context, dilution of the H3K27me3 mark may occur slowly, providing that de novo deposition by PcG is prevented.
A novel question thus arises on how PcG function is prevented, thus leading to H3K27me3 decrease over a longer period of activation. At least two molecular events could be involved, that were reported in previous studies. First, a cell-division dependent PcG eviction mechanism may take place, such as the one operated in the developing flower by the AG transcription factor at the KNU locus, [70]. Second, in mammalian cells, deposition of H3K27me3 by PcG proteins is hindered by the presence of H3K4me3 [71]. This should be the case in plants as well, deposition of H3K4me3 would hinder PcG proteins to access the respective histone and thus further lead to decrease in H3K27me3. Because H3K4me3 marks are required for active transcriptional elongation in plants [72], we propose a scenario along which, for gene activation at the flower meristem, H3K4me3 marks are deposited first, thus leading to active transcription and preventing de novo deposition of repressive H3K27me3 marks over cell divisions (Figure 7).
Moreover, the fact that decrease in H3K27me3 may be a consequence of gene transcription could explain our observation that histone mark abundance changes over the complete gene body (even for a relatively short t0-t2 transition) and the fact that genes changing in expression show a stronger change in histone mark abundance, thus further supporting our model.
Interestingly, genes changing in one histone mark display higher abundance of the antagonistic mark at stages preceding the change. This could indicate that these genes are kept in an intermediate state to allow fast regulation during development and raises the question whether active and repressive marks co-exist at a locus in a same cell on the same chromosome, at the time of flower bud initiation. Such so-called bivalent loci were described in the specific case of animal embryonic stem cells [73] and more recently reported in plant studies using Arabidopsis seedlings [74,75]. However, thus far, we have not been able to confirm bivalent states in inflorescence meristems and early developing flowers from the ap1cal 35S::AP1-GR tissue, by re-ChIP experiments. Whether bivalency does not exist or exist only very sporadically in a restricted number of cells of the flower meristem, will need to be further investigated in studies using technologies specifically designed for this purpose.
Finally, integrative analysis of histone marks, transcriptomics, profiling of MADS-domain proteins and chromatin accessibility, indicates that the majority of genes changing in H3K27me3 and H3K4me3 at early steps of flower morphogenesis are targets of the floral organ identity regulators SEP3 and AP1 and display local chromatin conformation change. Moreover, binding of the MADS-domain proteins occurs at the same time as histone mark changes or slightly later. This observation brings H3K4me3 and H3K27me3 dynamics at the forefront of molecular switches for gene expression, together with TFs that have been proposed to act as pioneer factors during flower development [29].
4. Materials and Methods
4.1. Plant Material and Growth Conditions
All employed lines were in Landsberg erecta (Ler) accession. AP1 and CAL double mutants (ap1-1 cal-1) [76] and 35S::AP1:GR ap1-1 cal-1 [26] were described previously.
After 2–3 days of stratification at 4 °C, plants were germinated on Murashige and Skoog medium (Sigma-Aldrich, Lyon, France ) supplemented with 0.3% sucrose in growth cabinets (Percival, Percival Scientific Incorporated, provided by CLF Plant Climatics, Emersacker, Germany) in a controlled environment in long days (LD) of 16 h light at 18 °C and 8 h darkness at 16 °C. At about ten days after germination, plants were transferred to a 1:1 mixture of soil/vermiculite.
4.2. Dexamethasone Treatment and Harvesting of Plant Material
The ap1cal 35S::AP1-GR system employs double mutants of the floral meristem identity genes AP1 and CAL. In ap1cal plants, inflorescence-meristem-like tissue over-proliferates while flower organogenesis does not proceed. A 35S::AP1-GR construct, for induction of flower development upon dexamethasone treatment, allows harvesting uniform tissue for synchronized floral stages, in sufficient amounts for ChIP [27,28,29]. To induce AP1 translocation to the nucleus, primary inflorescences of ap1cal 35S::AP1:GR plants were treated with induction solution (10 µM dexamethasone, 0.015% Silwet) by pipetting the solution directly onto the cauliflower structures. This process was repeated once after 30 min.
For harvesting of ap1cal and ap1cal 35S::AP1:GR samples, only the cauliflower structures of the main inflorescence were harvested. Leaf and pedicel tissue contamination was minimized by dissection. Samples were collected from untreated ap1cal (ac) and ap1cal 35S::AP1:GR (t0) tissue and ap1cal 35S::AP1:GR tissue 2 days after induction (t2). For inflorescence samples (I), whole main inflorescences of wild type plants were harvested. Flowers older than stage 12 were removed. For leaf (L) samples, rosette leaves of the same wild type plants were harvested. About 15 plants were harvested for one ChIP sample resulting in around 0.2–0.3 g of material for cauliflower and inflorescence tissues and 1–1.3 g for leaf tissue. For RNA samples, a fraction of 0.05–0.06 g for cauliflower and inflorescence tissues and 0.25 g for leaf tissue was taken from the sampling event. Replicates for biological sampling were grown independently (at different times) and harvested from the same groups of plants for ChIP-seq (two replicates) and RNA-seq (three replicates).
4.3. Scanning Electron Microscopy
For scanning electron microscopy [77], untreated inflorescence-like meristems of ~4 week old ap1cal or ap1cal 35S:AP1-GR plants were harvested, as were 5 d-post dexamethasone-treated ap1cal 35S:AP1-GR inflorescence-like meristems, and fixed using 1% glutaraldehyde in 0.025 M sodium phosphate buffer. Samples were then washed three times with 0.025 M sodium phosphate and further dehydrated in gradually concentrated alcohol solutions. The material was then critical point dried in liquid carbon dioxide, mounted on stubs and coated with platinum. Imaging was performed using a Hitachi 5500 scanning electron microscope (Hitachi Ltd., Tokyo, Japan).
4.4. Chromatin Immunoprecipitation
Formaldehyde cross-linking and extraction of nuclei was performed as described in [78] with the following modifications: infiltration time was prolonged (6 min and 24 min), filtration of the extract after grinding was performed through 75 µm and 50 µm nylon meshes and sonication was done for 13 × 30 s on, 1 min off, position H (this longer sonication was only performed for biological replicate 2).
Immunoprecipitation reactions were set up as described in [78] except that 15 µg of cauliflower and 10 µg of wild type chromatin was used and the chromatin was incubated with 5 µL of the antibody alone for 5 h prior to addition of Protein A Agarose beads without salmon sperm DNA (Millipore, Molsheim, France).
Antibodies used were anti-H3K27me3 (07-449, Millipore) and anti-H3K4me3 (17-614, Millipore). Specificity of each lot used was checked via dot-Blot (Figure S1H(I)) using peptides containing mono-, di and tri-methylated versions of the respective residue (Abcam, ab1780-1782, ab1340, ab1768 and ab1742) according to recommendations provided by the manufacturer.
Washing of the ChIP reaction was performed using binding/wash buffer as described in [78] but for four times 10 min at 4 °C and once 10 min at room temperature (RT).
Elution and reverse cross-linking was performed as described in [79] except that the elution was done by vortex mixing for 1 min at RT and input samples (1/3 of the chromatin used for one ChIP sample) were brought to the same volume by adding elution buffer and Tris pH9 in the same ratio as to the ChIP samples.
