1. Introduction
Insect infertility techniques involve the introduction of sterile male insects to reduce pest reproduction, thereby providing an environmentally sustainable approach to pest control [
1,
2]. This method is widely applied, especially in the control of fruit flies, mosquitoes, and other pests [
3,
4]. With the continuous expansion of its application, genes involved in reproductive regulation—particularly key genes participating in spermatogenesis—have emerged as critical targets for breeding sterile male insects. Insect sterility technology holds promise for evolving into a core component of next-generation integrated pest management strategies.
The silkworm represents an established model organism for Lepidopteran studies. Multiple testis-enriched genes associated with spermatogenesis have been identified in this species, including
BmPMFBP1,
BmSer1, and
BmC/EBPZ, among others [
5,
6,
7]. Comprehensive transcriptome analysis of the testis of the silkworm reveals the expression of 1705 testis-specific genes [
8]. Additionally, genes encoding serine/threonine protein kinases, organic cation transporter proteins, tyrosine protein kinases, lncRNAs, and immune-related proteins have been identified as crucial contributors to spermatogenesis and testicular development [
9]. During fertilization, sperm motility serves as a crucial functional parameter, specifically quantified as the percentage of spermatozoa exhibiting progressive linear motility relative to the total sperm count, which significantly impacts fertilization success rates [
10]. Genetic defects in specific genes and various non-biological factors can impair sperm motility, potentially leading to fertility disorders [
11,
12].
Glutamine synthetases (GSs) are enzymes that catalyze the ATP-dependent conversion of glutamate and ammonia to glutamine, exhibiting tissue-specific and developmentally regulated expression patterns [
13]. In plants, GSs have been extensively studied, particularly for their roles in maintaining nitrogen flow, facilitating internal nitrogen sensing during critical developmental stages, and enhancing nitrogen-use efficiency in crops [
14]. In insects, GS participates in various physiological processes. Studies in
Drosophila melanogaster have shown that GS enhances neuronal survival, while research in
Bactrocera dorsalis has demonstrated its involvement in ovarian development through
GS gene knockdown experiments [
15,
16]. Current research on GS in reproductive systems remains limited. Immunofluorescence studies have localized GS to the head of human spermatozoa. Additionally, glutamine, a component of seminal plasma, has been demonstrated to influence mammalian sperm viability [
17]. Studies demonstrate that GS exhibits significantly higher enzymatic activity in the caput epididymis compared to other tissues in mice. This elevated activity plays a crucial role in maintaining the optimal microenvironment for sperm maturation by eliminating glutamate and ammonia, thereby enhancing sperm motility, restoring acid–base balance, and ultimately promoting spermatogenesis [
18,
19]. In
Bombyx mori, two distinct forms of GS isozymes have been identified: the mitochondrial GS1 and the cytoplasmic GS2. The GS1 isozyme consists of a single subtype and exhibits highly tissue-specific expression patterns, whereas the GS2 isozyme comprises 2 to 4 subtypes.
Virtual screening, a computational approach for identifying small-molecule ligands based on the structures of target proteins and ligands, has been instrumental in the discovery of small-molecule inhibitors for therapeutic targets [
20,
21]. The use of virtual screening techniques to predict possible potential drugs plays an important role in identifying inhibitors targeting protease active sites. Molecular dynamics (MD) simulation holds significant value in molecular biology and drug discovery, serving as an effective method to predict ligand–target interactions while accounting for target flexibility [
22]. As a key enzyme in the glutamine metabolic pathway, inhibition of GS1 may compromise the functional integrity of sperm, leading to reduced motility. In this study, we utilized MD and virtual screening to identify potential inhibitors of glutamine synthetase 1 (BmGS1) in the silkworm,
Bombyx mori, and explored its role as a target for inducing male sterility.
2. Materials and Methods
2.1. Insects and Cells
The silkworms (D9L) were provided by the State Key Laboratory of Resource Insects (Southwest University, Chongqing, China) and were routinely bred with a diet of mulberry leaves at 25 ± 1 °C with 70 ± 5% humidity. The cell line used in this study was the Bombyx mori embryonic cell line (BmE), which was preserved by the State Key Laboratory of Silkworm Genome Biology and cultured in Grace Medium supplemented with 10% fetal bovine serum (FBS, Gibco, Waltham, MA, USA) and 200,000 units of penicillin–streptomycin solution per liter. During the culture process, the cells were passaged approximately every 4 days, depending on their growth status, and maintained in a constant temperature incubator at 27 °C.
2.2. Real-Time Quantitative PCR
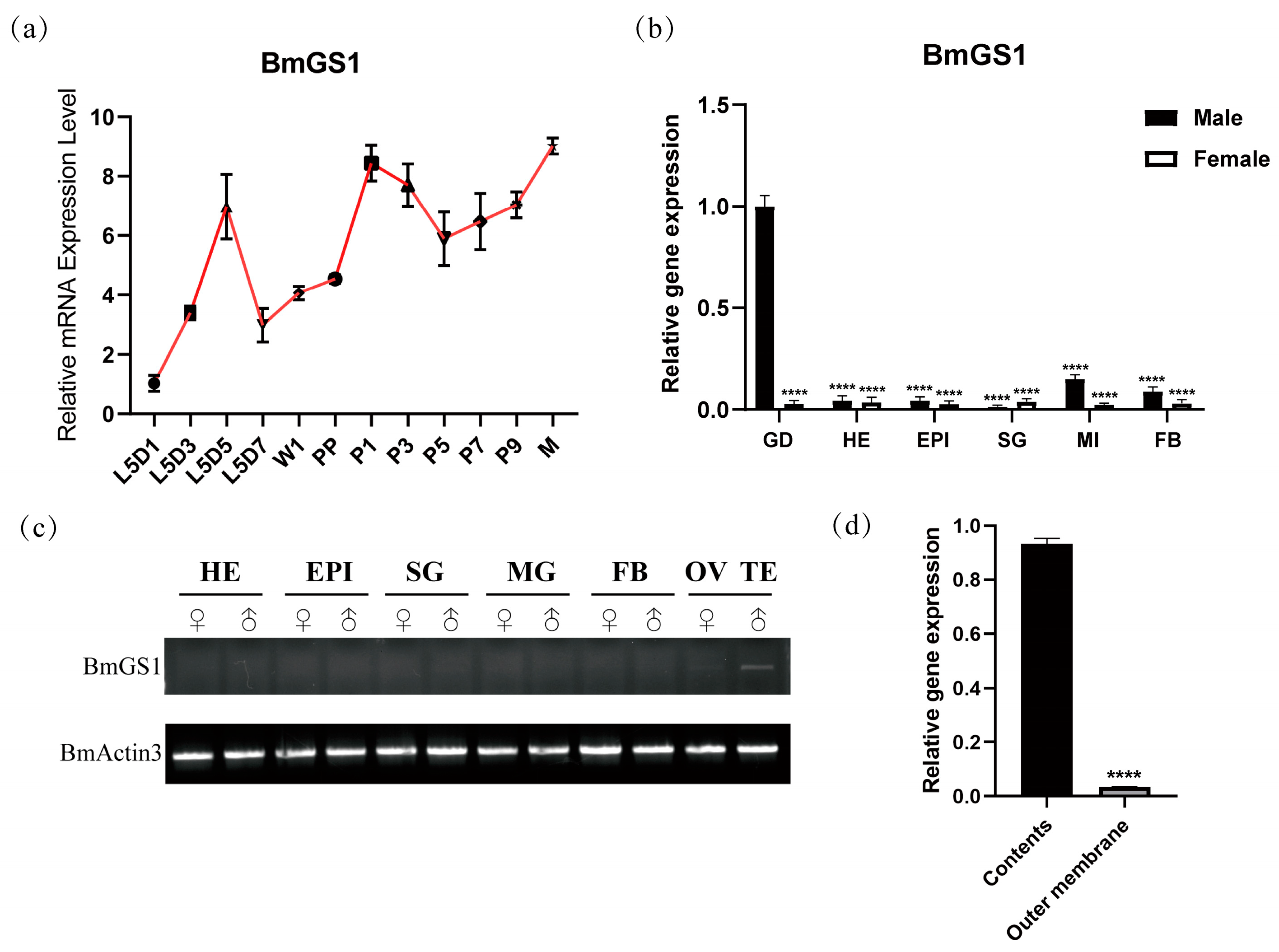
The silkworms were reared until the third day of the fifth instar, and the gonads, head, epidermis, silk glands, midgut, and fat body tissues were taken from males and females, respectively. We selected the testis of male silkworms from the first (L5D1), third (L5D3), fifth (L5D5), and seventh (L5D7) days of the fifth instar; wandering stage (W1); pre-pupae stage (PP); first (P1), third (P3), fifth (P5), seventh (P7), and ninth (P9) days of the pupae stage; and moth stage (M), and males with the same state of growth in P3. The testis were removed, and the contents and outer membrane were separated from the testis. The tissues were washed in PBS, blotted dry with high-pressure filter paper, collected into RNase-free 1.5 mL centrifuge tubes, and stored at −80 °C after liquid nitrogen flash-freezing. RNA was extracted using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA) and reverse-transcribed according to the instructions of PrimeScriptTM RT reagent Kit (TaKaRa, Otsu, Japan). Quantitative analysis was performed with the Novo Start SYBR Qpcr SuperMix Plus kit (Invitrogen, Carlsbad, CA, USA). The silkworm sw gene was used as the internal reference. qPCR was performed using an ABI 7500 Real-Time PCR System with the default thermal cycling protocol. Gene expression levels were calculated using the 2−ΔΔCT method, and data visualization was conducted using GraphPad Prism 8.0. Three biological replicates and three technical replicates were conducted for each qPCR run.
