Simple Summary
Diaeretiella rapae is a widespread aphid parasitoid that targets various aphids, especially those found on cruciferous plants and cereals. Since 1855, the species changed its taxonomic position many times. In this study, specimens of Diaeretiella were analyzed using both molecular and morphological methods. The analysis of mitochondrial COI region revealed 23 different haplotypes within the genus. Phylogenetic analysis placed D. rapae within the Aphidius clade, with genetic distances that are within the range of intraspecific genetic distances of Aphidius. Morphological comparisons showed that D. rapae shares key morphological characters with species in the genus Aphidius. Based on the results, we synonymize D. rapae as A. rapae and designate genus Diaeretiella as junior synonym of Aphidius.
Abstract
Diaeretiella rapae (McIntosh, 1855) is a cosmopolitan koinobiont endoparasitoid of aphids, occurring mainly on crucifers and cereals. From description, it has changed several genera and has about 20 synonyms. The specimens for this study were collected between 1989 and 2023 from sites across Europe and the Middle East. For molecular analysis, the barcode mitochondrial gene COI was used, and morphological analysis was conducted with other Aphidius species. Morphologically, D. rapae falls within the determined variability of the same characters of the genus Aphidius. Dieretiella rapae comprised 23 haplotypes with a mean genetic distance between haplotypes of 0.8%. Phylogenetically, D. rapae is nested within Aphidius species with a genetic distance of 2.1% to 11.3%, which is within the range of other Aphidius species. Our results on morphological and molecular level confirm that D. rapae belongs to the genus Aphidius.
1. Introduction
Diaeretiella rapae (McIntosh, 1855) is a koinobiont endoparasitoid that parasitizes around 100 species of aphids found on more than 180 plants [1,2]. The most common hosts are Brevicoryne brassicae (Linnaeus, 1758), Myzus persicae (Sulzer, 1776), Lipaphis erysimi (Kaltenbach, 1843), and Diuraphis noxia (Mordvilko, 1913), preferentially on cruciferous plants and cereals [1,2,3]. Although it is assumed that the place of origin of D. rapae is the Mediterranean part of Europe, it is almost cosmopolitan in distribution, inhabiting all continents except Antarctica [2]. Due to its role as a biological control agent, this species is considered to be of high economic importance and has been well studied compared to other members of the Aphidiinae subfamily; there are numerous studies which examined its biocontrol potential [4,5,6,7,8], phylogeography [9] and its biology [10,11,12,13,14,15]. Despite broad knowledge that was gathered over the last few decades, there are still some unsolved taxonomic problems. The authorship of this species has been problematic, as it was first described in Charles McIntosh’s “The Book of the Garden” [16] in 1855 and five years later in John Curtis’ “Farm insects” [17]. Historically, most authors considered Curtis to be the author of D. rapae in his book or in McIntosh’s book, as Curtis was a renowned entomologist in his time, while only some people ascribed authorship to McIntosh. The problem was solved by Mackauer [18], in which Charles McIntosh was given as the author of the species according to the International Code of Zoological Nomenclature.
Since its description, D. rapae has changed its taxonomic position several times. Gahan [19] placed the species in the genus Diaeretus and separated it from the other Trioxys Haliday, 1833 by the absence of prongs and from Lipolexis Förster, 1862 by the presence of a second discoidal cell. He also noted that Marshall [20] placed all species in the genus Aphidius, not accepting Foerster’s genus table. Starý [21] placed D. rapae in a separate monotypic genus Diaeretiella, differentiating it from the other Aphidiinae by the reduction of the wing venation and the shape of ovipositor sheath and separating it from Diaeretus by the narrow central areola of propodeum. Mackauer [22] initially did not accept Stary’s classification of the species and kept it in the genus Aphidius, but later in the same year, D. rapae was accepted as a valid name [23]. Hafez [10] agreed with Mackauer’s first statement that the species belongs to the genus Aphidius because the reduction of the wing venation cannot be a taxonomic character in the Aphidiinae (it is present in several independent lineages), and the female genitalia of D. rapae are typical of the genus Aphidius. Up to present times, its status was unchanged, and it has been re-described twice in the previous decade [24,25]. The diagram with synonyms and their publication time is given in Figure 1.
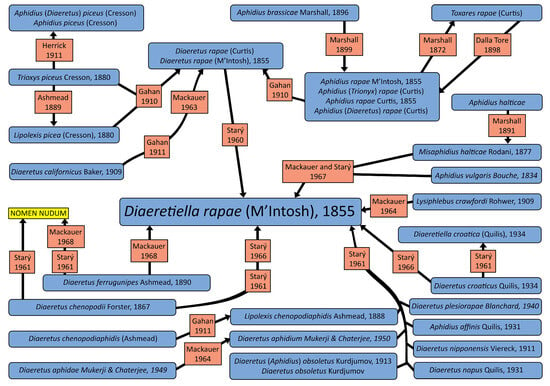
Figure 1.
Diagram of D. rapae synonyms (blue—synonyms, red—publications where taxon status changed, yellow—nomen nudum) [18,19,20,21,26,27,28,29,30,31,32,33,34,35,36].
The main goal of this research is to resolve the taxonomic position of D. rapae using morphological and molecular analysis.