Reverse cross-linked samples were purified using the MinElute Reaction Cleanup Kit (Qiagen, Hilden, Germany) and DNA was eluted in 30 µL Elution buffer.
For biological replicate 2, three technical replicates of ChIP on the same nuclear extract were performed and pooled on one MinElute column.
4.5. RNA Preparation, Reverse Transcription and Gene Expression Analysis by Semi-qPCR
Total RNA was isolated with the RNeasy Plant Mini Kit (Qiagen) and 8 µg of RNA were DNase treated with DNA-free Kit (Ambion, provided by ThermoFisher Scientific Inc., Illkirch, France). For semi-quantitative PCR (semi-qPCR), 2.2 µg of DNase treated RNA were used for reverse transcription with SuperScript® III RNase H—reverse transcriptase (Life Technologies, provided by ThermoFisher Scientific Inc., Illkirch, France) and an oligo dT primer (18 mer), according to the manufacturer’s instructions. From 20 μL of the reverse transcription (RT) product, 1 μL was used for each PCR reaction. The annealing temperature was 54 °C for all primer pairs and 25 cycles of PCR were performed for all genes. Primers for semi-qPCR were previously described [78]. Elongation factor 1α (EF1α) was used as an internal control.
4.6. Library Preparation and NGS
Libraries from ChIP samples of replicate 1 were prepared and sequenced on a GAIIx (Illumina, San Diego, CA, USA) sequencer as described previously [80,81].
For biological replicate 2, ChIP-seq library preparation was performed using the Truseq ChIP sample preparation kit (Illumina, ref. IP-202-1012) according to the manufacturer’s instructions except that PCR amplification was performed prior to size selection. Briefly, sonicated chromatin was repaired prior to 3′ ends adenylation. Illumina’s indexed adapters were ligated to the adenylated double-stranded DNA fragments. An 18-cycles PCR program was performed on the ligated DNA using Illumina’s PCR primers. The libraries were then size-selected between 300 bp and 700 bp on a 2% agarose gel. Each library was validated using high sensitivity chip (Agilent Technologies, Santa Clara, CA, USA, ref. 5067-4626) on an Agilent Bioanalyzer and quantified using the Kapa library quantification kit (Clinisciences, Nanterre, France, ref. KK4824). Equimolar amounts of four libraries were pooled, diluted to 10 nM, denatured, and diluted again to 7 pM. 100 µL of the diluted pool were hybridized on one lane of an Illumina’s Flow Cell. Clustering and 50 cycles sequencing were performed according to Illumina’s instructions on a Hiseq2000. Libraries were multiplexed by three (H3K4me3 samples) or four (H3K27m3 samples) and sequenced on a HiSeq2000 (Illumina) sequencer, yielding 50 bp single-end reads.
For RNA-seq, library preparation was performed using the Truseq stranded mRNA sample preparation kit (Illumina, ref. RS-122-2101) according to the manufacturer instructions. Briefly, polyadenylated RNAs were purified using oligo-d(T) magnetic beads, fragmented, and reverse transcribed using random hexamers, Super Script II (Life Technologies, ref. 18064-014) and Actinomycin D. During the second strand generation step, dUTP substituted dTTP. This prevented the second strand to be used as a matrix during the final PCR amplification. Double-stranded cDNAs were adenylated at their 3′ termini before ligation was performed using Illumia’s indexed adapters. Ligated cDNAs were amplified following 15 PCR cycles and PCR products were purified using AMPure XP Beads (Beckman Coulter Genomics, Bishop’s Stortford, UK, ref. A63881). Libraries were validated using a DNA1000 chip (Agilent, ref. 5067-1504) on a Agilent Bioanalyzer and quantified using the KAPA Library quantification kit (Clinisciences, Nanterre , France, ref. KK4824). Equimolar amounts of 4 libraries were pooled, diluted to 10 nM, denatured, and diluted again to 7 pM. 100 µL of the diluted pool were hybridized on one lane of an Illumina’s Flow Cell. Clustering and 50 cycles sequencing were performed according to Illumina’s instructions on a Hiseq2000.
For signal processing, image analysis and basecalling were performed using the HiSeq Control Software and Real-Time Analysis component. Demultiplexing was performed using Illumina’s sequencing analysis software (CASAVA 1.8.2). The quality of the data was assessed using FastQC from the Babraham Institute and the Illumina software SAV (Sequence Analysis Viewer).
4.7. ChIP-seq Analysis
Reads were mapped to the Columbia reference genome (TAIR10) using BWA [82], files were converted into bam format using samtools [82,83] and duplicated reads were removed using Picard-tools [84].
Pearson product-moment correlation coefficient (PCC) was calculated for sums of reads per million in 200 bp windows of the genome using a customized C++ script to assess similarity between samples. To determine ChIP enriched regions and differentially enriched regions we employed SICER V1.1 [85] with window size 200 bp, gap size 400 bp and effective genome size 90%. As controls, input samples of I were used for the wt samples and input samples of t2 were used for all cauliflower tissues. For all comparisons of samples (ChIP) and control (input), a FDR of 0.0001 was employed. Due to the lower number of reads in the first biological replicate, a more lenient FDR was used for the detection of the small quantitative changes between the samples (0.05). For replicate 2, FDR 0.0001 was used. For the H3K4me3 samples, a strong variation in the background levels was observed between samples. To eliminate biases in the normalisation process caused by this fact, SICER-df was applied to files containing only reads on union islands filtered by filter_raw_tags_by_islands.py from SICER as recommended in [85]. Genes were considered as significantly enriched (target genes) or significantly differentially enriched when overlapping with the respective regions with their gene body. Only genes found in both replicates were considered for further analysis.
4.8. RNA-seq Analysis
Mapping of reads to TAIR10 genome and determination of differentially expressed genes was performed using TopHat and Cufflinks with default parameters as described in [86]. Three biological replicates were analysed together.
4.9. Further Bioinformatics Analyses
4.9.1. Clustering
Heat map visualisation and clustering was performed with the software Genesis 1.7.6 [87]. Normalised expression values in each tissue/time point (denoted as Fragments Per Kilobase of Exon Per Million Fragments Mapped (FPKM)) were submitted to the analysis. To visualise patterns of expression dynamics over the time series rather than absolute values, z-scores were employed and calculated from FPKM values of a gene in a given sample as z = (FPKM-µ)/σ, where µ denotes the mean FPKM value for the gene over the whole tissue/time series and σ denotes the standard deviation of this mean. Hierarchical average linkage and k-means clustering were performed as described [88]. For the subset of floral regulators (Figure 1), 5 main branches are visible in the hierarchical clustering tree. Therefore, k-means clustering was performed with k = 5. For all DEGs, different numbers of clusters were tested and k = 8 yielded the highest diversity of patterns. Distance for all clustering analyses was Euclidean.
4.9.2. Gene Ontology Analysis
For a general overview of over-represented functions (GO slim) in gene sets, the Classification Super Viewer from the BAR website was employed [89]. For detailed functional analysis over-represented GO terms were determined with the FatiGO tool [90] of Babelomics 4 webservice [91].