2.3. Semi-Quantitative PCR
Specific primers were designed based on the coding sequence of the
BmGS1 gene using Primer Premier 5 software. Similarly, specific primers for the
BmActin3 reference gene were designed. PCR amplification was performed using 200 ng/μL cDNA as a template. Subsequently, normalized cDNA from various tissues was used as a template for PCR amplification, followed by nucleic acid electrophoresis analysis. All primer sequences are detailed in
Table S1.
2.4. Subcellular Localization
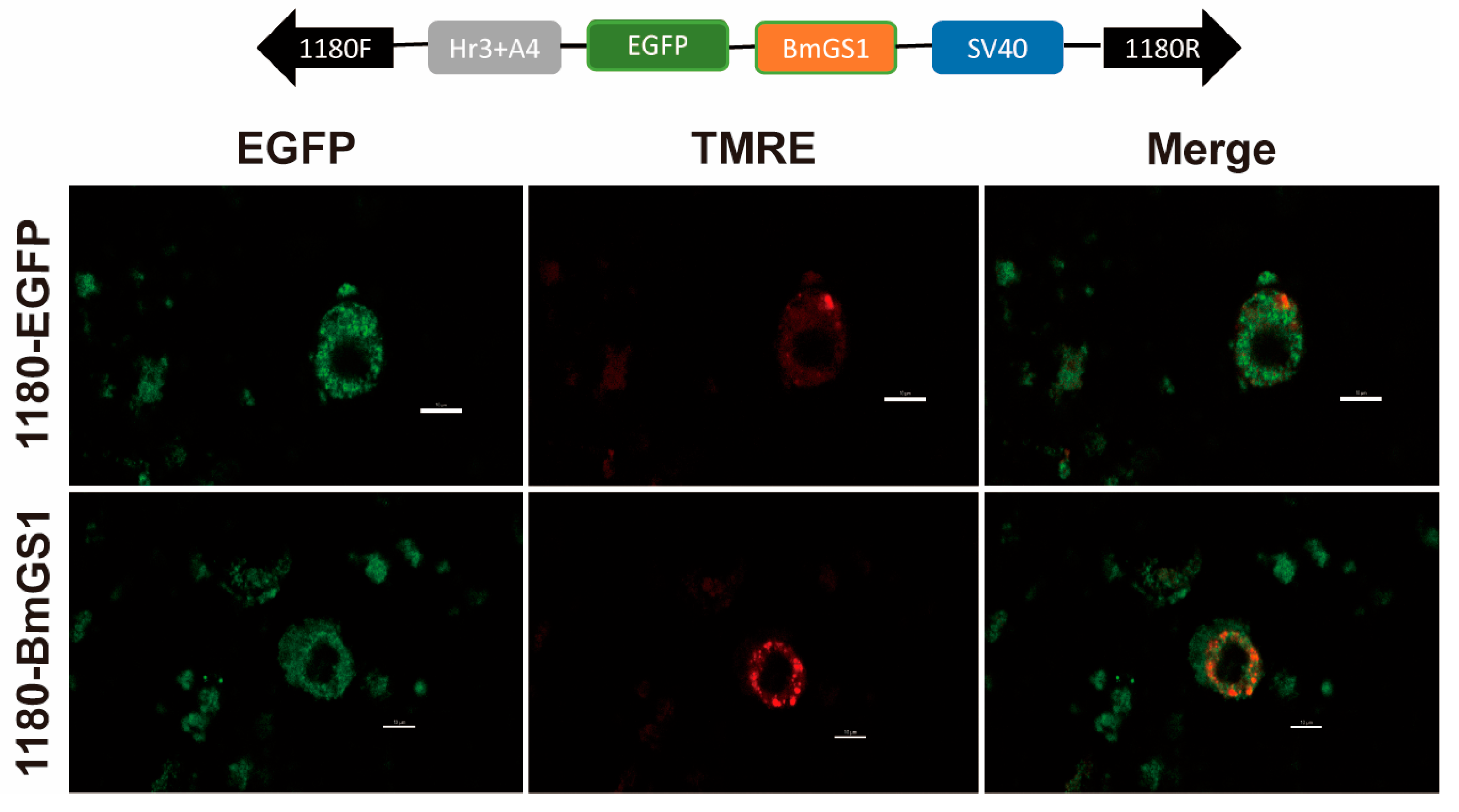
When the cell density reached 70%, the cells were seeded in cell-well plates and transfected with either pSL1180-BmGS1 or a control plasmid. After 48 h, the medium was discarded, and the cells were washed three times with PBS. The cells were then stained using the Mitochondrial Membrane Potential Detection Kit (Beyotime, Shanghai, China) in an incubator at 27 °C for 15 min, followed by another three washes with PBS. The cells were observed and photographed using the laboratory EVOS FL automated imaging system, with a scale bar of 10 µm.
2.5. Dual-Luciferase Reporter Experiment
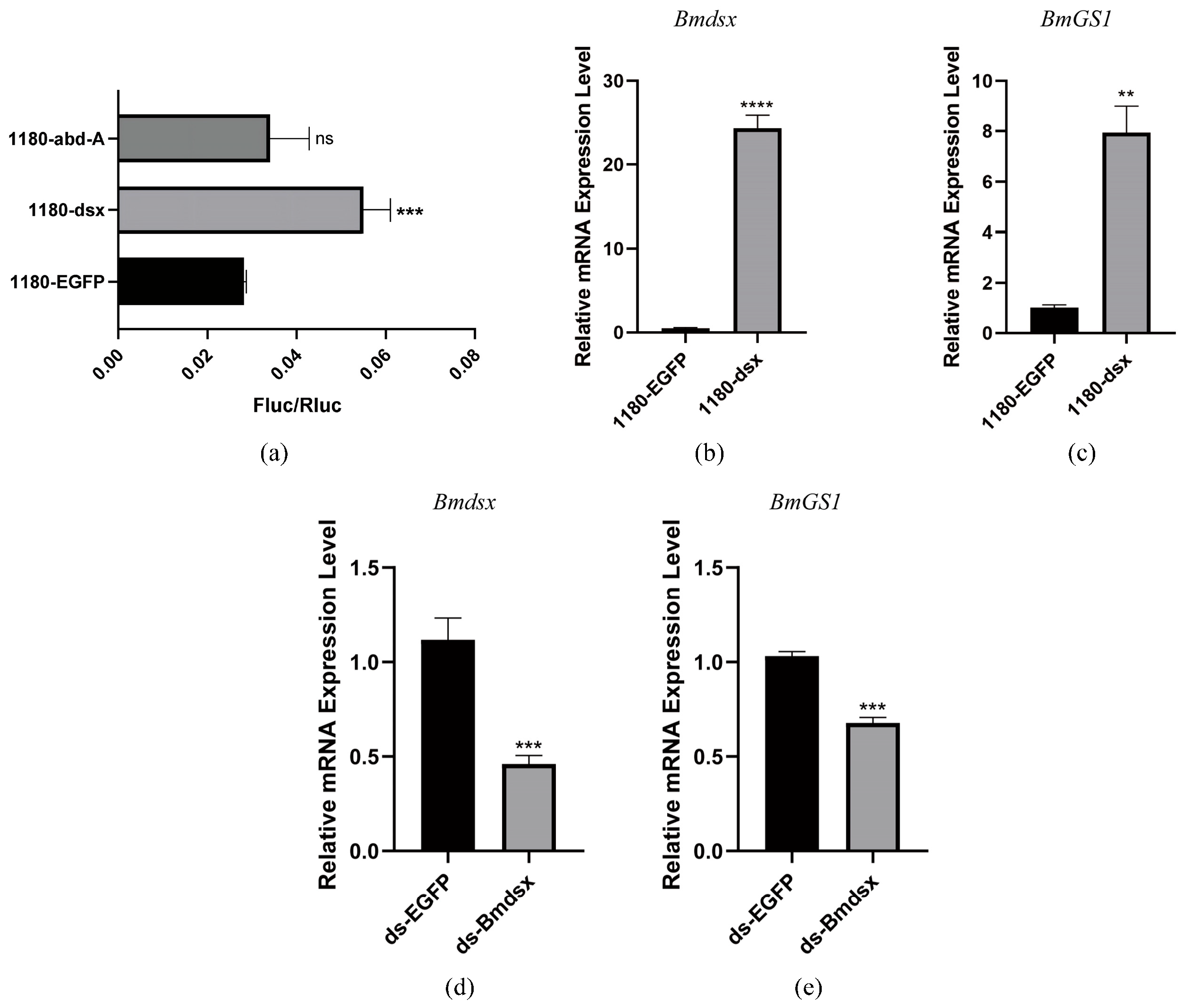
Information related to the
BmGS1 gene was retrieved from the NCBI database, and its promoter region was localized in GenBank. In this study, a fragment spanning from 2000 bp upstream to 100 bp downstream of the translation initiation site of this gene was selected as the promoter sequence for subsequent functional verification experiments. Potential transcription factor binding sites within the promoter sequence were predicted and analyzed using the JASPAR online platform (
https://jaspar.genereg.net/ accessed on 21 January 2026). The genes
dsx and
abd-A, which are closely associated with reproductive functions in
Drosophila, were screened out as candidate targets for further research to explore their roles in regulatory mechanisms.