2. Materials and Methods
2.1. Sample Collection and Morphological Analysis
Specimens were collected between 1989 and 2023 from sites across Europe and the Middle East (Serbia, Montenegro, Slovenia, Greece, Italy, Belgium, Russia and Turkey). Parasitoids were collected by sampling plant parts infested with aphids. Plants with aphid colonies were placed in plastic containers covered with a nylon mesh to allow ventilation. The samples were transported to the laboratory and kept under controlled conditions (22 °C, 65% relative humidity, 16 h light/8 h dark). After emergence, the parasitoids were preserved in 96% ethanol or dry mounted. Aphid and plant samples were identified to genus or species level to establish tri-trophic interactions. In total 1741♀ and 1145♂ belonging to D. rapae were examined under ZEISS SteREODiscovery.V12 (ZEISS Microscopy, Jena, Germany). Specimens were photographed using the Leica DM LS phase contrast microscope (Leica Microsystems GmbH, Wetzlar, Germany), while relevant measurements were taken using the ImageJ 1.50i software [37].
List of all D. rapae specimens is given in Supplementary Table S1. In addition, we examined specimens belonging to 43 Aphidius species listed in Table 1, as those exhibit the entire morphological variability in the genus Aphidius. Several Diaeretiella and Aphidius specimens were sputter coated with gold and examined using a Jeol JSM-6460LV scanning electron microscope (Jeol Ltd., Tokyo, Japan). For inter-generic comparison, we analyzed the following characters commonly used for Aphidius species [38]: antennae—number of antennomeres, shape of antennae, length of flagellomere 1 (ratio between length and width of flagellomere 1 at median level), color of flagellomere 1, number of longitudinal placodes on flagellomeres 1 and 2; number of labial and maxillary palpomeres; tentorial index (ratio between tentoriocular line/intertentorial line); forewing venation (length/width of pterostigma, ratio between length of vein R1 (= metacarpus) and length of pterostigma); petiole-dorsal and anterolateral sculpturation, length of petiole (ratio between length and width of petiole at the spiracle level); propodeal areola (closed or open); shape of ovipositor sheath (Figure 2). The terminology of morphological characters follows Sharkey and Wharton [39]. The parasitoids examined in this study are deposited in the collection of the Institute of Zoology, Faculty of Biology, University of Belgrade (FBUB).

Table 1.
List of Aphidius species used in this study.
Table 1.
List of Aphidius species used in this study.
| Aphidius absinthii Marshall, 1896 | Aphidius matricariae Haliday, 1834 |
| Aphidius aquilus Mackauer, 1961 | Aphidius microlophii Pennachio & Tremblay, 1987 |
| Aphidius areolatus Ashmead, 1906 | Aphidius phalangomyzi Starý, 1963 |
| Aphidius arvensis (Starý, 1960) | Aphidius platensis Brethes, 1913 |
| Aphidius asiaticus Kim & Tomanović, 2021 | Aphidius plocomaphidis (Starý, 1973) |
| Aphidius avenae Haliday, 1834 | Aphidius rhopalosiphi de Stefani-Perez, 1902 |
| Aphidius avenaphis (Fitchk, 1861) | Aphidius ribis Haliday, 1834 |
| Aphidius balcanicus Tomanović & Petrović, 2011 | Aphidius rosae Haliday, 1834 |
| Aphidius banksae Kittel, 2016 | Aphidius salicis Haliday, 1834 |
| Aphidius cingulatus Ruthe, 1859 | Aphidius schimitscheki (Starý, 1960) |
| Aphidius colemani Viereck, 1912 | Aphidius setiger (Mackauer, 1961) |
| Aphidius eadyi Starý, Gonzáles & Hall, 1980 | Aphidius smithi Sharma & Subba Rao, 1959 |
| Aphidius ericaphidis Pike & Starý, 2011 | Aphidius sonchi Marshall, 1896 |
| Aphidius ervi Haliday, 1834 | Aphidius staticobii Tomanović & Petrović, 2012 |
| Aphidius funebris Mackauer, 1961 | Aphidius sussi Pennachio& Tremblay, 1989 |
| Aphidius gerani Tomanović & Kavallieratos, 2009 | Aphidius tanacetarius Mackauer, 1962 |
| Aphidius gifuensis Ashmead, 1906 | Aphidius tarsalis van Achterberg, 2006 |
| Aphidius hieraciorum Starý, 1962 | Aphidius transcaspicus Telenga, 1958 |
| Aphidius hortensis Marshall, 1896 | Aphidius urticae Haliday, 1834 |
| Aphidius leclanti Tomanović & Chaubet, 2013 | Aphidius uzbekistanicus Luzhetzki, 1960 |
| Aphidius longipetiolus Takada, 1968 | Aphidius viaticus (Sedlag, 1968) |
| Aphidius longistigmus Kim & Tomanović, 2021 |
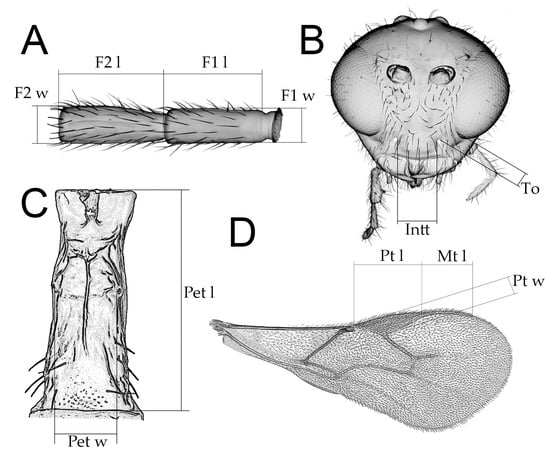
Figure 2.
Schematic drawing of character measurements: (A) First and second flagellar segments, F1 l—length of F1; F1 w—width of F1; F2 l—lenght of F2; F2 w—width of F2; (B) Head, frontal view, Intt—lenght of intertentorial line; To—length of tentoriocular line; (C) Petiole, dorsal view, Pet l—length of petiole; Pet w—width of petiole; (D) Fore wing, Pt l—length of pterostigma; Pt w—width of pterostigma; Mt l—length of vein R1 (= metacarpus).