4.9.3. Visualisation of Data in a Genome Browser
For visualisation, bam files were converted to bigwig files and normalised by the number of reads in the respective library using awk, bedtools [92] and bedGraphToBigWig [93]. Bigwig files were visualised in a locally running version of the Generic Genome Browser (GBrowse 2.54) [94].
4.9.4. Calculation of average histone mark abundance patterns and heat maps over a locus
For upstream and downstream regions of a locus, normalised histone mark abundance (expressed in reads per million) was calculated with base resolution and then averaged either in 200 bp bins for heat maps or over all loci of interest with base pair resolution for average patterns. Transcribed regions were partitioned in 1500 bins (10 bins for heat maps) to account for variation in gene length and average normalised histone mark abundance was calculated for each bin.
4.9.5. Pearson Product-Moment Correlation Coefficient
Pearson product-moment correlation coefficient (PCC) between expression changes and histone mark changes was determined by ranking DEGs by their difference in expression and correlating these ranks with differences in average normalised read count for the two histone marks over the gene body of the respective DEG. Averaged normalised read counts over the gene body were calculated from the values of the 10 bins generated to draw the heat maps.
4.10. Availability of Supporting Data
ChIP-seq and RNA-seq data were deposited at the NCBI Gene Expression Omnibus (GEO) under the SuperSeries GSE71583.
Normalised H3K27me3 and H3K4m3 profiles expressed as reads per million can be viewed at https://gbrowse.cea.fr.
5. Conclusions
Epigenomic analyses with temporal and tissue-specific resolution have not been reported yet during flower development, since most studies were performed at one developmental stage and often in a mixture of tissues, thus providing static and average views of chromatin landscapes. The reason for this lack of temporal dynamics was the challenge of harvesting tissue- or stage-specific chromatin in sufficient amounts for comparison between samples. To overcome these limitations, we employed the inducible “cauliflower” system that allows production of flower meristems and their synchronous development over time in large quantities.
Combination of the floral induction system with quantitative ChIP-seq analysis enabled us to detect changes in histone mark abundance with temporal resolution within the inflorescence, during early flower development. We found that quantitative changes in H3K27me3 and H3K4me3 marks take place within the first two days of flower initiation, affecting several hundreds of genes. During this period, we observed that H3K4me3 is a stronger predictor of expression changes than H3K27me3, for both activation and repression. Even for PcG target genes, increases in H3K4me3 prevail at early gene activation, and H3K27me3 remains present while H3K4me3 is elevated.
Although it was observed that many floral organ identity genes are up-regulated in the constitutive absence of H3K27me3 depositing enzymes, in the natural context H3K27me3 decline rather occurs after prolonged expression change. This indicates that presence of repressive H3K27me3 marks may not prevent early activation of genes, provided that active H3K4me3 marks are deposited.
In conclusion, we could establish the temporal resolution of chromatin events during the first steps of flower morphogenesis, and identified prevailing changes in H3K4me3 over H3K27me3.
Supplementary Materials
The following are available online at www.mdpi.com/link, Figure S1: Validation of the ap1cal AP1-GR induction system for the analysis of early events of gene activation, in situ; Figure S2: Comparison of expression changes from t0 to t2 in this study and in Wellmer et al. [26]; Figure S3: Major expression patterns present among differentially expressed genes (DEG); Figure S4: Functional characterisation of up-regulated DEGs; Figure S5: Functional characterisation of down-regulated DEGs; Figure S6: Overlap between target genes in the four considered time points/tissues for H3K27me3 (K27) and H3K4me3 (K4); Figure S7: Overlap between differentially marked genes for H3K27me3 and H3K4me3 for changes from leaf to meristematic tissue (L-t0) and from meristematic tissue to inflorescences (t0-I); Supplementary Figure 8. H3K4me3 signal shift between early and later expression changes; Figure S9: Expression changes of early DEGs during flower morphogenesis at t1; Figure S10: Expression of early DMGs during the time series; Figure S11: Correlation of genome-wide changes in histone marks from t0 to t2 and binding of MADS TFs or DNase hypersensitivity during early flower development; Table S1: List of genes that are differentially expressed in at least one condition; Table S2: Functional enrichment analysis for DEGs; Table S3: Pearson correlation coefficient analysis, ChIP-seq read numbers, average expression values for H3K27me3 and H3K4me3 target genes and significantly enriched genes for H3K27me3 and H3K4me3; Table S4: Differentially marked genes for H3K27me3 and H3K4me3 in the tissue/time series; Table S5: Functional enrichment analysis for H3K4me3 and H3K27me3 targets; Table S6: Functional enrichment analysis for H3K4me3 and H3K27me3 DMGs.
Acknowledgments
We thank Jean-Luc Putaux and Christine Lancelon for access to critical point dryer equipment and Pierre-Henri Jouneau for assistance with scanning-electron microscopy imaging. We thank Béatrice Berthet for help with dissection of ap1cal tissue and Armelle Donglois for her investment in re-ChIP experiments. We are grateful to Laura Gregoire for critical discussion and input on data visualisation and to Leor Eshed Williams for critical reading of the manuscript. This work was funded by the French National Agency Young Researcher grant for the ChromFlow project (ANR JCJC, project SVSE2-1206 01 to C.C.C., http://www.agence-nationale-recherche.fr/projet-anr/?solr=run&tx_lwmsuivibilan_pi2[4]=ANR-10-JCJC-1206), the Centre National de la Recherche Scientifique for a CNRS-Higher Education Chair (position 0428-64 to C.C.C.), the University of Grenoble Alpes for an UGA-UJF Initiative Chair (to C.C.C.), a research fellowship of the French embassy in Germany (to J.E.) and a Marie Curie Intra European Fellowship within the European Union’s Seventh Framework Programme (FP7/2007–2013) under REA grant agreement n◦ 327377 (http://cordis.europa.eu/project/rcn/108315_en.html) (to J.E.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
C.C.C. conceived and coordinated the study. J.E., R.B. and C.C.C. planned and performed all experiments except for sequencing library preparation, which was done by D.P. and H.P.. F.O. and M.R. performed base-calling and demultiplexing of sequencing reads. J.E. and R.B. analysed the ChIP-seq data, J.E. and C.K. analysed the RNA-seq data and were responsible for all other bioinformatics analyses. J.E. and C.C.C. wrote the manuscript with the help of R.B. and M.S. All authors carefully read, edited and approved the final version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Abbreviations
| AG | AGAMOUS |
| AGL15 | AGAMOUS-Like 15 |
| AIL6 | AINTEGUMENTA-LIKE 6 |
| AP1-3 | APETALA 1-3 |
| BXL1 | BETA-XYLOSIDASE 1 |
| CAL | CAULIFLOWER |
| ChIP | chromatin-immunoprecipitation |
| CUP-SHAPED COTYLEDON 1 | CUC1 |
| DEGs | differentially expressed genes |
| DHS | DNase I hypersensitive |
| DMGs | differentially marked genes |
| EF1α | Elongation factor 1α |
| FACS | fluorescence-activated cell sorting |
| FLC | FLOWERING LOCUS C |
| FT | FLOWERING LOCUS T |
| GO | Gene Ontology |
| IM | inflorescence meristem |
| LAC10 | LACCASE 10 |
| LBD39 | LOB DOMAIN-CONTAINING PROTEIN 39 |
| LFY | LEAFY |
| LSH | LIGHT SENSITIVE HYPOCOTYLS |
| PcG | Polycomb Group |
| PI | PISTILLATA |
| PRCs | Polycomb repressive complexes |
| PUP4 | PURINE PERMEASE 4 |
| SAM | shoot apical meristem |
| SDG4 | SET-DOMAIN-GROUP4 |
| SEP1-4 | SEPALLATA 1-4 |
| -seq | high-throughput sequencing |
| SHP2 | SHATTERPROOF 2 |
| SOC1 | SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 |
| SPT | SPATULA |
| SVP | SHORT VEGETATIVE PHASE |
| TBP | TATA-box binding protein |
| TF | transcription factor |
| trxG | trithorax Group |
| TSS | transcription start site |
| UFO | UNUSUAL FLORAL ORGANS |
| ULT1 | ULTRAPETALA 1 |
Appendix A. Additional Text
Appendix A.1. Description of Expression Clusters among Floral Regulator Genes
Cluster 1 corresponds to regulators of floral transition and inflorescence meristem identity. Their expression is highest in leaves, remains at almost the same level in t0 inflorescence meristem like tissue and then gradually decreases toward I. Cluster 2 correspond to genes regulating inflorescence meristem identity and specifying boundaries. Their expression is low in leaves, high at t0 and then gradually decreases. Cluster 3 comprises genes involved in floral transition, inflorescence meristem and boundaries, which also regulate floral organ development. Expression of these genes gradually rises from L to t2 and decreases in I. Clusters 4 and 5 carry late and early floral organ and meristem identity genes, respectively. Their expression rises from L to I with a main elevation of expression from t0 to t2 (Cluster 5) or from t2 to I (Cluster 4).