The BmGS1 promoter fragment was cloned into the dual-luciferase reporter vector pGL3. When BmE cells grew to an appropriate density, they were seeded in 24-well culture plates. The transcription factor expression vectors (pSL-1180-Bmdsx or pSL-1180-Bmabd-A), the promoter-pGL3 reporter vector, and the internal reference plasmid were co-transfected into the cells. After 48 h, we tested the fluorescence activity with a dual-luciferase reporter kit (Promega, Madison, WI, USA).
2.6. Electrophoretic Mobility Shift Assay (EMSA) Binding Experiment
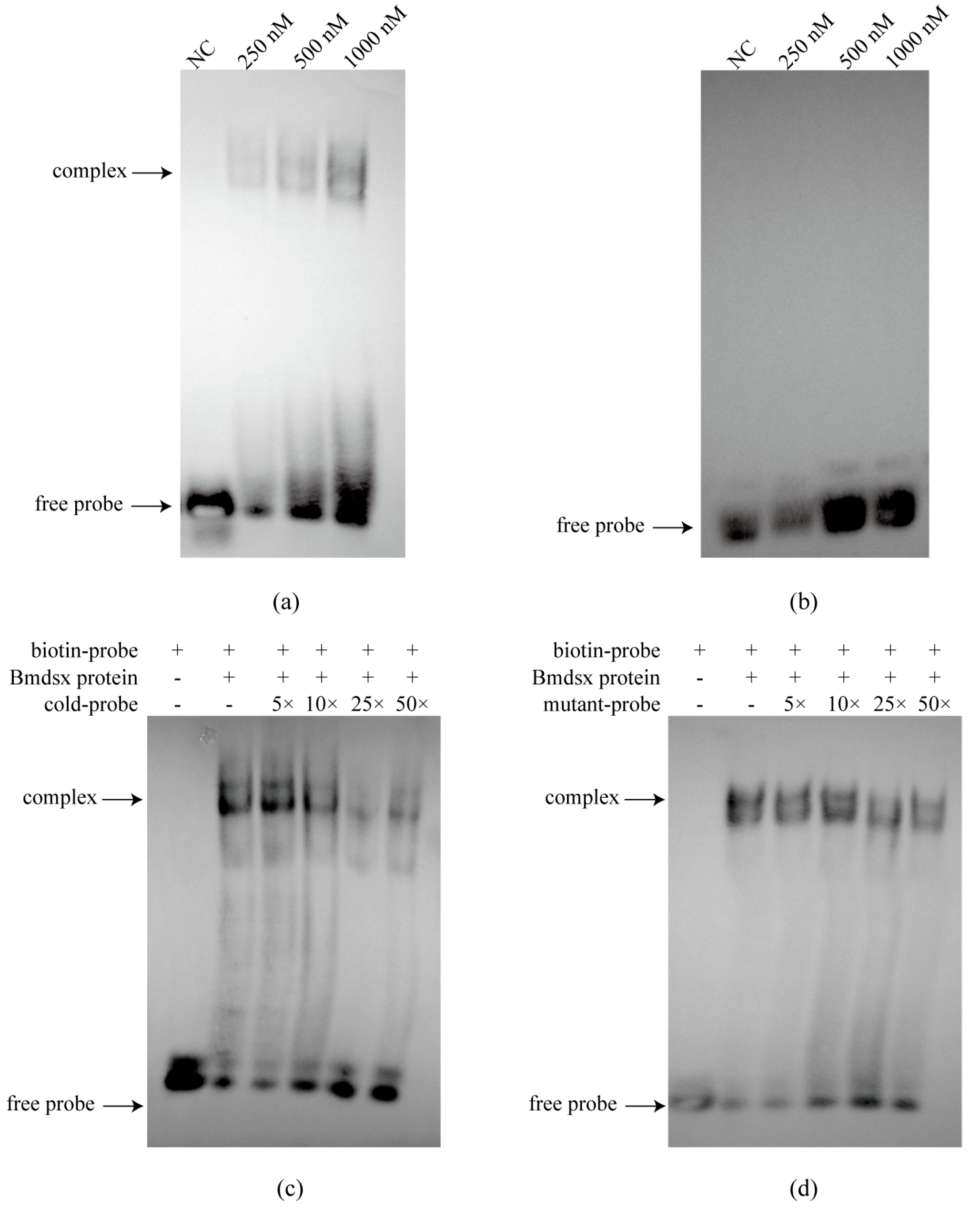
EMSA can be used to verify whether transcription factors interact with promoters. In this study, we first purified Bmdsx and Bmabd-A proteins, and then synthesized biotin-labeled probes based on the predicted binding sites. The binding of protein and nucleic acid was verified with a chemiluminescence kit (Beyotime, Shanghai, China).
2.7. Target Protein Acquisition and Preprocessing
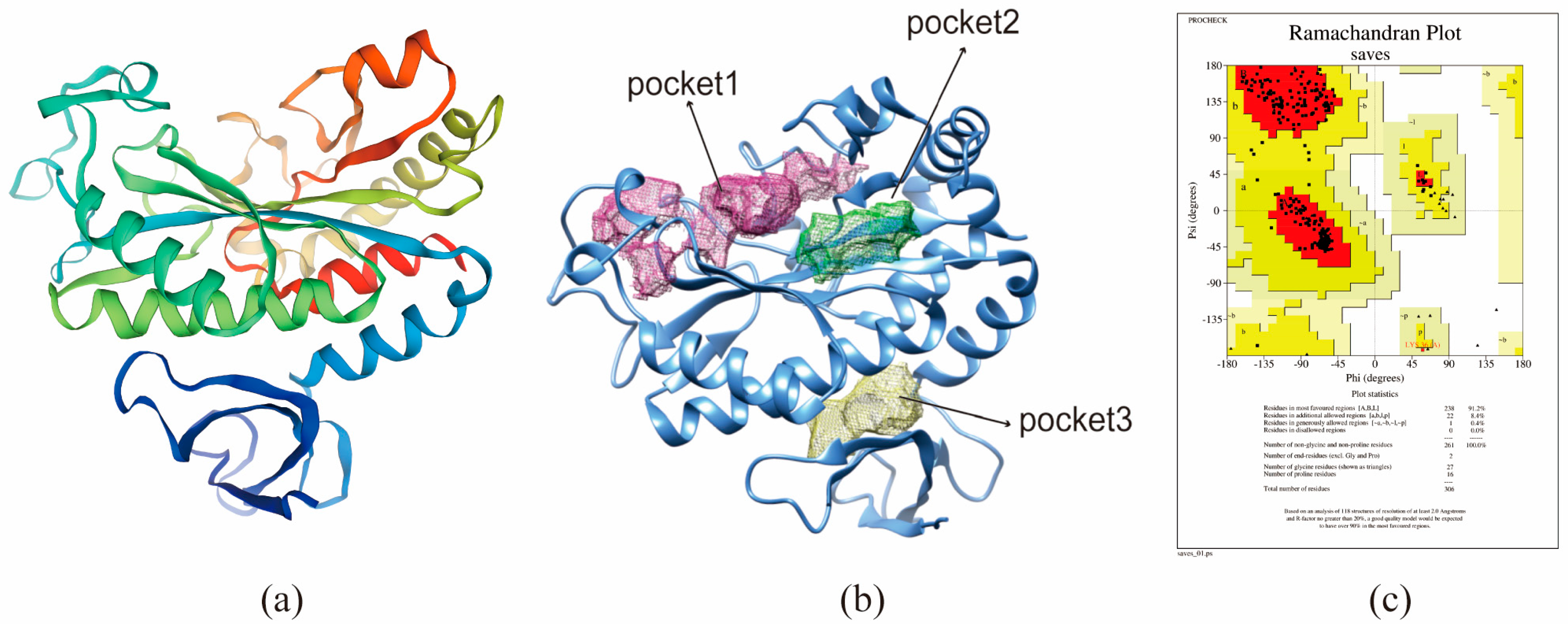
The protein sequence of BmGS1, was used for template searches in SWISS-MODEL (
https://swissmodel.expasy.org/ accessed on 21 January 2026). Templates with closely related enzyme species, high sequence homology, and comprehensive sequence coverage were selected. Homology modeling was subsequently conducted based on GMQE and QMEAN scores. After constructing the homology model, its structural rationality was evaluated using SAVES v6.0 (
https://saves.mbi.ucla.edu/ accessed on 21 January 2026).