2.2. Molecular Analysis
For 25 specimens, total genomic DNA was extracted non-destructively with Qiagen Dneasy® Blood & Tissue Kit (Qiagen Inc., Valencia, CA, USA), following the manufacturers’ protocol. The universal primers LCO1490 and HCO2198 were used for amplification of the cytochrome oxidase subunit I region [40]. The amplification mixture contained 32.6 μL of nuclease free water, 10 μL of amplification buffer, 1 μL of nucleotide solution, 1 μL of primers (each), 0.4 μL of taq polymerase and finally 4 μL of extracted DNA, adding to the final volume of 50 μL. The PCR amplification profile was: 5 min of initial denaturation, 37 cycles of 60 s (94 °C), 60 s annealing (56 °C) and 90 s extension (68 °C) and 7 min of final extension (72 °C). Amplification products were purified and sequenced by Macrogen Inc. (Seoul, Republic of Korea).
Electropherograms were visualized in Finch TV Geospiza Inc. (Seattle, WA, USA) and manually edited and aligned using BioEdit software 7.2.5 [41]. The analysis of evolutionary divergence for Diaeretiella sequences was conducted in MEGA 11 software [42] using the Kimura 2-parameter distance model. The evolutionary history of Diaeretiella rapae was inferred by using the Maximum Likelihood (ML) method and Tamura–Nei model [43], which was indicated by MEGA 11 as the best fitting model. The final dataset for the reconstruction of phylogenetic relationships between Diaeretiella samples consisted of 315 COI sequences: 25 newly acquired sequences, additional 290 sequences acquired from GenBank, and one outgroup sequence—Toxares deltiger (Haliday, 1833) (Table S2). Diaeretiella rapae haplotype diversity was estimated by the software DNAsp v6.10.04 [44], and a haplotype network was constructed by Network (version 10.2.0.0).
To establish the phylogenetic position of Diaeretiella within subfamily Aphidiinae, additional 68 sequences were acquired from GenBank, representing 13 Aphidiinae genera. The final dataset for this phylogenetic reconstruction consisted of 73 COI sequences (5 newly acquired sequences).
In order to determine phylogenetic relationships between Diaeretiella and Aphidius species, three outgroup species were used, Venturia canescens (Gravehorst, 1829), Ephedrus niger Gautier, Bonnamour & Gaumont, 1929 and Toxares deltiger. The final dataset for this phylogenetic reconstruction consisted of 138 COI sequences (3 newly acquired sequences, additional 135 sequences acquired from GenBank, and 3 outgroup sequences) (Table S3).
In both phylogenetic reconstructions, Bayesian evolutionary analysis was performed with BEAST 2.5 [45] software employing the initial data set constructed in BEAUti v1.10.4 [45], with designated strict clock type and Yule process of speciation. The analysis ran for 10 million generations, and the sampling was conducted every 1000 generations, while the first million trees were discarded as a burn in. The effective sample size (ESS) of the parameters of the Markov chain Monte Carlo was estimated by Tracer v1.7.1 [46]. The saturation level for the third codon position was inspected in DAMBE 7.2.133 software [47] using the Xia model test [48]. The phylogenetic tree was visualized using FigTree 1.4.3 software [49].
3. Results
3.1. Morphology
All analyzed morphological characters of D. rapae fall within the determined variability of the same characters of the genus Aphidius. Number of antennomeres corresponds to Aphidius species that possess short antennae with only 13–14 segments (A. setiger, A. aquilus, A. salicis). Species of the genus Aphidius possess variable number of maxillar and labial palpomeres (Table 2, MP, LP values), whereas D. rapae possesses maxillar and labial palps constituted out of 3–4 and 2 palpomeres, respectively, which is a value that is characteristic for most Aphidius species. Propodeum possesses a visible central narrow pentagonal areola that is also present in most Aphidius species. Its shape is also characteristic to most of the other Aphidius species. The lateral side of the petiole has curved costulae, also present in most Aphidius species. The shape of D. rapae ovipositor sheaths is typical for Aphidius. Table 2 gives the values of characters in D. rapae as well as the minimum and maximum range for the genus Aphidius. The state of some characters in both D. rapae and Aphidius spp. is shown in Figure 3, Figure 4, Figure 5 and Figure 6. For every character we chose five Aphidius species which reflect total variability of character states.

Table 2.
Value of morphological characters important for taxonomy of genus Aphidius.
Table 2.
Value of morphological characters important for taxonomy of genus Aphidius.
| D. rapae | Aphidius (Minimal Value) | Aphidius (Maximal Value) | |||
|---|---|---|---|---|---|
| Antennae | (12) 13–14 | 12–13 | (A. salicis) | (19) 20–21 | (A. eadyi) |
| F1 l/w | 2.5–3.5 | 2.11–2.52 | (A. longistigmus) | 4–5 | (A. gifuensis) |
| F2 l/w | 2–3 | 1.63–1.76 | (A. areolatus) | 3–4,3 | (A. banksae) |
| MPS F1 | 0–1 | 0 | (A. ribis) | 3–6 | (A. rosae) |
| MPS F2 | 2–4 | 0 | (A. leclanti) | 4–7 | (A. cingulatus) |
| F1/F2 | 1 | 0.85–0.93 | (A. banksae) | 1.06 | (A. leclanti) |
| MP | 3–4 | 2 | (A. arvensis) | 4 | (A. absinthii) |
| LP | 2 | 1 | (A. arvensis) | 3 | (A. uzbekistanicus) |
| Ti | 0.29–0.36 | 0.3–0.4 | (A. schimitscheki) | 0.74–0.8 | (A. cingulatus) |
| Pt l/w | 2.9–4 | 2.8–3 | (A. plocomaphidis) | 4.96–5.46 | (A. longistigmus) |
| Ptl/Mtl | 1.39–2.63 | 0.93–1.05 | (A. urticae) | 1.5–2 | (A. avenae) |
| Pet l/w | 1.93–2.65 | 2.17 | (A. areolatus) | 3.88–4 | (A. geranii) |
Antennae—number of antennomeres; F1 l/w—ratio between length and width of flagellomere 1 at median level; F2 l/w—ratio between length and width of flagellomere 2 at median level; MPS F1—number of longitudinal placodes on flagellomere 1; MPS F2—number of longitudinal placodes on flagellomere 2; F1/F2—ratio between lengths of flagellomere 1 and 2; MP—number of maxillary palpomeres; LP—number of labial palpomeres; Ti—ratio between tentoriocular line/intertentorial line; Pt l/w—ratio between length and width of pterostigma; Ptl/Mtl—ratio between length of pterostigma and length of vein R1; Pet l/w—ratio between length and width of petiole at level of spiracles.