For most of the floral regulator genes, changes in expression from t0 to t2 are similar to those observed by Wellmer et al. [26] in a micro-array experiment (Figure S2).
Appendix A.2. K-Means Clustering of DEGs
K-means clustering of all DEGs (14,459) with k = 8 (Figure S3) revealed that the most abundant pattern observed (cluster VII, more than one third of the DEGs) contains genes with a higher expression in leaves than in all other tissues, where their expression is uniformly lower. Three patterns include genes that are lower expressed in leaves than in all other tissues. This is to be expected as leaf tissue fundamentally differs from the three other tissue types (no floral organs, no meristematic tissue, fully differentiated). However, we also find a large number of genes that display elevated expression in inflorescence tissue compared to all other tissues (cluster VI, around one fifth of the DEGs), thus likely to be genes involved in later steps of flower development. The third most abundant group contains genes that are up-regulated in the t0 and t2 tissues and lower in both L and I, where L is lower than I (cluster V).
Appendix A.3. GO Analysis of DEGs
To functionally characterize the DEGs, we performed a Gene Ontology (GO) analysis, starting with a broad GO slim analysis (Figures S4 and S5) followed by a detailed, full GO analysis of categories overrepresented in the DEGs lists (Table S2). This analysis revealed that the changes in expression pattern correlate with expected functionality of the tissues on a genome-wide scale.
Genes activated during the series (from L to I) display over-representation for “developmental processes” in all comparisons except t0 to I. An over-representation for the “TFs” molecular function and the “transcription” biological process in t0 to t2 DEGs indicates that the 2-day induction of flower development affected gene regulatory mechanisms in a broad fashion. Terms related to floral organ development were expectedly enriched when considering genes up-regulated from L to t0 or L to I. Importantly, those related to floral whorl, carpel or gynoecium development, already appeared during early induction of flower development (t0 to t2) further confirming the suitability of the chosen time points, while genes up-regulated later (t0 to I) are enriched for terms mainly referring to processes for later stages of development (e.g., pollen development).
Plastid-localised proteins were over-represented in leaves and repressed elsewhere or present in “green” inflorescences compared to the white meristem structures present in t0 and t2, (Figure S5). Many plastid-localised proteins encoded by genes down-regulated from L to other tissues refer to terms concerning light perception, photosynthesis, sugar metabolism and transport, reflecting a true difference in the photosynthetic capacity and yield. Similarly, terms referring to the response to stimuli are over-represented when comparing down-regulated DEGs in L vs. t0 or vs. I, but also t0 vs. I, indicating that biotic and abiotic responses mainly occur in fully expanded tissues rather than in meristematic tissues.
Appendix A.4. Quality Control for ChIP-seq Experiments
We observed a high correlation between our two biological replicates, both at the read level (Pearson correlation coefficient > 0.75 for all time points, >0.9 for t0 and t2) and at the target gene level (overlap >77% for all time points comprising the most distant L and I, >90% for most) (Table S3).
Appendix A.5. DMGs during Early Flower Morphogenesis
Among the 13 genes that change in both marks towards activation, three are key regulators of flower development (SEP1, SEP2 and LFY), one is involved in flower organogenesis (SPATULA, SPT) and one participates in SAM development (AINTEGUMENTA-LIKE 6 (AIL6)) (Table 2) [42,43,45,46,48,49,50,51,52].
The 15 genes that change in both marks towards repression (Table 2), encode two negative regulators of floral transition, one putative negative growth regulator, three members of the LIGHT SENSITIVE HYPOCOTYLS (LSH) family (members involved in organ boundary specification and light regulation of seedling development), a leaf development regulator and a promoter of pseudo-etiolation in light [53,54,55,56,59,61,62,63,64].
Appendix A.6. GO Analysis of DMGs
To functionally characterize all DMGs, we determined GO terms that are significantly enriched among DMGs compared to all genes carrying the respective mark (Tables S5 and S6).
The over-represented functions among H3K4me3-elevated genes vary strongly between the different comparisons: L-t0 and L-I are enriched in development-related functions including flower and meristem development, but also in DNA metabolism and cell cycle as expected for a comparison between a differentiated tissue and a meristematic (t0) or a meristem-containing tissue (I). In the two comparisons from t0 or t2 to I (more advanced in flower development), we found enriched terms related to response to light and other stimuli, photosynthesis and sugar metabolism, while during early flower morphogenesis (t0 to t2 comparison), DMGs changing towards activation are especially enriched in transcription and floral development (carpel and gynoecium development) specific terms. This result is in line with the fact that many flower regulators are TFs.
Genes showing reduced H3K4me3 for L to t0 or L to I, are enriched in terms related to response to stimulus and depleted in terms related to development, DNA metabolism and transport. Genes with reduced H3K4me3 for t0 to I or t2 to I are enriched in terms related to development, including flower and floral organ development, suggesting that several genes involved in early flower development significantly loose activation marks at later stages.
H3K27me3-changing targets mainly represent all types of H3K27me3-related functions (e.g., developmental processes, Table S5), for H3K27me3 reduction, only the t0-to-t2 DMGs are strongly enriched in several specific terms for floral organ development. Genes displaying elevated H3K27me3 in L to t0 or L to I comparisons are enriched in metabolic processes, L-to-t0 genes are depleted in multicellular-organismal-development, while t2 to I is in flower development.
