Reduced Diversity in the Bacteriome of the Phytophagous Mite Brevipalpus yothersi (Acari: Tenuipalpidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection, Identification, and Laboratory Rearing of Mite Species
2.2. Genomic DNA Extraction and Pyrosequencing
2.3. Data Processing and Quality Control
2.4. Analysis of Bacterial Diversity
2.5. Phylogenetic Analysis of Cardinium Strains
3. Results
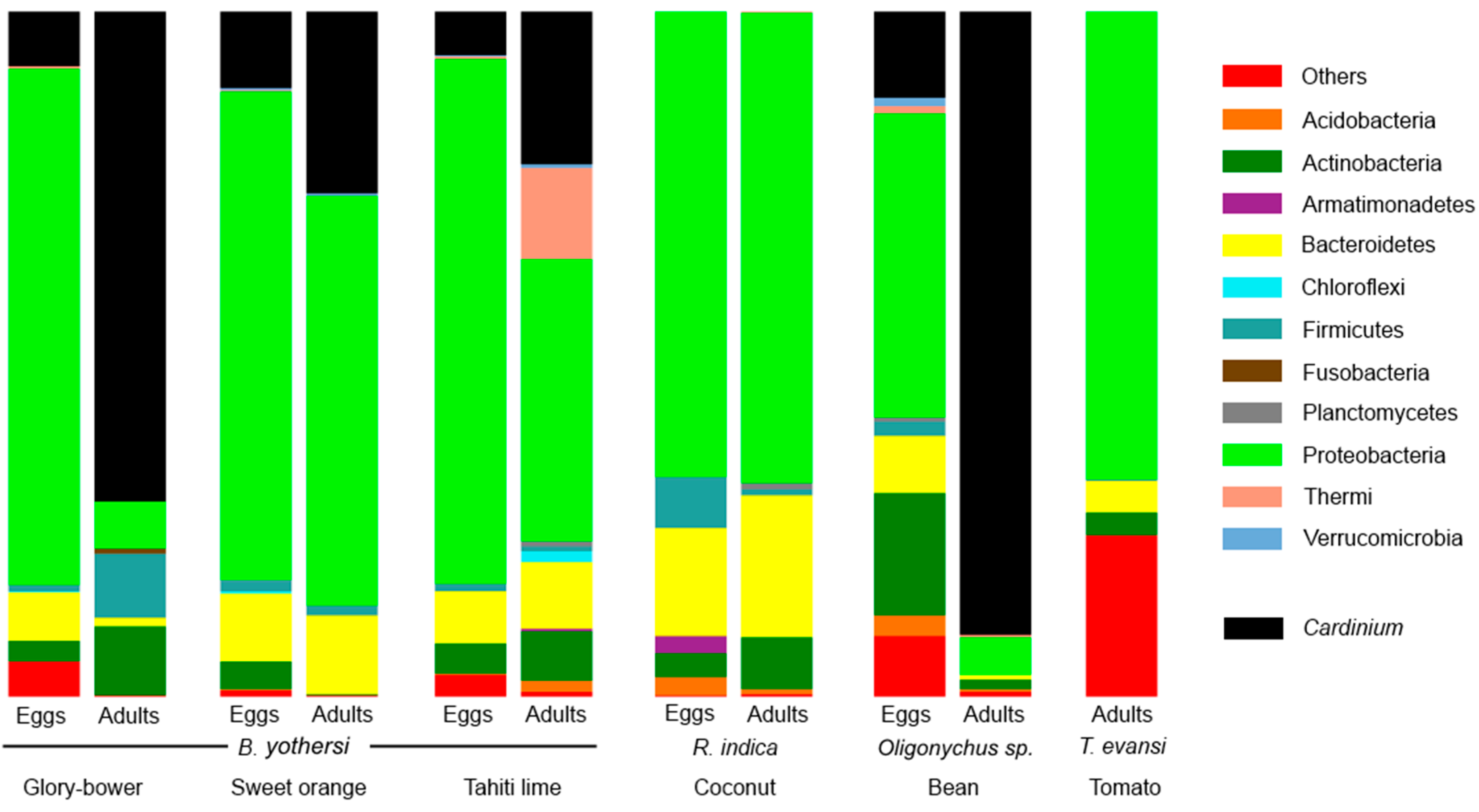
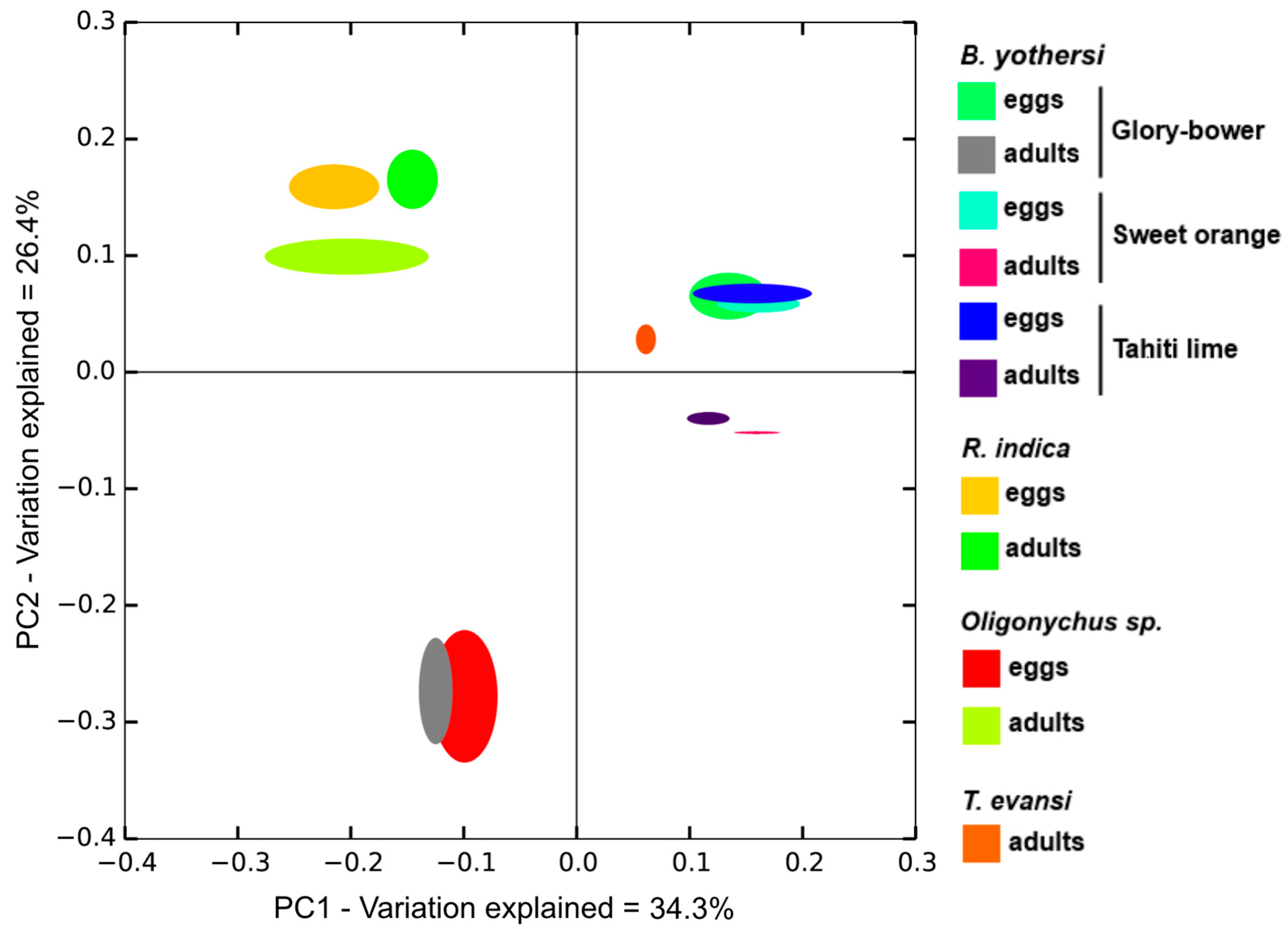
3.1. Composition of the Bacteriomes
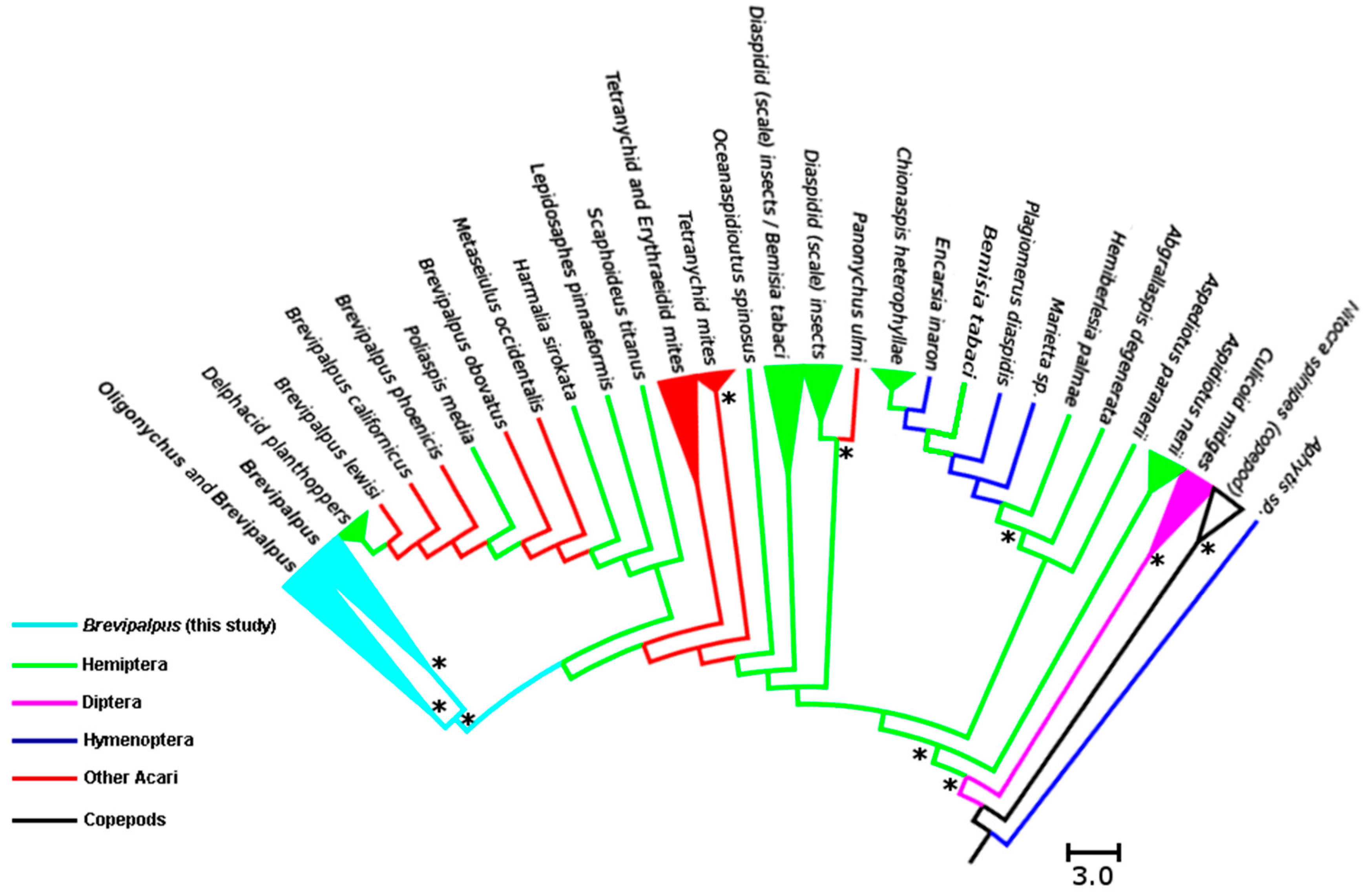
3.2. Analysis of Cardinium Endosymbionts
4. Discussion
4.1. Factors Affecting Diversity in Bacterial Communities
4.2. Phylogenetic Relationship among Cardinium Strains in Different Systems
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kitajima, E.W.; Rezende, J.A.M.; Rodrigues, J.C.V. Passion fruit green spot virus vectored by Brevipalpus phoenicis (Acari: Tenuipalpidae) on passion fruit in Brazil. Exp. Appl. Acarol. 2003, 30, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, E.W.; Rodrigues, J.C.V.; Freitas-Astua, J. An annotated list of ornamentals naturally found infected by Brevipalpus mite-transmitted viruses. Sci. Agric. 2010, 67, 348–371. [Google Scholar] [CrossRef]
- Rodrigues, J.C.V.; Childers, C.C. Brevipalpus mites (Acari: Tenuipalpidae): Vectors of invasive, non-systemic cytoplasmic and nuclear viruses in plants. Exp. Appl. Acarol. 2013, 59, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Beard, J.J.; Ochoa, R.; Bauchan, G.R.; Trice, M.D.; Redford, A.J.; Walters, T.W.; Mitter, C. Flat Mites of the World. Available online: http://idtools.org/id/mites/flatmites/ (accessed on 8 September 2016).