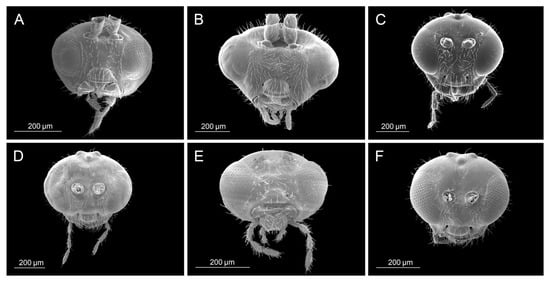
Figure 3.
Head: (A) Diaeretiella rapae, (B) Aphidius cingulatus, (C) Aphidius balcanicus, (D) Aphidius banksae, (E) Aphidius ericaphidis, (F) Aphidius platensis.
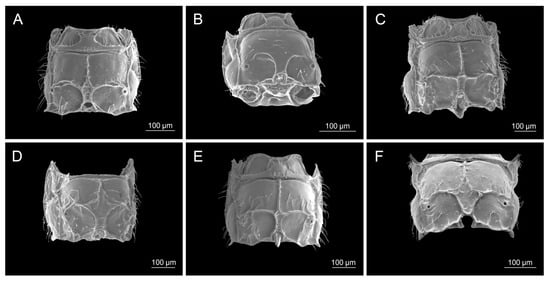
Figure 4.
Propodeum: (A) Diaeretiella rapae, (B) Aphidius staticobii, (C) Aphidius eadyi, (D) Aphidius banksae, (E) Aphidius ervi, (F) Aphidius colemani.
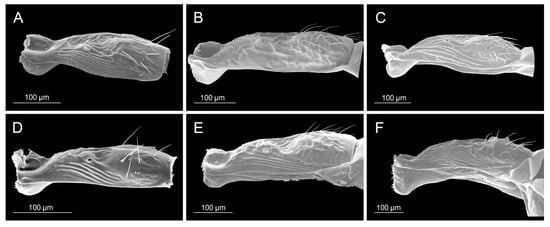
Figure 5.
Petiole: (A) Diaeretiella rapae, (B) Aphidius ervi, (C) Aphidius avenae, (D) Aphidius colemani, (E) Aphidius smithi, (F) Aphidius eadyi.
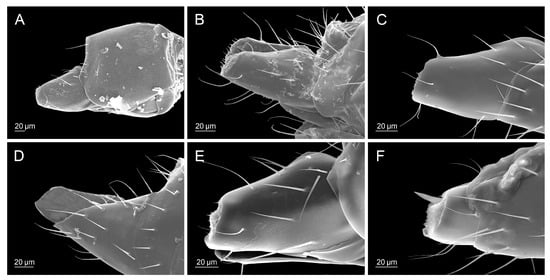
Figure 6.
Ovipositor sheaths: (A) Diaeretiella rapae, (B) Aphidius ericaphidis, (C) Aphidius geranii, (D) Aphidius staticobii, (E) Aphidius balcanicus, (F) Aphidius urticae.
The most important character that currently discriminates genus Diaeretiella from Aphidius is reduced wing venation, with the median (M + m − cu) and r − m veins absent in D. rapae, which is also the case for some Aphidius species. For example, in A. aquilus it is very common to have specimens with and without M + m − cu and r − m veins present within the same population (Figure 7). Also, some populations of A. salicis show similar venation pattern as A. aquilus. Species of the subgenus Lysaphidus have developed only a small part of M + m − cu vein under a developed r − m vein [49].
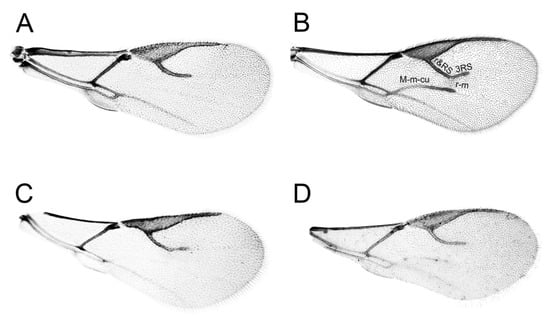
Figure 7.
Fore wings: (A) Diaeretiella rapae, (B) Aphidius ervi, (C,D) Aphidius aquilus.
3.2. Molecular Analysis
The 315 COI sequences obtained from D. rapae comprised 23 haplotypes (H1–H23) with mean genetic distance of 0.8% between haplotypes. The highest recorded intraspecific genetic distance was 1.4% between haplotypes H9 and H20 (Table S4).
The obtained ML phylogenetic tree of D. rapae (presented as a single sequence haplotype tree in Figure 8) is in congruence with determined low genetic diversity between haplotypes.