Appendix A.7. Analysis of DMGs That Are Not DEGs
The observed delay in H3K27me3 changes for DEGs suggests that expression changes might be rather a cause than a consequence of H3K27me3 dynamics. This raises the question why several genes show significant changes in H3K27me3 during early flower morphogenesis but not in expression (Figure 3). It might be that changes in those genes were just below the threshold for differential expression. We therefore considered the absolute expression values and early expression changes for H3K27me3 DMGs (Figure S10). We found that expression changes in the expected direction are indeed limited to a small proportion of the H3K27me3 DMGs. Interestingly, the majority of genes that do not change in expression are expressed at very low levels or not at all during the complete series. We hypothesise that these genes might gain or lose H3K27me3 maybe through the action of other repressive chromatin features such as DNA methylation or H3K9me3 marking.
For H3K4me3, at least half of the DMGs show at least some change in the expected direction. However, also few changes in the opposite direction are observed, maybe due to interplay with repressive TFs.
Appendix B.
Appendix B.1. Supplementary Figure Legends
Supplementary Figure 1. Validation of the material used for the analysis of early events of gene activation, in situ. (A–G) Validation of the ap1cal AP1-GR induction system. To verify the morphological effect of AP1 induction and assess its blockage by the GR system, we compared inflorescence tissue of ap1cal plants and ap1cal 35S::AP1-GR plants, pre- and post-induction. (A–D) Floral induction in ap1cal double mutants by AP1-GR activation. Top viewed inflorescences of (A) ap1cal, (B–C) ap1cal 35S::AP1-GR and (D) Ler wild type (WT). Pictures were taken 12 days after single treatment with (A, C, D) or (B) without 1 μM of dex solution. Activation of the AP1-GR protein in the ap1cal led to the production of WT looking flowers. (E)–(H) Scanning electron micrographs of inflorescence-like meristems of (E) ap1-1 cal-1, (F) untreated ap1cal 35S::AP1-GR and (G) ap1cal 35S::AP1-GR, five days after a single treatment with 1 μM of dex solution. Scale bars: 10 μm. While ap1cal and non-induced ap1cal 35S::AP1-GR plants show over-proliferated inflorescence meristems with no difference in floral organ formation, ap1cal 35S::AP1-GR plants had formed full flower buds with clearly visible sepals five days after induction. (H–I) Specificity assessment of antibody lots used in the chromatin immuno-precipitation experiments. Dot-Blots using anti-H3K4me3 and anti-H3K27me3 antibodies were performed on peptides containing mono- (me1), di (me2) and tri (me3)-methylated versions of the respective residue.
Supplementary Figure 2. Comparison of expression changes from t0 to t2 in this study and in Wellmer et al. [26]. Fold changes (FC) in expression are depicted by a heat map reaching from genes expressed 5 times lower in t2 than t0 (blue) to genes expressed 10 times higher in t2 than t0 (red).
Supplementary Figure 3. Major expression patterns present among differentially expressed genes (DEG). K-means clustering with k = 8 was performed for all DEGs yielding an overview of expression profiles in the 8 clusters. Relative expression values are expressed as z-scores to reveal similarities in expression patterns. Averages of z-scores in each cluster are depicted in red. The grey lines represent the single genes in the cluster.
Supplementary Figure 4. Functional characterisation of up-regulated DEGs. Panels were generated with the Classification Super Viewer from the BAR website (http://bar.utoronto.ca) using default parameters. The expected background is calculated by bootstrapping 100 sets of the same size from the whole genome. The y-axis displays normed frequencies with the expected frequency in the background distribution (whole genome) set to 1 (red line).
Supplementary Figure 5. Functional characterisation of down-regulated DEGs. Panels were generated with the Classification Super Viewer from the BAR website (http://bar.utoronto.ca) using default parameters. The expected background is calculated by bootstrapping 100 sets of the same size from the whole genome. The y-axis displays normed frequencies with the expected frequency in the background distribution (whole genome) set to 1 (red line).
Supplementary Figure 6. Overlap between target genes in the four considered time points/tissues for H3K27me3 (K27) and H3K4me3 (K4).
Supplementary Figure 7. Overlap between differentially marked genes for H3K27me3 and H3K4me3 for changes from leaf to meristematic tissue (L-t0) and from meristematic tissue to inflorescences (t0-I).
Supplementary Figure 8. H3K4me3 signal shift between early and later expression changes. Only genes that are differentially expressed in both the t0 to t2 and t0 to I comparison, i.e., are activated or repressed early and stay activated/repressed after t2, are considered here (131 genes for activation and 105 genes for repression). Average differences in H3K4me3 signal (in reads per million) for genes are calculated in 200 bp windows/10% bins over genes from −200 bp to 200 bp downstream for each gene. For each bin, the percentage of the change on the whole change over the gene is displayed for t2 minus t0 and t2 minus I for both up-regulated (A) and down-regulated genes (B).
Supplementary Figure 9. Expression changes of early DEGs during flower morphogenesis at t1. Heat maps showing the difference in expression from t0 to t1 as measured in a microarray experiment [26]. Each line represents a DEG (from t0 to t2 comparison in our study). Genes are sorted by their average H3K27me3 change (t2-t0) as shown in the first column of the heat map with marks changing in the expected direction at the top. Note that only genes present on the microarray are shown and thus not all DEGs are considered.
Supplementary Figure 10. Expression of early DMGs during the time series. Heat maps showing expression values (FPKM) for significantly changing genes for H3K27me3 and H3K4me3. Each line represents a gene sorted by the expression changes during early flower morphogenesis (t2 minus t0, indicated as a column of the heatmap for orientation), with the highest negative expression change (down-regulation) at the top.
Supplementary Figure 11. Correlation of genome-wide changes in histone marks from t0 to t2 and binding of MADS TFs or DNase hypersensitivity during early flower development. (A) Fraction of DMGs for H3K27me3, H3K4me3 and both marks from t0 to t2 (K27 down: H3K27me3 reduced t0-t2, K4 up: H3K4me3 elevated t0-t2, both: H3K27me3 reduced and H3K4me3 elevated t0-t2) that are bound by AP1 and/or SEP3 during early flower development. TF binding at three time points after dex-induction was considered: t2, t4 and t8 (Pajoro et al., 2014). Numbers above each bar indicate the total number of DMGs for each respective comparison. (B) Fractions of same DMGs as in (A) that overlap with DNase I hypersensitive sites (DHS) during the same three time points as in (A) and at t0 (Pajoro et al., 2014). Numbers on each bar indicate the total number of DMGs in the respective comparison.
Appendix B.2. Supplementary Table Legends
Supplementary Table 1. List of genes that are differentially expressed in at least one condition. For each comparison of conditions, a number code indicates if genes are not changed from condition 1 to condition 2 (0), up-regulated in condition 2 compared to condition 1 (1) or down-regulated (−1). Genes are considered as differentially expressed when determined as significantly differentially expressed by Cufflinks (p < 0.05).