- Beard, J.J.; Ochoa, R.; Braswell, W.E.; Bauchan, G.R. Brevipalpus phoenicis (Geijskes) species complex (Acari: Tenuipalpidae)—A closer look. Zootaxa 2015, 3944, 1–67. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.C.V.; Kitajima, E.W.; Childers, C.C.; Chagas, C.M. Citrus leprosis virus vectored by Brevipalpus phoenicis (Acari: Tenuipalpidae) on citrus in Brazil. Exp. Appl. Acarol. 2003, 30, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Carstens, E.B. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009). Arch. Virol. 2010, 155, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Weeks, A.R.; Marec, F.; Breeuwer, J.A.J. A mite species that consists entirely of haploid females. Science 2001, 292, 2479–2482. [Google Scholar] [CrossRef] [PubMed]
- Zchori-Fein, E.; Gottlieb, Y.; Kelly, S.E.; Brown, J.K.; Wilson, J.M.; Karr, T.L.; Hunter, M.S. A newly discovered bacterium associated with parthenogenesis and a change in host selection behavior in parasitoid wasps. Proc. Natl. Acad. Sci. USA 2001, 98, 12555–12560. [Google Scholar] [CrossRef] [PubMed]
- Feldhaar, H.; Straka, J.; Krischke, M.; Berthold, K.; Stoll, S.; Mueller, M.J.; Gross, R. Nutritional upgrading for omnivorous carpenter ants by the endosymbiont Blochmannia. BMC Biol. 2007. [Google Scholar] [CrossRef] [PubMed]
- Sabree, Z.L.; Kambhampati, S.; Moran, N.A. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc. Natl. Acad. Sci. USA 2009, 106, 19521–19526. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.M.; Moran, N.A.; Hunter, M.S. Variation in resistance to parasitism in aphids is due to symbiosis not host genotype. Proc. Natl. Acad. Sci. USA 2005, 102, 12795–12800. [Google Scholar] [CrossRef] [PubMed]
- Scarborough, C.L.; Ferrari, J.; Godfray, H.C.J. Aphid protected from pathogen by endosymbiont. Science 2005. [Google Scholar] [CrossRef] [PubMed]
- Stouthamer, R.; Breeuwer, J.A.J.; Hurst, G.D. Wolbachia pipientis: Microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 1999, 53, 71–102. [Google Scholar] [CrossRef] [PubMed]
- Pais, R.; Lohs, C.; Wu, Y.; Wang, J.; Aksoy, S. The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 2008, 74, 5965–5974. [Google Scholar] [CrossRef] [PubMed]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. 2008, 6, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.M.; Degnan, P.H.; Burke, G.R.; Moran, N.A. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 2010, 55, 247–266. [Google Scholar] [CrossRef] [PubMed]
- Cordaux, R.; Bouchon, D.; Greve, P. The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends Genet. 2011, 27, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, M.P. Mutualistic endosymbiotic microbes: An underappreciated benefit of group living. Behav. Ecol. Sociobiol. 2008, 62, 479–497. [Google Scholar] [CrossRef]
- Wang, Y.; Gilbreath, T.M.; Kukutla, P.; Yan, G.; Xu, J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS ONE 2011, 6, e24767. [Google Scholar] [CrossRef] [PubMed]
- Vasanthakumar, A.; Handelsman, J.; Schloss, P.D.; Bauer, L.S.; Raffa, K.F. Gut microbiota of an invasive subcortical beetle, Agrilus planipennis Fairmaire, across various life stages. Environ. Entomol. 2008, 37, 1344–1353. [Google Scholar] [CrossRef]
- Yen, J.H.; Barr, A.R. New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiens L. Nature 1971, 232, 657–658. [Google Scholar] [CrossRef] [PubMed]