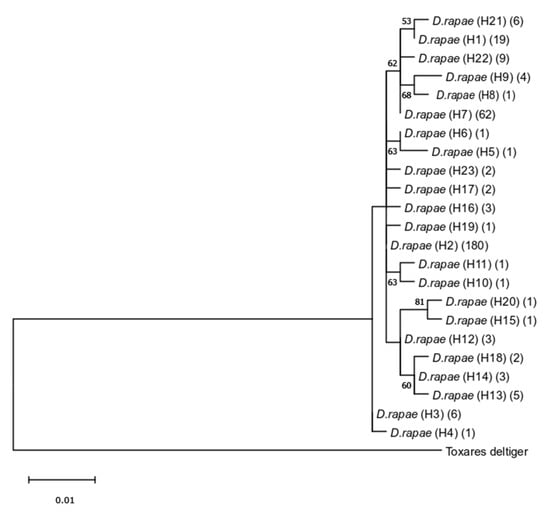
Figure 8.
Phylogenetic tree based on partial mtCOI sequences of Diaeretiella rapae obtained using the Maximum Likelihood method. Bootstrap values are indicated above/below branches. Numbers in parentheses refer to the number of sequences for each haplotype.
The reconstructed phylogenetic network shows a slight separation of haplotypes from the Indian subcontinent (Figure 9). Within this group of six haplotypes (Figure 9 right side), H7 is the most common with 62 sequences and the nodal point for the remaining haplotypes, while H8 is represented with a single sequence from India. The other 17 haplotypes (eight with a single sequence), were grouped together (Figure 9, left side) with H2 as the most abundant, containing 180 sequences from Holarctic, Neotropic and Afrotropic regions.
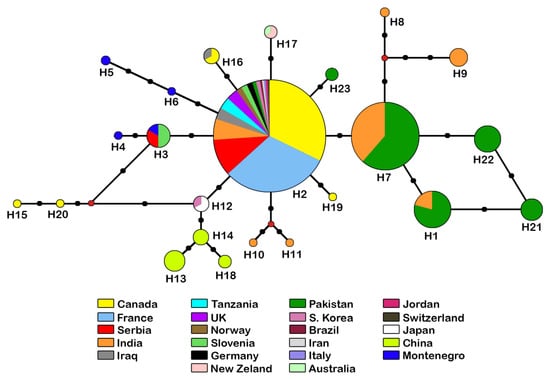
Figure 9.
Haplotype network for COI sequences from 315 specimens belonging to the D. rapae. The circle size indicates the number of specimens with a haplotype (not to scale); each black dot represents a nucleotide substitution; red dots represent median vectors.
Analysis of phylogenetic relationships between Diaeretiella rapae and other Aphidiinae genera (Figure 10) revealed that D. rapae is nested within Aphidius species.
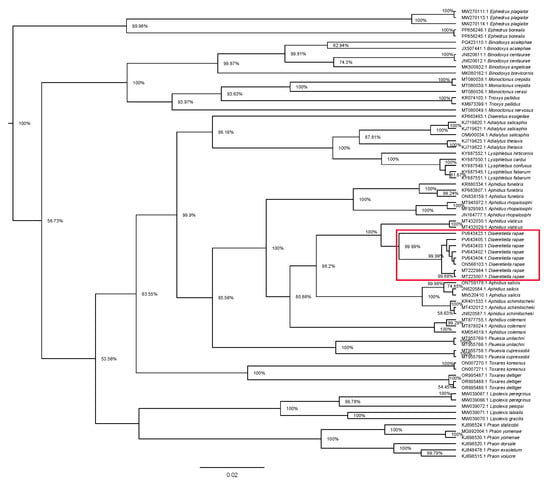
Figure 10.
Phylogenetic tree based on 73 cytochrome oxidase c subunit I (COI) sequences of Aphidiinae species, representing 13 genera. Red rectangle represents D. rapae sequences.
Analysis of phylogenetic relationships between Diaeretiella rapae and Aphidius species (Figure 11) revealed that D. rapae is nested within Aphidius species. According to COI sequences, D. rapae is a monophyletic species which together with all analysed Aphidius species forms a monophyletic clade. Diaeretiella rapae formed a separate clade with A. viaticus, A. schimitscheki, A. salicis and A. aquilus. Aphidius viaticus positioned as the closest relative to D. rapae, with a genetic distance of 2.1% between them. Those two species together form a sister clade to a clade consisting of A. schimitscheki, A. aquilus and A. salicis, with the mean genetic distance of 6.3% between the two clades. Aphidius platensis showed the highest mean genetic distance (11.3%) from D. rapae. Mean genetic distances within analysed Aphidius species varied from 0.0% to 11.7%.
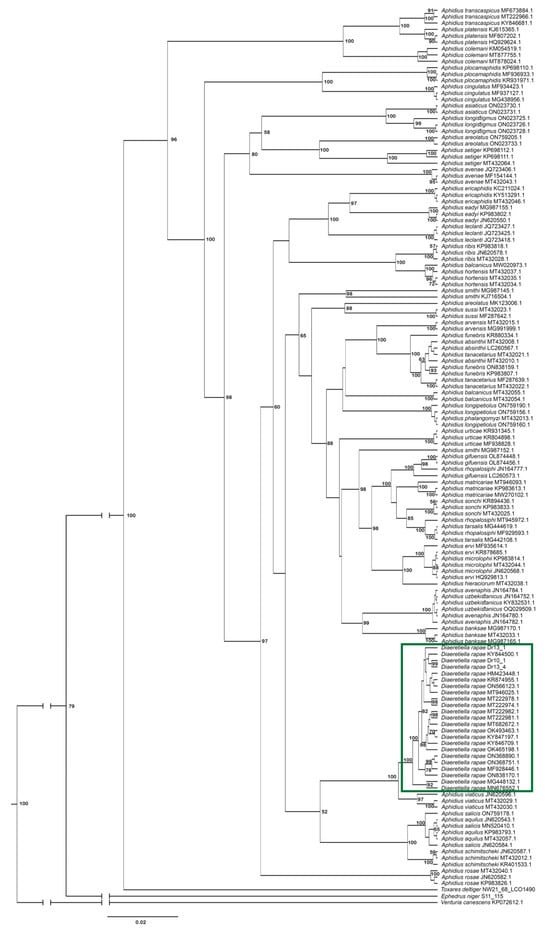
Figure 11.