Supplementary Table 2. Functional enrichment analysis for DEGs. Significantly enriched GO terms among DEGs were determined for the three most relevant comparisons (L-I, t0-t2, t0-I) compared to the whole genome. False discovery rate (FDR) corrected p-values (adj_pvalue) are output of the FatiGO web tool, terms are considered significantly enriched for FDR corrected p-values lower than 0.05.
Supplementary Table 3. Pearson correlation coefficient analysis, ChIP-seq read numbers, average expression values for H3K27me3 and H3K4me3 target genes and significantly enriched genes for H3K27me3 and H3K4me3. Sheet 1: Read numbers for ChIP-seq samples and Pearson correlation coefficients for comparisons between replicates and between samples and inputs and average expression values of target genes (FPKM values). Read numbers given are reads that map to the TAIR10 genome annotation after removal of duplicated reads. Sheet 2: Target genes of H3K27me3 and H3K4me3: genes were considered as target genes when overlapping with a significantly enriched region determined by SICER. Only genes that were detected in both replicates were considered as target genes of the respective mark for further analysis. A comparison of all conditions and with previously published target gene lists (Roudier et al., 2011 ; Farrona et al., 2011 ; Luo et al., 2012) is shown. Sheet 3–10: Target gene lists for the different conditions.
Supplementary Table 4. Differentially marked genes for H3K27me3 and H3K4me3 in the tissue/time series. Sheet 1: Gene numbers of comparisons for all combinations of DMGs. Changes with elevated H3K4me3 (K4) and reduced H3K27me3 (K27) are highlighted in green, changes with reduced K4 and elevated K27 are highlighted in red. The grey diagonal indicates comparisons between the same time points/tissues. Numbers in parenthesis indicate the number of genes in the respective lists. For comparisons in the same direction and for the same mark, these numbers can be read out from the grey diagonal. Sheet 2: List of changes in all comparisons for all DMGs: 1 denotes elevation of the respective mark, −1 reduction, 0 denotes no change.
Supplementary Table 5. Functional enrichment analysis for H3K4me3 and H3K27me3 targets. Significantly enriched GO terms were determined for the four samples L, t0, t2 and I compared to the whole genome. FDR corrected p-values (adj_pvalue) are output of the FatiGO web tool, terms are considered significantly enriched for FDR corrected p-values lower than 0.05 (highlighted in green). Blank fields indicate that the term is not enriched in the respective sample.
Supplementary Table 6. Functional enrichment analysis for H3K4me3 and H3K27me3 DMGs. Significantly enriched or depleted GO terms among DMGs were determined for the L-t0, L-I, t0-t2, t0-I and t2-I comparisons for changes in both directions for both marks (separately and common DMGs). All targets of the respective mark were employed as background distribution. FDR corrected p-values (adj_pvalue) are output of the FatiGO web tool, terms are considered significantly enriched (highlighted in green) or depleted (highlighted in red) compared to all targets of the mark for FDR corrected p-values lower than 0.05. Blank fields indicate that the term is not enriched in the respective sample. If a comparison is not listed for one direction, no significant terms were found for this specific set of DMGs.
References and Note
- Alvarez-Buylla, E.R.; Benitez, M.; Corvera-Poire, A.; Chaos Cador, A.; de Folter, S.; Gamboa de Buen, A.; Garay-Arroyo, A.; Garcia-Ponce, B.; Jaimes-Miranda, F.; Perez-Ruiz, R.V.; et al. Flower development. Arabidopsis Book 2010, 8, e0127. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Pajoro, A.; Angenent, G.C. Regulation of transcription in plants: Mechanisms controlling developmental switches. Nat. Rev. Genet. 2010, 11, 830–842. [Google Scholar] [CrossRef] [PubMed]
- Chahtane, I.; Denay, G.; Engelhorn, J.; Monniaux, M.; Moyroud, E.; Moreau, F.; Carles, C.; Tichtinsky, G.; Zubieta, C.; Parcy, F. Flower development: An integrated view. In From Molecules to Living Organisms: An Interplay Between Biology and Physics; Pebay-Peyroula, E., Nury, H., Parcy, F., Ruigrok, R.W.H., Ziegler, C., Cugliandolo, L.F., Eds.; Oxford University Press: Oxford, UK, 2017; in press. [Google Scholar]
- Engelhorn, J.; Blanvillain, R.; Carles, C.C. Gene activation and cell fate control in plants: A chromatin perspective. Cell. Mol. Life Sci. CMLS 2014, 71, 3119–3137. [Google Scholar] [CrossRef] [PubMed]
- Pirrotta, V. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell 1998, 93, 333–336. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Chourrout, D.; Vervoort, M.; Leblanc, B.; Cavalli, G. Genome regulation by polycomb and trithorax proteins. Cell 2007, 128, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Morey, L.; Helin, K. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 2010, 35, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Clarenz, O.; Cokus, S.; Bernatavichute, Y.V.; Pellegrini, M.; Goodrich, J.; Jacobsen, S.E. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007, 5, e129. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Park, S.; van Nocker, S. Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 2008, 4, e1000077. [Google Scholar] [CrossRef] [PubMed]
- Roudier, F.; Ahmed, I.; Berard, C.; Sarazin, A.; Mary-Huard, T.; Cortijo, S.; Bouyer, D.; Caillieux, E.; Duvernois-Berthet, E.; Al-Shikhley, L.; et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011, 30, 1928–1938. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Martinez, A.M.; Iovino, N.; Cavalli, G. Trithorax group proteins: Switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 2011, 12, 799–814. [Google Scholar] [CrossRef] [PubMed]
- Kingston, R.E.; Tamkun, J.W. Transcriptional regulation by trithorax-group proteins. Cold Spring Harb. Perspect. Biol. 2014, 6, a019349. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Venegas, R. Regulation by polycomb and trithorax group proteins in Arabidopsis. Arabidopsis Book 2010, 8, e0128. [Google Scholar] [CrossRef] [PubMed]
- Kohler, C.; Hennig, L. Regulation of cell identity by plant Polycomb and trithorax group proteins. Curr. Opin. Genet. Dev. 2010, 20, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Muller, R.; Goodrich, J. Sweet memories: Epigenetic control in flowering. F1000 Biol. Rep. 2011, 3. [Google Scholar] [CrossRef]
- De la Paz Sanchez, M.; Aceves-Garcia, P.; Petrone, E.; Steckenborn, S.; Vega-Leon, R.; Alvarez-Buylla, E.R.; Garay-Arroyo, A.; Garcia-Ponce, B. The impact of Polycomb group (PcG) and Trithorax group (TrxG) epigenetic factors in plant plasticity. New Phytol. 2015, 208, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Pu, L.; Sung, Z.R. PcG and trxG in plants—Friends or foes. Trends Genet. TIG 2015, 31, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Weinhofer, I.; Hehenberger, E.; Roszak, P.; Hennig, L.; Kohler, C. H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Lafos, M.; Kroll, P.; Hohenstatt, M.L.; Thorpe, F.L.; Clarenz, O.; Schubert, D. Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 2011, 7, e1002040. [Google Scholar] [CrossRef] [PubMed]