- Morse, S.F.; Bush, S.E.; Patterson, B.D.; Dick, C.W.; Gruwell, M.E.; Dittmar, K. Evolution, multiple acquisition, and localization of endosymbionts in bat flies (Diptera: Hippoboscoidea: Streblidae and Nycteribiidae). Appl. Environ. Microbiol. 2013, 79, 2952–2961. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Dong, P.; Xiao, L.S.; Dou, W. Effects of removal of Cardinium infection on fitness of the stored-product pest Liposcelis bostrychophila (Psocoptera: Liposcelididae). J. Econ. Entomol. 2008, 101, 1711–1717. [Google Scholar] [CrossRef]
- Rodrigues, J.C.V.; Gallo-Meagher, M.; Ochoa, R.; Childers, C.C.; Adams, B.J. Mitochondrial DNA and RAPD polymorphisms in the haploid mite Brevipalpus phoenicis (Acari: Tenuipalpidae). Exp. Appl. Acarol. 2004, 34, 275–290. [Google Scholar] [CrossRef] [PubMed]
- Navajas, M.; Lagnel, J.; Gutierrez, J.; Boursot, P. Species-wide homogeneity of nuclear ribosomal ITS2 sequences in the spider mite Tetranychus urticae contrasts with extensive mitochondrial COI polymorphism. Heredity 1998, 80, 742–752. [Google Scholar] [CrossRef] [PubMed]
- Hori, M.; Fukano, H.; Suzuki, Y. Uniform amplification of multiple DNAs by emulsion PCR. Biochem. Biophys. Res. Commun. 2007, 352, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; Desantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Baumann, P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 2005, 59, 155–189. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A.; McCutcheon, J.P.; Nakabachi, A. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 2008, 42, 165–190. [Google Scholar] [CrossRef] [PubMed]
- Mcneill, M.R.; Richards, N.K.; White, J.A.; Laugraud, A. Hidden arsenal: Endosymbionts in arthropods, their role and possible implications for biological control success. N. Z. Plant Prot. 2014, 67, 204–212. [Google Scholar]
- Moran, N.A.; Hansen, A.K.; Powell, J.E.; Sabree, Z.L. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE 2012, 7, e36393. [Google Scholar] [CrossRef] [PubMed]
- Osei-Poku, J.; Mbogo, C.M.; Palmer, W.J.; Jiggins, F.M. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol. Ecol. 2012, 21, 5138–5150. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.T.; Sanchez, L.G.; Fierer, N. A cross-taxon analysis of insect-associated bacterial diversity. PLoS ONE 2013, 8, e61218. [Google Scholar] [CrossRef] [PubMed]
- Pernice, M.; Simpson, S.J.; Ponton, F. Towards an integrated understanding of gut microbiota using insects as model systems. J. Insect Physiol. 2014, 69, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Juchault, P.; Rigaud, T.; Mocquard, J.-P. Evolution of sex-determining mechanisms in a wild population of Armadillidium vulgare Latr. (Crustacea, Isopoda): Competition between two feminizing parasitic sex factors. Heredity 1992, 69, 382–390. [Google Scholar] [CrossRef]
- Macaluso, K.R.; Sonenshine, D.E.; Ceraul, S.M.; Azad, A.F. Rickettsial infection in Dermacentor variabilis (Acari: Ixodidae) inhibits transovarial transmission of a second Rickettsia. J. Med. Entomol. 2002, 39, 809–813. [Google Scholar] [CrossRef] [PubMed]
- Kondo, N.; Shimada, M.; Fukatsu, T. Infection density of Wolbachia endosymbiont affected by co-infection and host genotype. Biol. Lett. 2005, 1, 488–491. [Google Scholar] [CrossRef] [PubMed]
- Hughes, G.L.; Dodson, B.L.; Johnson, R.M.; Murdock, C.C.; Tsujimoto, H.; Suzuki, Y.; Patt, A.A.; Cui, L.; Nossa, C.W.; Barry, R.M.; et al. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc. Natl. Acad. Sci. USA 2014, 111, 12498–12503. [Google Scholar] [CrossRef] [PubMed]