The BmGS1 protein activity pockets were predicted using DoGSiteScorer (
https://proteins.Plus/ accessed on 21 January 2026) and ranked based on their size, surface area, and druggability score. Chimera software (1.18) was used to visualize the predicted active pockets of BmGS1. The top three ranked pockets were selected and displayed along with the BmGS1 structure.
Proteins were prepared for virtual screening and molecular docking by hydrogenation, charge assignment, and removal of water molecules using AutoDockTools-1.5.7.
2.8. Establishment of a Small-Molecule Database
Small-molecule compounds for virtual screening were obtained from the PubChem database (
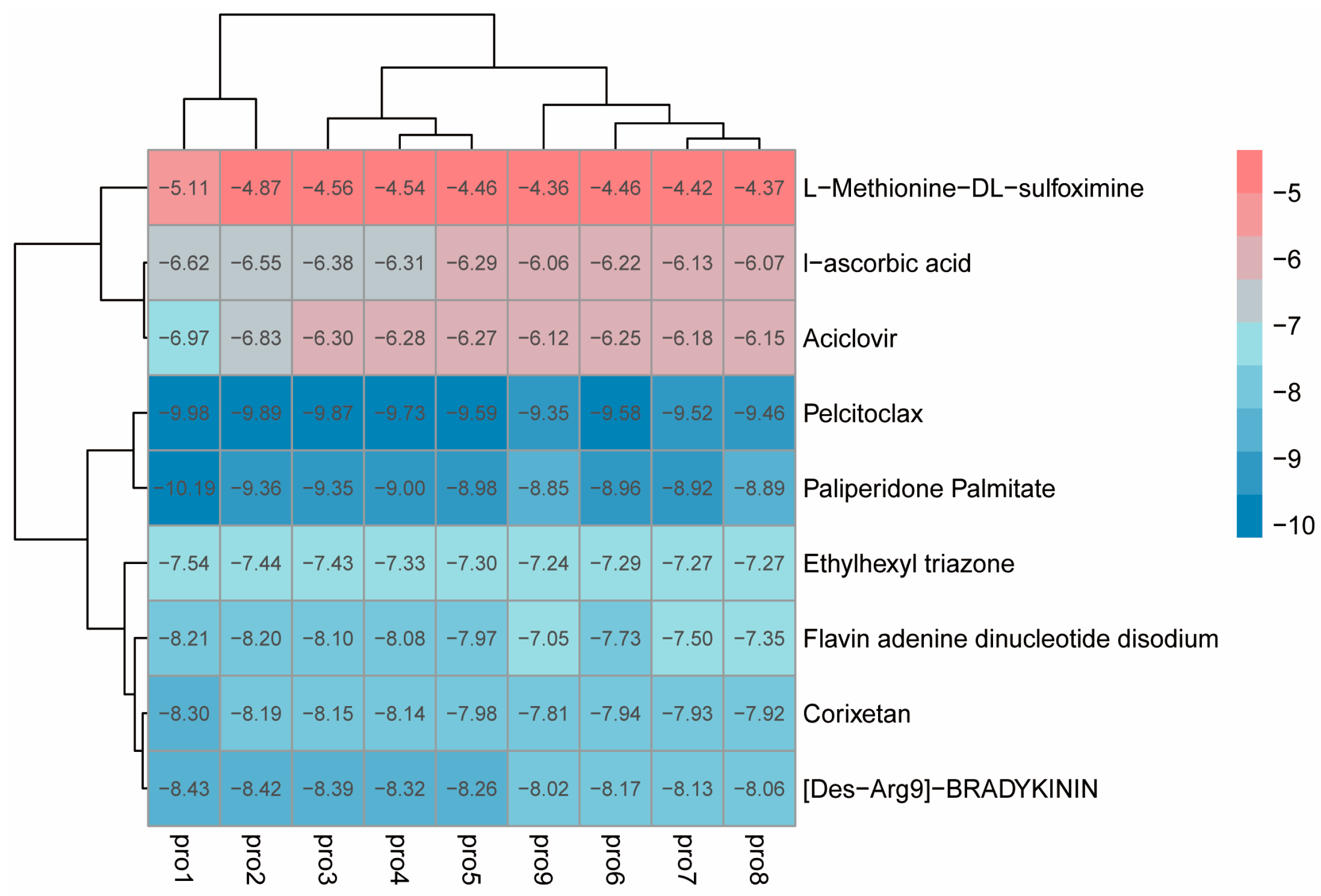
https://pubchem.ncbi.nlm.nih.gov accessed on 21 January 2026). The FDA Global Substance Registry library was selected, resulting in a dataset of 5764 three-dimensional small-molecule structures. OpenBabel software (2.4.1) was used for the conformational conversion of the compounds. Additionally, the literature reports that L-Methionine-DL-sulfoximine (MSX) is a specific inhibitor of glutamine synthetase, widely applied in medicine, pharmacology, and biology [
23,
24]. We therefore selected it as one of the candidate small molecules.
2.9. Virtual Screening and Molecular Docking
The preprocessed small molecules were subjected to batch docking using a screening software. The first active pocket position predicted by DoGSiteScorer was selected, and the top-ranked small molecules in terms of affinity were obtained. Then, AutoDock Vina was used for molecular docking, and the vina docking file config was exported. AutoDock Vina was run, and relevant studies indicate that the more negative the binding energy, the stronger the binding [
25,
26]. Therefore, by considering both binding energy and practicality, small molecules with a free binding energy lower than −4.0 kcal/mol and reasonable binding conformations were selected as potential binders of BmGS1.