- Brusslan, J.A.; Rus Alvarez-Canterbury, A.M.; Nair, N.U.; Rice, J.C.; Hitchler, M.J.; Pellegrini, M. Genome-wide evaluation of histone methylation changes associated with leaf senescence in Arabidopsis. PLoS ONE 2012, 7, e33151. [Google Scholar] [CrossRef] [PubMed]
- Brusslan, J.A.; Bonora, G.; Rus-Canterbury, A.M.; Tariq, F.; Jaroszewicz, A.; Pellegrini, M. A Genome-Wide Chronological Study of Gene Expression and Two Histone Modifications, H3K4me3 and H3K9ac, during Developmental Leaf Senescence. Plant Physiol. 2015, 168, 1246–1261. [Google Scholar] [CrossRef] [PubMed]
- Charron, J.B.; He, H.; Elling, A.A.; Deng, X.W. Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 2009, 21, 3732–3748. [Google Scholar] [CrossRef] [PubMed]
- He., C.; Chen, X.; Huang, H.; Xu, L. Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet. 2012, 8, e1002911. [Google Scholar] [CrossRef] [PubMed]
- Adrian, J.; Farrona, S.; Reimer, J.J.; Albani, M.C.; Coupland, G.; Turck, F. cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 2010, 22, 1425–1440. [Google Scholar] [CrossRef] [PubMed]
- Buzas, D.M.; Robertson, M.; Finnegan, E.J.; Helliwell, C.A. Transcription-dependence of histone H3 lysine 27 trimethylation at the Arabidopsis polycomb target gene FLC. Plant J. Cell Mol. Biol. 2011, 65, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Wellmer, F.; Alves-Ferreira, M.; Dubois, A.; Riechmann, J.L.; Meyerowitz, E.M. Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet. 2006, 2, e117. [Google Scholar] [CrossRef]
- Kaufmann, K.; Wellmer, F.; Muino, J.M.; Ferrier, T.; Wuest, S.E.; Kumar, V.; Serrano-Mislata, A.; Madueno, F.; Krajewski, P.; Meyerowitz, E.M.; et al. Orchestration of floral initiation by APETALA1. Science 2010, 328, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.; Muino, J.M.; Blanvillain, R.; Busscher, M.; Busscher-Lange, J.; Dinh, Q.D.; Liu, S.; Westphal, A.H.; Boeren, S.; et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 2012, 109, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Pajoro, A.; Madrigal, P.; Muino, J.M.; Matus, J.T.; Jin, J.; Mecchia, M.A.; Debernardi, J.M.; Palatnik, J.F.; Balazadeh, S.; Arif, M.; et al. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014, 15, R41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Bernatavichute, Y.V.; Cokus, S.; Pellegrini, M.; Jacobsen, S.E. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009, 10, R62. [Google Scholar] [CrossRef] [PubMed]
- Gan, E.S.; Huang, J.; Ito, T. Functional roles of histone modification, chromatin remodeling and microRNAs in Arabidopsis flower development. Int. Rev. Cell Mol. Biol. 2013, 305, 115–161. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Fromm, M.; Avramova, Z. H3K27me3 and H3K4me3 chromatin environment at super-induced dehydration stress memory genes of Arabidopsis thaliana. Mol. Plant 2014, 7, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Gillissen, B.; Burkle, L.; Andre, B.; Kuhn, C.; Rentsch, D.; Brandl, B.; Frommer, W.B. A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in Arabidopsis. Plant Cell 2000, 12, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Madrid, R.; Mestres, D.; Marinho, P.; Jacquot, J.P.; Decottignies, P.; Miginiac-Maslow, M.; Meyer, Y. Evidence for five divergent thioredoxin h sequences in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1995, 92, 5620–5624. [Google Scholar] [CrossRef] [PubMed]
- Laloi, C.; Mestres-Ortega, D.; Marco, Y.; Meyer, Y.; Reichheld, J.P. The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol. 2004, 134, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- Sarry, J.E.; Kuhn, L.; Ducruix, C.; Lafaye, A.; Junot, C.; Hugouvieux, V.; Jourdain, A.; Bastien, O.; Fievet, J.B.; Vailhen, D.; et al. The early responses of Arabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics 2006, 6, 2180–2198. [Google Scholar] [CrossRef] [PubMed]
- Sweat, T.A.; Wolpert, T.J. Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis. Plant Cell 2007, 19, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Mena, C.; de Folter, S.; Costa, M.M.; Angenent, G.C.; Sablowski, R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 2005, 132, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Dinant, S.; Clark, A.M.; Zhu, Y.; Vilaine, F.; Palauqui, J.C.; Kusiak, C.; Thompson, G.A. Diversity of the superfamily of phloem lectins (phloem protein 2) in angiosperms. Plant Physiol. 2003, 131, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Yanofsky, M.F.; Meyerowitz, E.M. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991, 5, 484–495. [Google Scholar]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kim, M.R.; Bedgar, D.L.; Moinuddin, S.G.; Cardenas, C.L.; Davin, L.B.; Kang, C.; Lewis, N.G. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 1455–1460. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.; Smyth, D.R. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 1999, 126, 2377–2386. [Google Scholar] [PubMed]
- Heisler, M.G.; Atkinson, A.; Bylstra, Y.H.; Walsh, R.; Smyth, D.R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 2001, 128, 1089–1098. [Google Scholar] [PubMed]
- Turlapati, P.V.; Kim, K.W.; Davin, L.B.; Lewis, N.G. The laccase multigene family in Arabidopsis thaliana: Towards addressing the mystery of their gene function(s). Planta 2011, 233, 439–470. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A. Aintegumenta and Aintegumenta-Like6 regulate auxin-mediated flower development in Arabidopsis. BMC Res. Notes 2011, 4, 176. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A.; Eaddy, M. AINTEGUMENTA-LIKE6 regulates cellular differentiation in flowers. Plant Mol. Biol. 2012, 78, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Alvarez, J.; Smyth, D.R.; Yanofsky, M.F.; Meyerowitz, E.M. LFY controls floral meristem identity in Arabidopsis. Cell 1992, 69, 843–859. [Google Scholar] [CrossRef]
- Blazquez, M.A.; Soowal, L.N.; Lee, I.; Weigel, D. LEAFY expression and flower initiation in Arabidopsis. Development 1997, 124, 3835–3844. [Google Scholar] [PubMed]
- Parcy, F.; Nilsson, O.; Busch, M.A.; Lee, I.; Weigel, D. A genetic framework for floral patterning. Nature 1998, 395, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.X.; Wang, Y.X.; Liu, X.F.; Li, J.Y. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 2004, 14, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Fowler, S.G.; Cook, D.; Thomashow, M.F. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005, 137, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Petricka, J.J.; Clay, N.K.; Nelson, T.M. Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2008, 56, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Hanano, K.; Kariya, A.; Shimizu, S.; Zhao, L.; Matsui, M.; Tasaka, M.; Aida, M. CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. Cell Mol. Biol. 2011, 66, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.; Zambryski, P.C. Organ boundary1 defines a gene expressed at the junction between the shoot apical meristem and lateral organs. Proc. Natl. Acad. Sci. USA 2011, 108, 2154–2159. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Nakazawa, M.; Takase, T.; Manabe, K.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Matsui, M. Overexpression of LSH1, a member of an uncharacterised gene family, causes enhanced light regulation of seedling development. Plant J. Cell Mol. Biol. 2004, 37, 694–706. [Google Scholar] [CrossRef]