- Indiragandhi, P.; Anandham, R.; Madhaiyan, M.; Kim, G.H.; Sa, T. Cross-utilization and expression of outer membrane receptor proteins for siderophore uptake by Diamondback moth Plutella xylostella (Lepidoptera: Plutellidae) gut bacteria. FEMS Microbiol. Lett. 2008, 289, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Broderick, N.A.; Raffa, K.F.; Goodman, R.M.; Handelsman, J. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl. Environ. Microbiol. 2004, 70, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Pornwiroon, W.; Kearney, M.T.; Husseneder, C.; Foil, L.D.; Macaluso, K.R. Comparative microbiota of Rickettsia felis-uninfected and -infected colonized cat fleas, Ctenocephalides felis. ISME J. 2007, 1, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Hawlena, H.; Rynkiewicz, E.; Toh, E.; Alfred, A.; Durden, L.A.; Hastriter, M.W.; Nelson, D.E.; Rong, R.; Munro, D.; Dong, Q.; et al. The arthropod, but not the vertebrate host or its environment, dictates bacterial community composition of fleas and ticks. ISME J. 2013, 7, 221–223. [Google Scholar] [CrossRef] [PubMed]
- Levy, R.; Borenstein, E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc. Natl. Acad. Sci. USA 2013, 110, 12804–12809. [Google Scholar] [CrossRef] [PubMed]
- Zindel, R.; Gottlieb, Y.; Aebi, A. Arthropod symbioses: A neglected parameter in pest- and disease-control programmes. J. Appl. Ecol. 2011, 48, 864–872. [Google Scholar] [CrossRef]
- Lee, J.; Shin, S.C.; Kim, S.J.; Kim, B.K.; Hong, S.G.; Kim, E.H.; Park, H.; Lee, H. Draft genome sequence of a Sphingomonas sp., an endosymbiotic bacterium isolated from an arctic lichen Umbilicaria sp. J. Bacteriol. 2012, 194, 3010–3011. [Google Scholar] [CrossRef] [PubMed]
- Estes, A.M.; Hearn, D.J.; Bronstein, J.L.; Pierson, E.A. The olive fly endosymbiont, “Candidatus Erwinia dacicola”, switches from an intracellular existence to an extracellular existence during host insect development. Appl. Environ. Microbiol. 2009, 75, 7097–7106. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.T.; Knight, R.; Martin, A.P. Bacterial communities of disease vectors sampled across time, space, and species. ISME J. 2010, 4, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Costello, E.K.; Stagaman, K.; Dethlefsen, L.; Bohannan, B.J.M.; Relman, D.A. The application of ecological theory toward an understanding of the human microbiome. Science 2012, 336, 1255–1262. [Google Scholar] [CrossRef] [PubMed]
- Horner-Devine, M.C.; Bohannan, B.J.M. Phylogenetic clustering and overdispersion in bacterial communities. Ecology 2006, 87, S100–S108. [Google Scholar] [CrossRef]
- Ceja-Navarro, J.A.; Nguyen, N.H.; Karaoz, U.; Gross, S.R.; Herman, D.J.; Andersen, G.L.; Bruns, T.D.; Pett-Ridge, J.; Blackwell, M.; Brodie, E.L. Compartmentalized microbial composition, oxygen gradients and nitrogen fixation in the gut of Odontotaenius disjunctus. ISME J. 2014, 8, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Koeppel, A.F.; Wu, M. Species matter: The role of competition in the assembly of congeneric bacteria. ISME J. 2014, 8, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Emerson, B.C.; Gillespie, R.G. Phylogenetic analysis of community assembly and structure over space and time. Trends Ecol. Evol. 2008, 23, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Zchori-Fein, E.; Perlman, S.J. Distribution of the bacterial symbiont Cardinium in arthropods. Mol. Ecol. 2004, 13, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- Chaisiri, K.; McGarry, J.W.; Morand, S.; Makepeace, B.L. Symbiosis in an overlooked microcosm: A systematic review of the bacterial flora of mites. Parasitology 2015, 142, 1152–1162. [Google Scholar] [CrossRef] [PubMed]