Phylogenetic tree based on 138 cytochrome oxidase c subunit I (COI) sequences of Aphidius species. The green rectangle represents D. rapae sequences.
There are no significant differences between Diaeretiella rapae and Aphidius species, based on both morphological and molecular analysis that would discriminate between these two genera.
3.3. Re-Description of Aphidius Rapae McIntosh, 1855
Type material: Original type specimen is not available and is probably lost. Mackauer [18] designated a single female of Lysiphlebus crawfordi as a lectotype of D. rapae (Phoenix, Ariz.; “Myzus persicae”; U.S.N.M., No. 66797). The whole specimen is slide mounted. The authors of this study examined lectotype photographs provided by Robert Kula.
Diagnosis: In most cases, Aphidius rapae differs from other Aphidius species by the absence of the M + m − cu and r − m veins. However, based only on wing venation, specimens of several species could be confused with A. rapae. This is because they may not have these veins or they are transparent or very pale. It is most similar to A. schimitscheki from which it differs by the ratio of the petiole at spiracles level (A. rapae 1.9–2.6; A schimitscheki 3.0–3.5). It can be differentiated from species of subgenus Lysaphidus (e.g., A. viaticus, A. arvensis, A. erysimi) by a developed r − m vein in Aphidius (Lysaphidus) spp. [50]. A. aquilus and A. salicis can be differentiated from A. rapae when specimens show slight traces of the M + m − cu vein. If this vein is not visible, these two species often exhibit a visible point of connection between the r + Rs and 3 RS veins. Additionally, the aphid hosts of A. aquilus and A. salicis typically feed on trees.
Description: Head rounded, moderately setose. Face moderately setose. Tentorial index 0.29–0.36. Malar space 0.21–0.3 as long as longitudinal eye diameter. Mandible bidentate, covered with setae (Figure 13A). Antenna filiform, with (12) 13–14 antennomeres (Figure 13B) and with semierect setae 2/3 length of flagellomere diameter. Flagellomere 1 (F1) as long as F2 (F1/F2 1.0). F1 2.5–3.5 times as long as wide at the middle. Flagellomere 2 2.0–3.0 as long as wide at middle. F1 without or with one longitudinal placode, F2 with two to four longitudinal placodes (Figure 13C). Maxillary palp with four palpomeres, sometimes 3 (if apical segment is undivided), labial palp with two short palpomeres. Clypeus with 9–18 setae.
Mesosoma. Mesoscutum with notaulices only in short ascending part of its anterolateral area and outlined by 2 rows of long scattered setae extending almost to scutellum (Figure 13D). Scutellum with 4–10 setae laterally. Propodeum areolated, central areola pentagonal and very narrow (Figure 13F). Fore wing pterostigma 2.9–4.0 times as long as wide. Ratio between length of pterostigma and R1 vein (=metacarpus) 1.39–2.63 (Figure 13E).
Metasoma. Petiole 1.93–2.65 times as long as wide at spiracle level, with 14–18 curved costulae at anterolateral area. Petiole slightly rugose dorsally (Figure 13G).
Color. Head brown to dark brown. Mouth parts yellow. Scape and pedicel brown. Annelus and base of F1 yellowish, remainder of antenna brown. Mesoscutum dark brown to almost black. Propodeum brown. Petiole yellowish to light brown. Abdomen and ovipositor sheath dark brown. Legs yellowish to light brown. Wing hyaline.
Morphologically similar to female.Tentorial index 0.4–0.45, malar space 0.35–0.4 as long as longitudinal eye diameter (Figure 14A). Antenna filiform, with 15–16 antennomeres (Figure 14B) and with semierect setae 1/3 length of flagellomere diameter. Flagellomere 1 (F1) as long as F2. F1 almost two times long as wide at the middle (1.93–1.95). F2 1.7–1.8 times as long as wide at middle. F1 with 4 longitudinal placodes, F2 with 5 longitudinal placodes (Figure 14C). Genitalia as in Figure 14H.
Color: Head dark brown to black. Mouth parts yellowish. Antenna brown. Mesoscutum dark brown to almost black. Propodeum and petiole brown. Abdomen dark brown. Legs light brown to brown. Wing hyaline.
Body length: 2.0 mm.

Figure 12.
Aphidius rapae: (A) female, (B) male.
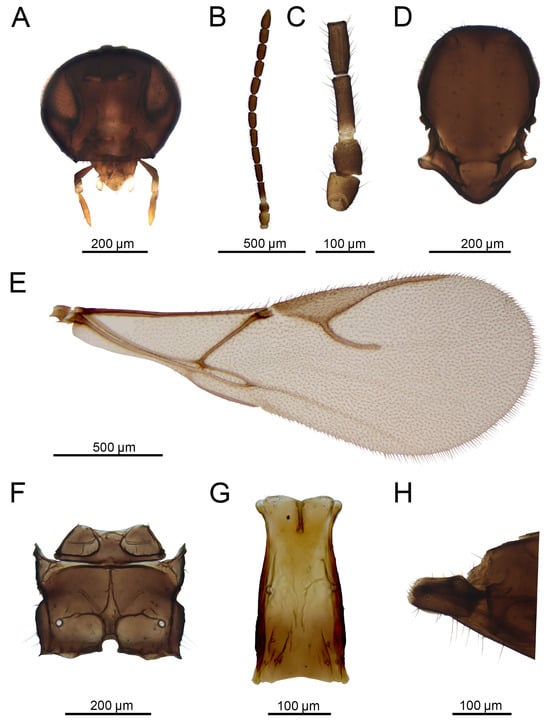
Figure 13.