2.10. Molecular Dynamics Simulations
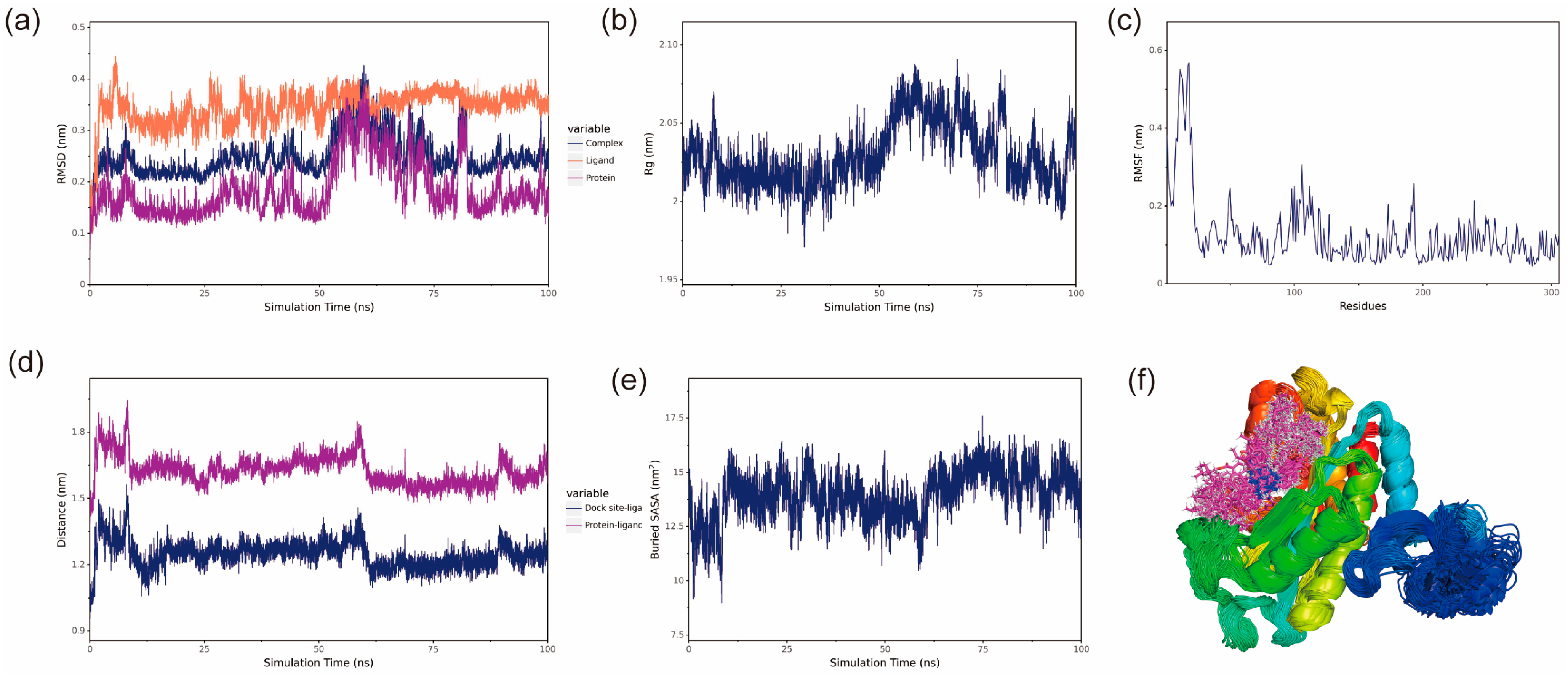
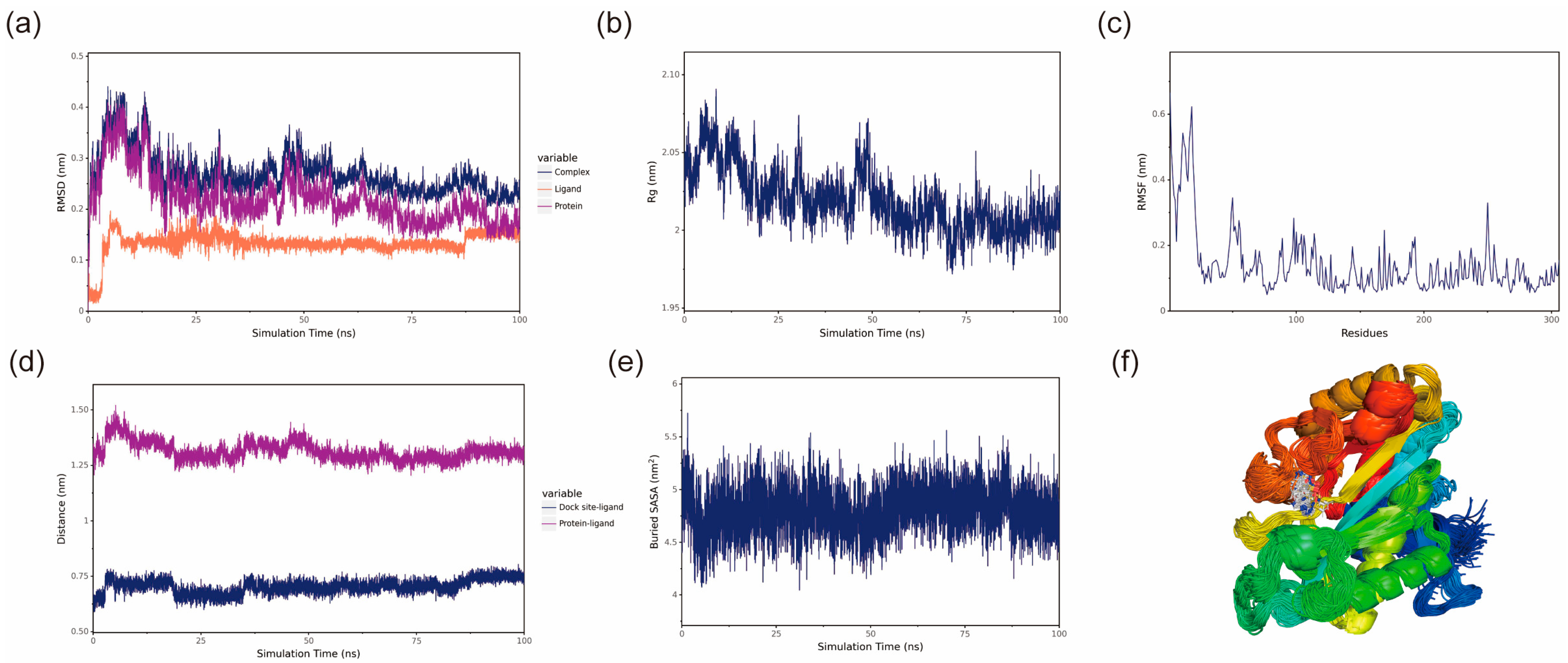
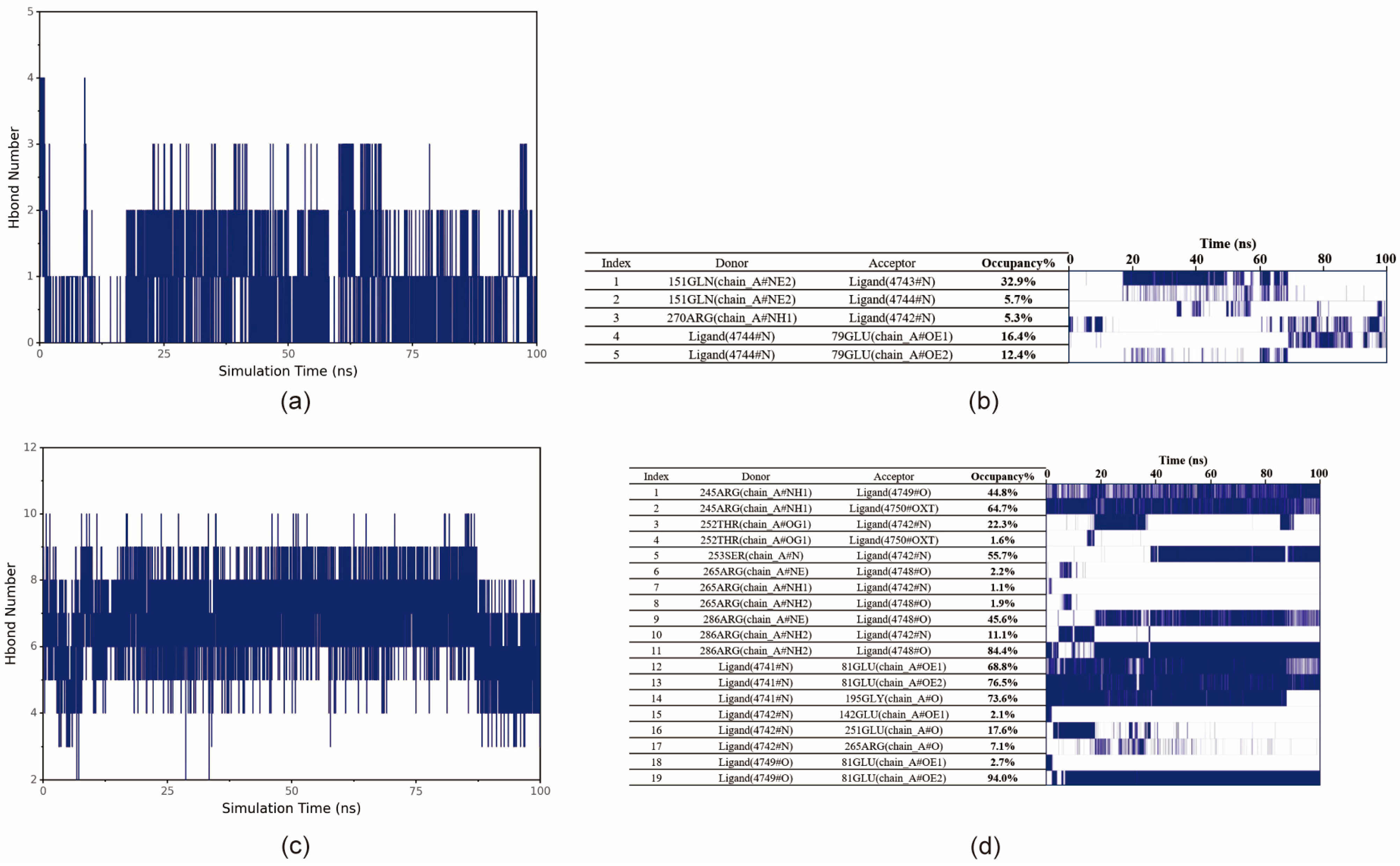
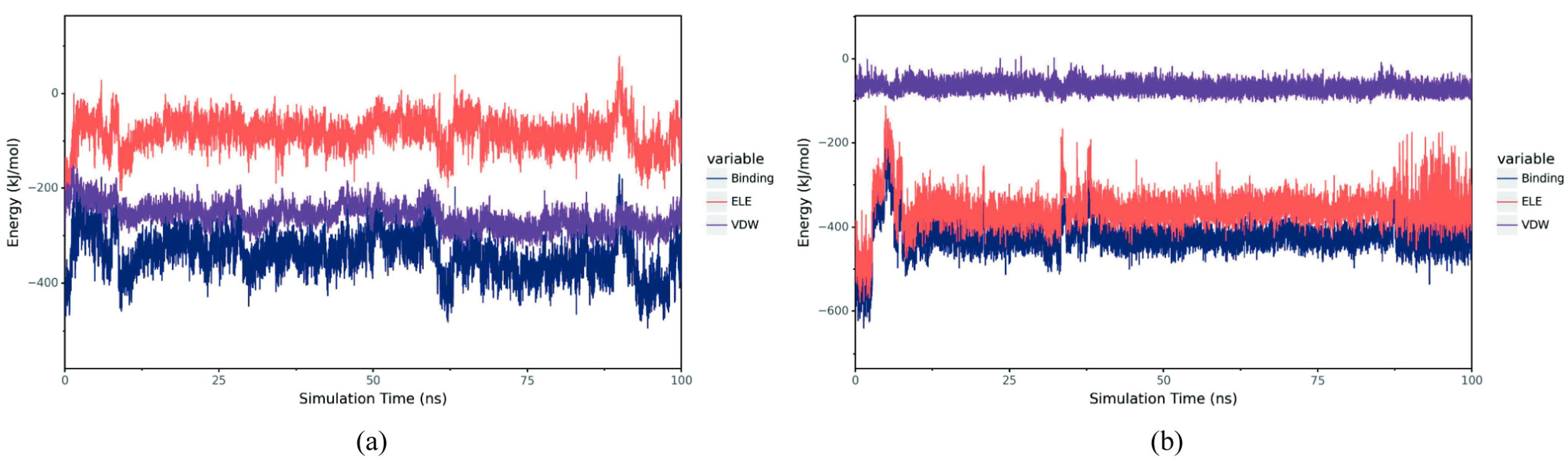
To investigate the binding modes between small molecules and BmGS1, 100 ns MD simulations were conducted to explore their interactions. The protein and optimal ligands were separated from the docking results, and ligand force field files were generated using the Antechamber tool in AmberTools and converted to GROMACS-compatible formats using the ACPYPE tool. The General Amber Force Field (GAFF) was applied to the ligands, while the AMBER14SB force field and TIP3P water model were used for the protein. The protein and ligand files were merged to construct the simulation system. MD simulations were performed using GROMACS under periodic boundary conditions at constant temperature (298 K) and pressure (1 bar). Bonds involving hydrogen atoms were constrained with the LINCS algorithm, using a 2 fs integration time step. Electrostatic interactions were calculated with the Particle-Mesh Ewald (PME) method (cutoff distance: 1.2 nm), and the non-bonded interaction cutoff was set to 10 Å, updated every 10 steps. System equilibration was carried out with 100 ps of NVT and NPT