- Maynard-Smith, J. The Evolution of Sex; Cambridge University Press: Cambridge, UK, 1978. [Google Scholar]
- Lynch, M.; Burger, R.; Butcher, D.; Gabriel, W. The mutational meltdown in asexual populations. J. Hered. 1993, 84, 339–344. [Google Scholar] [PubMed]
| Species | Host Plant | Stage | Tag |
|---|---|---|---|
| Brevipalpus yothersi | Glory-bower | eggs | GAGATCAG |
| adults | GAGTAGAC | ||
| Sweet orange | eggs | GAGATCTC | |
| adults | GAGATGAC | ||
| Tahiti lime | eggs | GAGTACTC | |
| adults | GAGTACAG | ||
| Raoiella indica | Coconut | eggs | GAGTCACT |
| adults | GATGCAAG | ||
| Oligonychus sp. | Bean | eggs | GATGAGCA |
| adults | GATGAGGT | ||
| Tetranychus evansi | Tomato | adults | GAGTAGTG |
| Mite Species | Host | Stage | Cardinium | Portiera | Tremblaya | Wolbachia |
|---|---|---|---|---|---|---|
| Brevipalpus yothersi | Glory-bower | Eggs | 7.9 | - | - | - |
| Adults | 71.5 | - | - | - | ||
| Sweet orange | Eggs | 11.1 | - | - | 0.1 | |
| Adults | 26.5 | - | - | - | ||
| Tahiti lime | Eggs | 6.4 | 0.3 | - | - | |
| Adults | 22.2 | - | - | - | ||
| Raoiella indica | Coconut | Eggs | - | - | - | - |
| Adults | - | - | <0.1 | - | ||
| Oligonychus sp. | Bean | Eggs | 12.6 | - | - | - |
| Adults | 90.9 | - | - | - | ||
| Tetranychus evansi | Tomato | Adults | - | - | - | - |
| All the sequences | 22.25 | 0.03 | <0.01 | <0.01 | ||
| Species | Host Plant | Stage | Chao1 | Shannon |
|---|---|---|---|---|
| Brevipalpus yothersi | Glory-bower | eggs | 100.88 ± 16.95 | 3.32 ± 0.13 |
| adults | 94.75 ± 16.26 | 2.79 ± 0.09 | ||
| Sweet orange | eggs | 95.20 ± 11.62 | 2.73 ± 0.09 | |
| adults | 55.93 ± 9.46 | 2.12 ± 0.07 | ||
| Tahiti lime | eggs | 97.66 ± 18.88 | 2.36 ± 0.10 | |
| adults | 146.02 ± 21.25 | 4.24 ± 0.09 | ||
| Raoiella indica | Coconut | eggs | 82.31 ± 15.13 | 4.41 ± 0.09 |
| adults | 115.49 ± 15.59 | 4.74 ± 0.10 | ||
| Oligonychus sp. | Bean | eggs | 79.46 ± 7.19 | 4.42 ± 0.09 |
| adults | 47.17 ± 13.36 | 1.00 ± 0.07 | ||
| Tetranychus evansi | Tomato | adults | 54.52 ± 4.45 | 3.66 ± 0.05 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ospina, O.E.; Massey, S.E.; Verle Rodrigues, J.C. Reduced Diversity in the Bacteriome of the Phytophagous Mite Brevipalpus yothersi (Acari: Tenuipalpidae). Insects 2016, 7, 80. https://doi.org/10.3390/insects7040080
Ospina OE, Massey SE, Verle Rodrigues JC. Reduced Diversity in the Bacteriome of the Phytophagous Mite Brevipalpus yothersi (Acari: Tenuipalpidae). Insects. 2016; 7(4):80. https://doi.org/10.3390/insects7040080
Chicago/Turabian StyleOspina, Oscar E., Steven E. Massey, and Jose Carlos Verle Rodrigues. 2016. "Reduced Diversity in the Bacteriome of the Phytophagous Mite Brevipalpus yothersi (Acari: Tenuipalpidae)" Insects 7, no. 4: 80. https://doi.org/10.3390/insects7040080
APA StyleOspina, O. E., Massey, S. E., & Verle Rodrigues, J. C. (2016). Reduced Diversity in the Bacteriome of the Phytophagous Mite Brevipalpus yothersi (Acari: Tenuipalpidae). Insects, 7(4), 80. https://doi.org/10.3390/insects7040080