Aphidius rapae McIntosh, 1855 female: (A) head, (B) antenna, (C) scape, pedicel, annellus and first and second flagellar segments, (D) mesonotum (=mesoscutum + scutellum), dorsal view, (E) forewing, (F) propodeum, dorsal view, (G) petiole, dorsal view, (H) ovipositor sheath, lateral view.
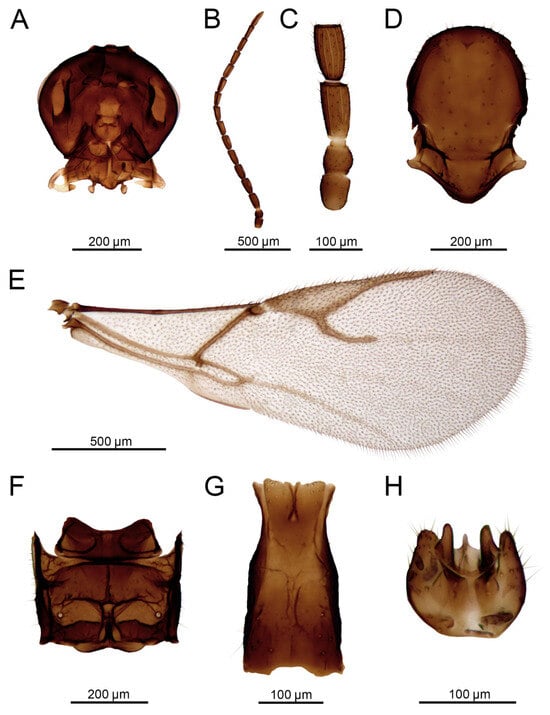
Figure 14.
Aphidius rapae McIntosh, 1855 male: (A) head, (B) antenna, (C) scape, pedicel, annellus and first and second flagellar segments, (D) mesonotum (=mesoscutum + scutellum), dorsal view, (E) forewing, (F) propodeum, dorsal view, (G) petiole, dorsal view, (H) aedeagus, ventral view.
4. Discussion
In this study, we analyzed the taxonomic status of A. rapae (D. rapae) within the subfamily Aphidiinae and proved that A. rapae belongs to the genus Aphidius according to morphological and molecular characters. This confirms statements of Hafez [10] and Mackauer [22] that A. rapae belongs to this genus, as the reduced wing venation within genus Aphidius could not be used as a character for taxonomic determination.
As already noted, A. rapae is a cosmopolitan species, which is confirmed by the specimens from all over the world used in this study. The haplotype network of A. rapae is organized in a star shape, with the central haplotype having the most sequences, suggesting that the population of A. rapae has recently expanded [51]. The combination of data from the haplotype network and the phylogenetic tree of A. rapae confirms that most branches are monophyletic. The central haplotype (H2) is the most important node in the haplotype network, linking nine other groups/haplotypes. It has a cosmopolitan distribution (17 countries around the world) and is the most common in all samples. Although the majority of specimens (≈ 75%) were sampled from three countries (Canada, France, Serbia) because the research groups are based in these countries or extensive research has been conducted there, it does not diminish the significance of this haplotype. In the haplotype network, some haplotype patterns reflect their geographical distribution: one cluster consists exclusively of Indian specimens (H10, H11), one of specimens from Montenegro (H5, H6), one of specimens from the Balkan Peninsula (Slovenia, Montenegro and Serbia, haplotypes H3 and H4) and one haplotype from Australasia (H17). The Chinese group (H13, H14, H18) is linked to the East Asian group (H12), probably because they are closely related. These clusters are linked to the Canadian group (H15, H20), which can be explained as the cause of the introduction of A. rapae to North America from China in the early 1990s [52]. There is a slight separation of the haplotypes from South Asia (Pakistan and India) (H1, H7, H8, H9, H21, H22) from the others, which is also confirmed by the phylogenetic tree, suggesting a common ancestor. The results of research on the phylogeography of this species from the Old and New World show, as in our research, that A. rapae has recently expanded its range. This study also shows that A. rapae does not show genetic isolation, even when the distribution of the samples is taken into account [9]. This indicates that the genetic differences between the analysed populations in our study are not significant.
It is known that A. rapae was introduced to North America to control the aphid D. noxia because the native parasitoid population was unsuccessful in controlling this pest [52,53,54]. It was also introduced from Queensland and New South Wales to Western Australia in 1902 and from Sri Lanka in 1909 for biological control of B. brassicae [55]. For New Zealand, there is no record of deliberate introduction of A. rapae, and the first finding dates back to 1930 [56]. The separation of haplotype 17, which consists only of specimens from Australia and New Zealand and no other haplotype from these areas, gives us an indication that A. rapae from this region may have a different migration history. It is also shown by research based on microsatellites where the populations from Australia, Kazakhstan, France and Morocco were compared [5]. The results of this research indicated that A. rapae from Australia probably originates from one population as it exhibited significant differences from three other countries.
In the phylogenetic tree, A. rapae is positioned in the central section. It is grouped with other species, such as A. viaticus, A. salicis, A. aquilus and A. schimitscheki. This species group has a smaller number of antennomeres and various degrees of forewing vein reduction, a morphological trait that is also characteristic for A. rapae. The genetic distance between A. rapae and other Aphidius species ranges from 2.1 to 12.6%, while the mean distance between other Aphidius species is 0.0–12.9%. This also confirms that A. rapae is no more genetically different from other Aphidius species than they are from each other.