simulations, followed by 100 ns MD simulations with snapshots saved every 10 ps. Simulation trajectories were analyzed using VMD (1.9.3) and PyMOL (3.0.4), and binding free energy was calculated using the g_mmpbsa tool based on the Molecular Mechanics Poisson Boltzmann Surface Area (MMPBSA) method.
2.11. Key Amino Acid Mutations and In Vitro Fluorescence Binding Validation
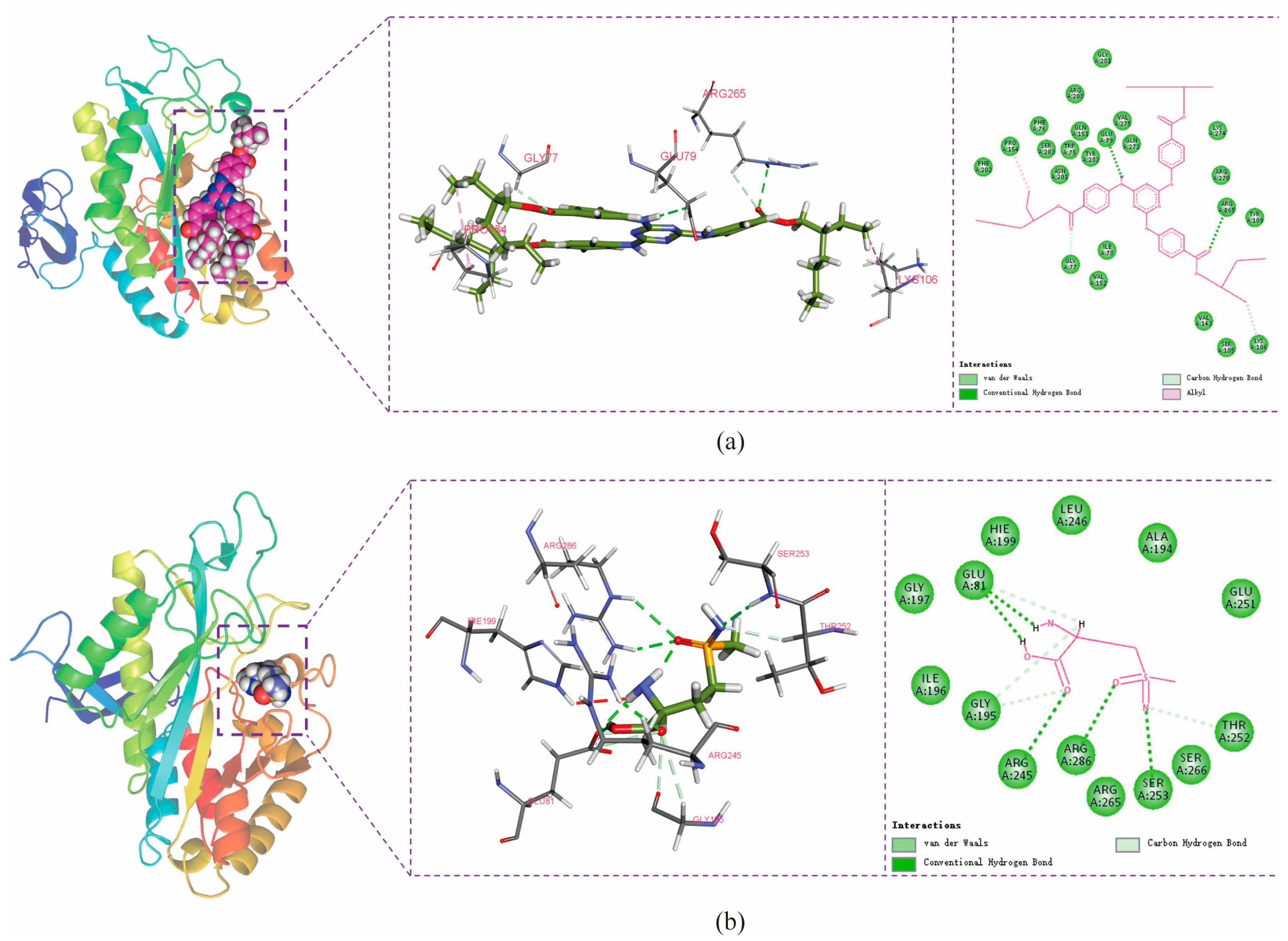
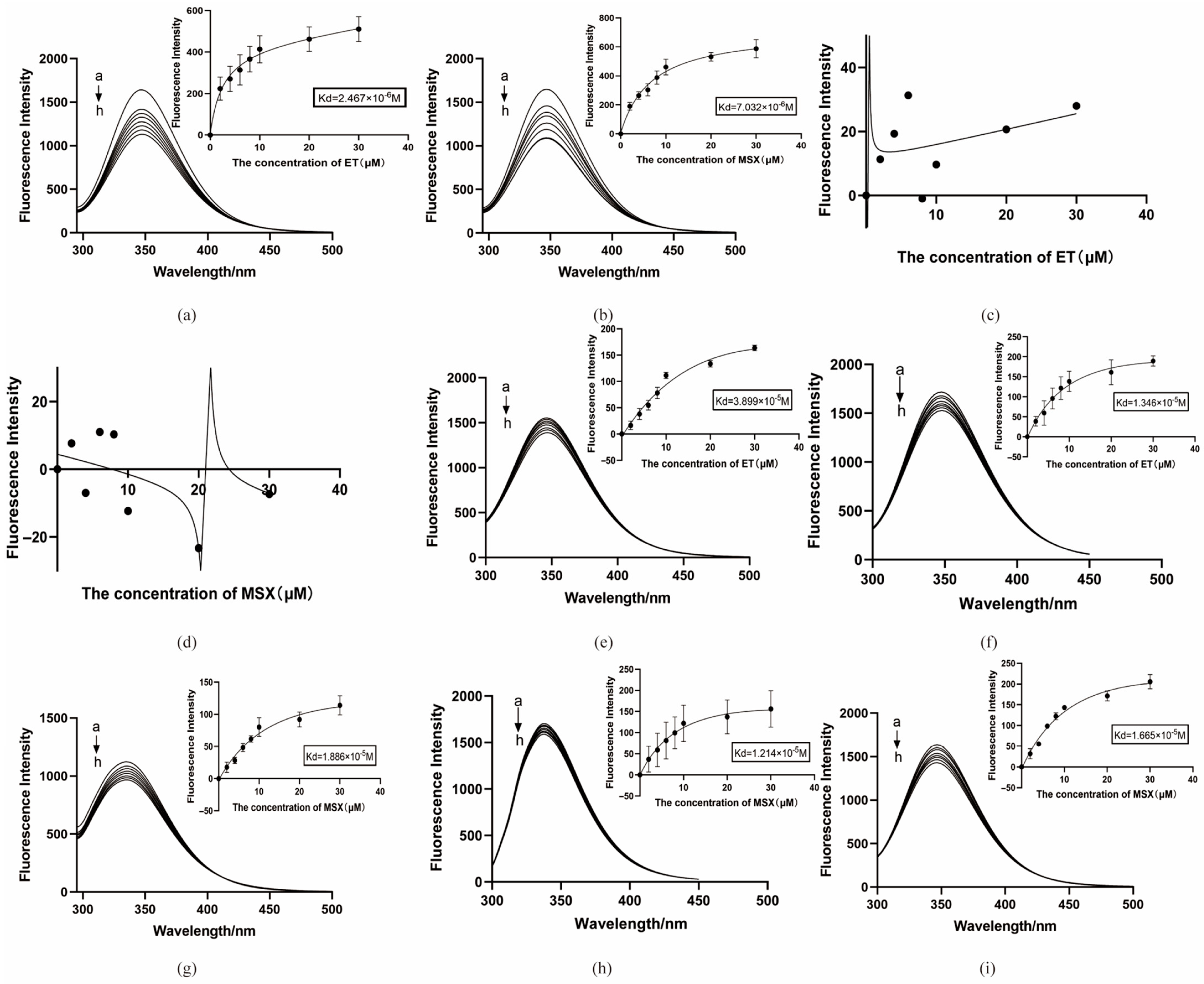
To confirm the binding of the two screened ligand molecules to BmGS1 and to investigate the roles of the putative key amino acids involved in this interaction, we performed targeted mutagenesis at positions Glu79, Arg265, Glu81, Arg245, and Arg286 based on molecular dynamics simulations. A Pcold-TF-BmGS1 prokaryotic expression vector was constructed for protein purification. The binding affinity was measured using a fluorescence binding assay, where the fluorescence intensity displayed a linear relationship with ligand concentration, allowing the determination of the dissociation constant (Kd). Additionally, the enzymatic activity of BmGS1 was assessed after the addition of the small molecules using the Glutamine Synthetase Activity Assay Kit (Boxbio, Beijing, China) to verify whether the ligands affected the protein’s activity.