- Joshi, V.; Laubengayer, K.M.; Schauer, N.; Fernie, A.R.; Jander, G. Two Arabidopsis threonine aldolases are nonredundant and compete with threonine deaminase for a common substrate pool. Plant Cell 2006, 18, 3564–3575. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, T.; Nakazawa, M.; Kawashima, M.; Iizumi, H.; Kuroda, H.; Kondou, Y.; Tsuhara, Y.; Suzuki, K.; Ishikawa, A.; Seki, M.; et al. The FOX hunting system: An alternative gain-of-function gene hunting technique. Plant J. Cell Mol. Biol. 2006, 48, 974–985. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.C.; Flematti, G.R.; Riseborough, J.A.; Ghisalberti, E.L.; Dixon, K.W.; Smith, S.M. Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 7095–7100. [Google Scholar] [CrossRef] [PubMed]
- Cartagena, J.A.; Matsunaga, S.; Seki, M.; Kurihara, D.; Yokoyama, M.; Shinozaki, K.; Fujimoto, S.; Azumi, Y.; Uchiyama, S.; Fukui, K. The Arabidopsis SDG4 contributes to the regulation of pollen tube growth by methylation of histone H3 lysines 4 and 36 in mature pollen. Dev. Biol. 2008, 315, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Moylan, E.C.; Langdale, J.A. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. Cell Mol. Biol. 2008, 56, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Arsovski, A.A.; Popma, T.M.; Haughn, G.W.; Carpita, N.C.; McCann, M.C.; Western, T.L. AtBXL1 encodes a bifunctional beta-D-xylosidase/alpha-L-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol. 2009, 150, 1219–1234. [Google Scholar] [CrossRef] [PubMed]
- Schubert, D.; Primavesi, L.; Bishopp, A.; Roberts, G.; Doonan, J.; Jenuwein, T.; Goodrich, J. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006, 25, 4638–4649. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Deng, S.; Wang, H.; Ye, J.; Wu, H.W.; Sun, H.X.; Chua, N.H. CURLY LEAF Regulates Gene Sets Coordinating Seed Size and Lipid Biosynthesis. Plant Physiol. 2016, 171, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Smyth, D.R.; Bowman, J.L.; Meyerowitz, E.M. Early flower development in Arabidopsis. Plant Cell 1990, 2, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, R.; Das, P.; Mirabet, V.; Moscardi, E.; Traas, J.; Verdeil, J.L.; Malandain, G.; Godin, C. Imaging plant growth in 4D: Robust tissue reconstruction and lineaging at cell resolution. Nat. Methods 2010, 7, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Looi, L.S.; Guo, S.; He, Z.; Gan, E.S.; Huang, J.; Xu, Y.; Wee, W.Y.; Ito, T. Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 2014, 343, 1248559. [Google Scholar] [CrossRef] [PubMed]
- Schmitges, F.W.; Prusty, A.B.; Faty, M.; Stutzer, A.; Lingaraju, G.M.; Aiwazian, J.; Sack, R.; Hess, D.; Li, L.; Zhou, S.; et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell. 2011, 42, 330–341. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ndamukong, I.; Xu, Z.; Lapko, H.; Fromm, M.; Avramova, Z. ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PLoS Genet. 2012, 8, e1003111. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Sidote, D.J.; Zhang, Y.; Kerstetter, R.A.; Michael, T.P.; Lam, E. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J. Cell Mol. Biol. 2013, 73, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Sequeira-Mendes, J.; Araguez, I.; Peiro, R.; Mendez-Giraldez, R.; Zhang, X.; Jacobsen, S.E.; Bastolla, U.; Gutierrez, C. The Functional Topography of the Arabidopsis Genome Is Organized in a Reduced Number of Linear Motifs of Chromatin States. Plant Cell 2014, 26, 2351–2366. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Alvarez, J.; Weigel, D.; Meyerowitz, E. M.; Smyth, D. R. Control of flower development in Arabidopsis thaliana by APETALA 1 and interacting genes. Development 1993, 119, 721–743. [Google Scholar]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genes directing flower development in Arabidopsis. Plant Cell 1989, 1, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Carles, C.C.; Fletcher, J.C. The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev. 2009, 23, 2723–2728. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Muino, J.M.; Osteras, M.; Farinelli, L.; Krajewski, P.; Angenent, G.C. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 2010, 5, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Yant, L.; Mathieu, J.; Dinh, T.T.; Ott, F.; Lanz, C.; Wollmann, H.; Chen, X.; Schmid, M. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 2010, 22, 2156–2170. [Google Scholar] [CrossRef] [PubMed]
- Immink, R.G.; Pose, D.; Ferrario, S.; Ott, F.; Kaufmann, K.; Valentim, F.L.; de Folter, S.; van der Wal, F.; van Dijk, A.D.; Schmid, M.; et al. Characterization of SOC1's central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol. 2012, 160, 433–449. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Picard-tools dfpsnin, website now moved to http://broadinstitute.github.io/picard, version used is 1.93.
- Zang, C.; Schones, D.E.; Zeng, C.; Cui, K.; Zhao, K.; Peng, W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 2009, 25, 1952–1958. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Sturn, A.; Quackenbush, J.; Trajanoski, Z. Genesis: Cluster analysis of microarray data. Bioinformatics 2002, 18, 207–208. [Google Scholar] [CrossRef] [PubMed]
- Engelhorn, J.; Turck, F. Metaanalysis of ChIP-chip data. Methods Mol. Biol. 2010, 631, 185–207. [Google Scholar] [CrossRef] [PubMed]
- Provart, N.; Zhu, T. A browser-based functional classification SuperViewer for Arabidopsis genomics. Curr. Comput. Mol. Biol. 2003, 2003, 271–272. [Google Scholar]
- Al-Shahrour, F.; Minguez, P.; Tarraga, J.; Medina, I.; Alloza, E.; Montaner, D.; Dopazo, J. FatiGO +: A functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 2007, 35, W91–W96. [Google Scholar] [CrossRef] [PubMed]
- Medina, I.; Carbonell, J.; Pulido, L.; Madeira, S.C.; Goetz, S.; Conesa, A.; Tarraga, J.; Pascual-Montano, A.; Nogales-Cadenas, R.; Santoyo, J.; et al. Babelomics: An integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 2010, 38, W210–W213. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Zweig, A.S.; Barber, G.; Hinrichs, A.S.; Karolchik, D. BigWig and BigBed: Enabling browsing of large distributed datasets. Bioinformatics 2010, 26, 2204–2207. [Google Scholar] [CrossRef] [PubMed]
- Stein, L.D. Using GBrowse 2.0 to visualize and share next-generation sequence data. Brief Bioinform. 2013, 14, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Farrona, S.; Thorpe, F.L.; Engelhorn, J.; Adrian, J.; Dong, X.; Sarid-Krebs, L.; Goodrich, J.; Turck, F. Tissue-specific expression of FLOWERING LOCUS T in Arabidopsis is maintained independently of polycomb group protein repression. Plant Cell 2011, 23, 3204–3214. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).