2.12. Injections of Inhibitors into Silkworms
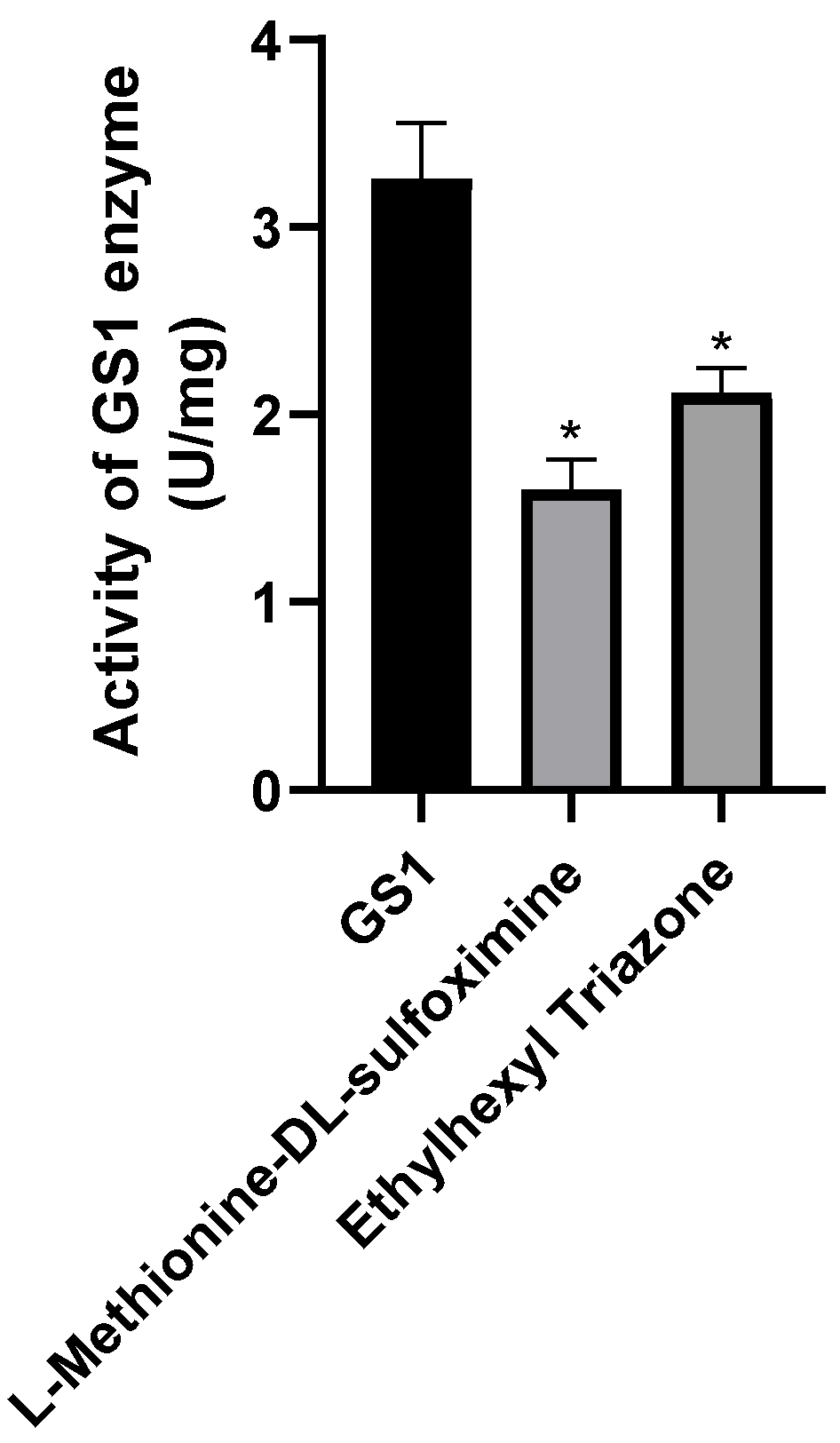
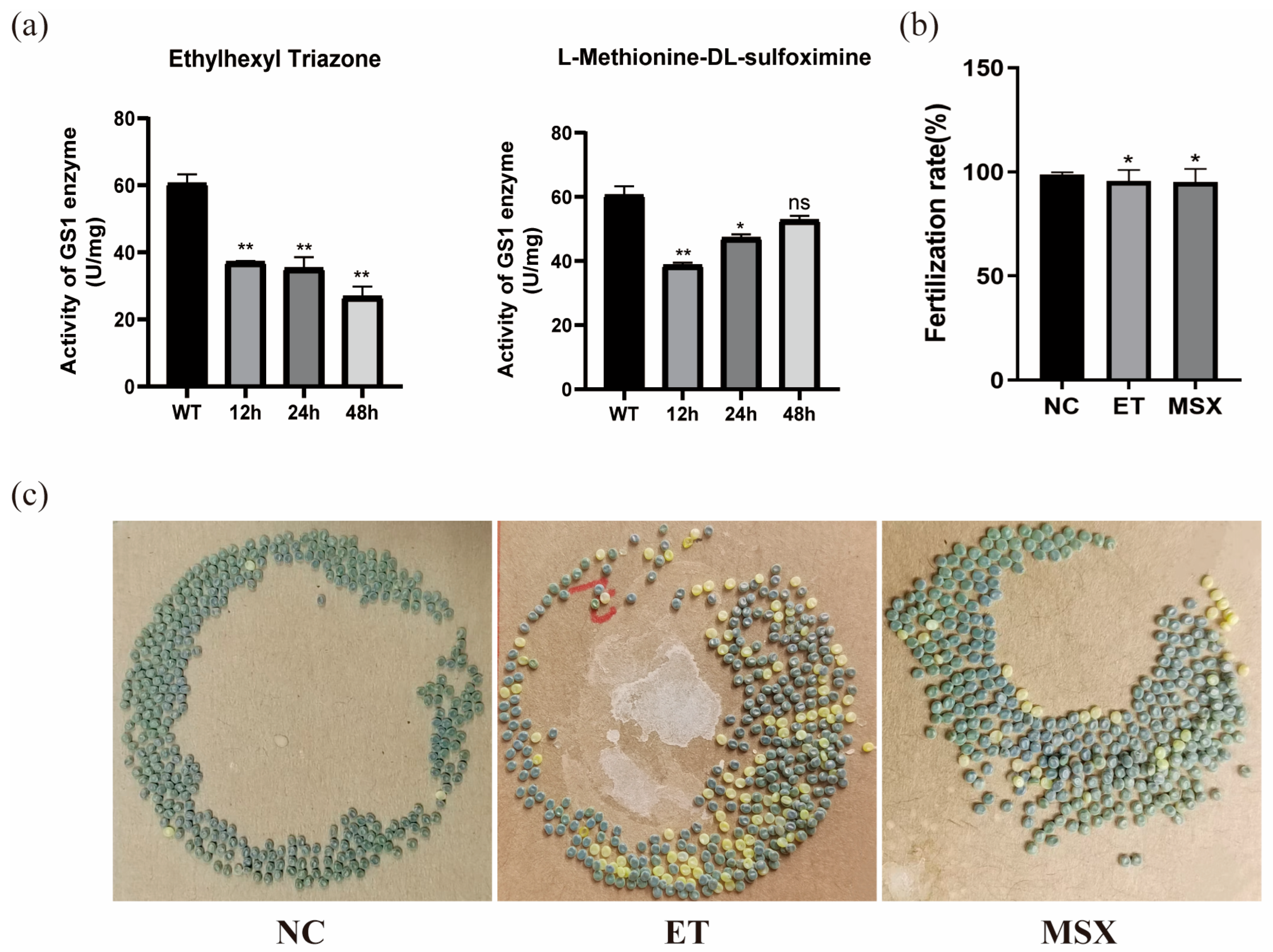
The results showed that the intensity of the protein absorption peak was significantly reduced upon the addition of 2 μM small molecules, indicating the optimal binding efficiency. Therefore, 2 μM of the small molecule was used for injection. Each group included three replicates, with 10 male silkworms in each group. And the testes were collected at 12 h, 24 h, and 48 h post-treatment for the measurement of glutamine synthetase enzymatic activity.
During the pupal stage, a timed injection protocol was initiated on the first day of pupation. Male silkworms with similar size and body weight were selected for injection. A total of three groups, each consisting of 15 individuals, received injections every 48 h. The inhibitors ET and MSX were dissolved in 0.1% DMSO to a final concentration of 2 μM. Each injection, with a volume of 2 µL, was administered into the testes of male silkworm pupae from the dorsal side of the fifth abdominal segment. Males that successfully molted to the adult stage were subjected to mating trials with untreated females, as well as with males treated with each of the two small molecules. After oviposition, the number of eggs laid and the number of fertilized eggs were recorded for 15 brood cycles in each treatment group to calculate fertilization rates.
4. Discussion
In this study, we investigated the
glutamine synthetase 1 (
BmGS1) gene in the model lepidopteran species
Bombyx mori. Expression profiling of
BmGS1 via RT-PCR and qPCR revealed its abundant expression in the testis, with significantly higher transcript levels in sperm cells compared to the outer membrane tissue. During testis development,
BmGS1 was expressed throughout the fifth instar, pupal, and moth stages, peaking at the moth stage. These results suggest that BmGS1 may play a functional role in sperm development and maturation. Studies have revealed that glutamate serves as one of the essential amino acids required for sperm immobilization and storage in the murine epididymis, where it constitutes the most abundant free amino acid. The presence of glutamate likely plays a pivotal role in osmoregulation. During sperm maturation, the conversion of glutamate to glutamine significantly enhances sperm motility by eliminating excess glutamate [
29,
30]. The specific high expression of
BmGS1 in
Bombyx mori spermatozoa suggests that GS may plays an important role in maintaining sperm function. However, due to differences in insect and mammalian reproductive systems, this could reflect functional convergence. Alternatively, it might be a conserved, homologous function across many species that is not yet clearly defined. Subcellular localization confirmed that BmGS1 is expressed in mitochondria and cytoplasm. The findings suggest that BmGS1 may also be functionally linked to mitochondrial metabolic processes. The glutamine synthesized by BmGS1 can be converted into α-ketoglutarate via the glutaminolysis pathway, subsequently participating in mitochondrial aerobic respiration and anabolic metabolism. This process not only provides cellular energy but also maintains redox homeostasis and protects cells from oxidative damage by scavenging reactive oxygen species [
31]. We therefore hypothesize that BmGS1 may indirectly influence sperm quality by modulating mitochondrial physiological functions.
To elucidate the upstream regulatory mechanism of the BmGS1 gene, we conducted a comprehensive analysis of the BmGS1 promoter. First, we predicted the upstream regulatory factors of the BmGS1 promoter. Based on the characteristic that BmGS1 is highly preferentially expressed in sperm, we selected the reproduction-related transcription factors Bmdsx and Bmabd-A as candidate upstream regulatory factors. A dual-luciferase activity assay showed that Bmdsx had a promoting effect on the BmGS1 promoter, while Bmabd-A had no significant effect. Furthermore, the EMSA experiment revealed that the transcription factor Bmdsx could specifically bind to the BmGS1 promoter. The detection of changes in BmGS1 expression after overexpression of Bmdsx at the cellular level showed that the expression level of the BmGS1 gene was upregulated with the overexpression of Bmdsx. The detection of changes in BmGS1 expression after interference with Bmdsx at the individual level showed that the expression level of the BmGS1 gene was downregulated with the interference of Bmdsx. These results indicate that the BmGS1 gene is positively regulated by the transcription factor Bmdsx. The regulation of BmGS1 expression by reproduction-related transcription factors suggests that BmGS1 plays an important role in spermatogenesis in Bombyx mori.
Potential small-molecule inhibitors (ET and MSX) against BmGS1 activity were screened using virtual screening, molecular docking, and MD simulation techniques. MD simulation results showed that the binding of the two small molecules to the protein remained stable throughout the simulation, with no significant displacement in the center of mass and relatively stable hydrogen bond counts. The van der Waals forces and electrostatic interactions during binding were also stable. Further analysis revealed that the amino acids GLU79 and ARG265 in the BmGS1 protein formed key hydrogen bond interactions with ET, while GLU81, ARG245, ARG286, and SER253 formed hydrogen bond interactions with MSX. These results not only confirm the binding stability of the inhibitors but also identify amino acid residues that may play critical roles in binding in the active site of BmGS1.
Fluorescence binding assays confirmed that the BmGS1 protein can bind to the small-molecule compounds ET and MSX in vitro. The binding sites of the small molecules to the protein were both located in the catalytic domain of the protein. Enzymatic activity assays also showed that both small molecules could inhibit the enzyme activity. Subsequently, based on the results of molecular dynamics simulations, five amino acids that may play critical roles in binding, Glu79, Arg265, Glu81, Arg245, and Arg286, were selected for site-directed mutagenesis. After specifically mutating these amino acid sites, the dissociation constants of both small molecules with the mutated target proteins increased, and the binding ability decreased, indicating that these amino acid sites may play an important role in the binding of small-molecule compounds to the BmGS1 protein. To investigate the effects of ET and MSX on BmGS1 protease activity, small-molecule administration experiments were conducted at the individual level of silkworms. Enzymatic activity assays showed that both ET and MSX treatments effectively reduced BmGS1 activity in the testes of silkworms. Additionally, results from small-molecule injection experiments revealed that when male moths treated with ET or MSX mated with wild-type female moths, the fertilization rate of the laid eggs exhibited a downward trend. It is important to note that this study has several limitations. First, the characterization of the inhibitors was performed qualitatively, and their half-maximal inhibitory concentration as well as dose–response relationships have not yet been determined, preventing the identification of their effective concentration range. Future work should address this through quantitative pharmacological experiments. Second, although the observed reduction in fertilization strongly correlates with BmGS1 inhibition, definitive proof of causality will require future direct sperm functional analyses. Finally, even though the inhibitors used in this study did not cause individual mortality, they may still exert certain effects on other organisms or the environment. Therefore, in considering their potential application as pest control agents, we must reasonably assess their specificity and environmental safety. Although this study only statistically analyzed the fertilization rate of the contemporary generation, and the observed downward trend was not significant—and future work could include statistics on fertilization rates across different reproductive cycles—this still preliminarily indicates that the identified small molecules ET and MSX can inhibit BmGS1 activity in silkworm testes, providing valuable clues and potential targets for the study of sperm activity inhibitors and male sterility.
In summary, we analyzed the expression profile, subcellular localization, and upstream regulatory mechanisms of the glutamine synthetase gene
BmGS1 in the silkworm. Two small-molecule inhibitors, ET and MSX, were screened and verified for their specific binding and inhibitory effects on BmGS1. Based on the functional conservation of GS1 across species (
Figure S2), ET and MSX may exhibit broad application potential. Using the silkworm as a model organism, this study provides a foundational framework and conceptual basis for developing male sterility strategies in pest populations. In the future, such small-molecule inhibitors could be employed in sterility techniques targeting pests such as
Manduca sexta or
Pectinophora gossypiella by acting on orthologs of
BmGS1 in these species, thereby contributing to the development of more sustainable and environmentally friendly alternatives to broad-spectrum insecticides.