There is a general trend within the Parasitica group of Hymenoptera toward a reduction in body size and wing venation [57,58,59]. Wing venation reduction at a degree which is present in Aphidius rapae is also present in several other genera (Adialytus Förster, 1862; Binodoxys Mackauer, 1960; Betuloxys Mackauer, 1960; Trioxys Haliday, 1833; Diaeretus Förster, 1863; Lipolexis Förster, 1863; Monoctonus Haliday, 1833) and this event has occurred several times in the Aphidiinae [57]. In the genus Aphidius, it has probably occurred only once, which is also indicated by the results of our study. The genus Aphidius belongs to the more evolutionarily recent genera within the subfamily Aphidiinae, where the evolutionary trend is toward a more elongated body, a more slender petiole, reduced wing venation, and longer antennae with a greater number of antennal segments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects16070736/s1, Table S1: Specimens used for morphological analysis; Table S2: Data on specimens which sequences were used in phylogenetic analysis; Table S3: Accession numbers of Aphidius sequences; Table S4: Genetic distances between A. rapae haplotypes.
Author Contributions
N.P. and A.P. conceived and designed the experiments. N.P., K.K., Ž.T. and A.P. provided specimens that were used for morphological and molecular analyses. K.K., N.P. and A.P. carried out laboratory work and data analysis. N.P. and K.K. provided species re-descriptions. N.P., K.K. and A.P. wrote the original draft of the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by taxpayers in Serbia through the Ministry of Science, Technological Development and Innovation of the Republic of Serbia (Grants No. 451-03-136/2025-03/200178 and 451-03-137/2025-03/200178).
Data Availability Statement
The new specimen sequences analyzed in this study are deposited in the GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers PV643399–PV643423.
Acknowledgments
The authors would like to dedicate this paper to students and teachers who stood against corruption and the collapse of the educational system in Serbia during the academic 2024–2025 year. We would also like to thank all the taxpayers in Serbia for supporting science and education. A tiny fraction of tax revenue is distributed to us by the Ministry of Science, Technological Development and Innovation of the Republic of Serbia.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Pike, K.S.; Starý, P.; Miller, T.; Allison, D.; Graf, G.; Boydston, L.; Miller, R.; Gillespie, R. Host range and habitats of the aphid parasitoid Diaeretiella rapae (Hymenoptera: Aphidiidae) in Washington State. Environ. Entomol. 1999, 28, 61–71. [Google Scholar] [CrossRef]
- Singh, R.; Singh, G. Systematics, distribution and host range of Diaeretiella rapae (McIntosh) (Hymenoptera: Braconidae, Aphidiinae). Int. J. Res. Stud. Biosci. 2015, 3, 1–36. [Google Scholar]
- Kavallieratos, N.G.; Tomanović, Ž.; Starý, P.; Athanassiou, C.G.; Sarlis, G.P.; Petrović, O.; Niketić, M.; Veroniki, M.A. A survey of aphid parasitoids (Hymenoptera: Braconidae: Aphidiinae) of Southeastern Europe and their aphid-plant associations. Appl. Entomol. Zool. 2004, 39, 527–563. [Google Scholar] [CrossRef]
- Zhang, W.Q.; Hassan, S.A. Use of the parasitoid Diaeretiella rapae (McIntoch) to control the cabbage aphid Brevicoryne brassicae (L.). J. Appl. Entomol. 2003, 127, 522–526. [Google Scholar] [CrossRef]
- Baker, D.A.; Loxdale, H.D.; Edwards, O.R. Genetic variation and founder effects in the parasitoid wasp, Diaeretiella rapae (M’intosh) (Hymenoptera: Braconidae: Aphidiidae), affecting its potential as a biological control agent. Mol. Ecol. 2003, 12, 3303–3311. [Google Scholar] [CrossRef]
- Desneux, N.; Fauvergue, X.; Dechaume-Moncharmont, F.X.; Kerhoas, L.; Ballanger, Y.; Kaiser, L. Diaeretiella rapae limits Myzus persicae populations after applications of deltamethrin in oilseed rape. J. Econ. Entomol. 2005, 98, 9–17. [Google Scholar] [CrossRef]
- Neuville, S.; Le Ralec, A.; Outreman, Y.; Jaloux, B. The delay in arrival of the parasitoid Diaeretiella rapae influences the efficiency of cabbage aphid biological control. BioControl 2016, 61, 115–126. [Google Scholar] [CrossRef]
- Soni, S.; Kumar, S. Biological control potential of an aphid parasitoid, Diaeretiella rapae (McIntosh) (Hymenoptera: Braconidae) against Brevicoryne brassicae (Linnaeus) (Hemiptera: Aphididae), a pest of oilseed brassicas in India. Int. J. Trop. Insect Sci. 2021, 41, 2361–2372. [Google Scholar] [CrossRef]
- Baer, C.F.; Tripp, D.W.; Bjorksten, T.A.; Antolin, M.F. Phylogeography of a parasitoid wasp (Diaeretiella rapae): No evidence of host-associated lineages. Mol. Ecol. 2004, 13, 1859–1869. [Google Scholar] [CrossRef]
- Hafez, M. Seasonal fluctuations of population density of the cabbage aphid, Brevicoryne brassicae (L.), in the Netherlands, and the role of its parasite, Aphidius (Diaeretiella) rapae (Curtis). Tijdschr. Plantenziekten 1961, 67, 345–548. [Google Scholar] [CrossRef]
- Sedlag, U. Zur Biologie und Bedeutung von Diaeretiella rapae (McIntosh) als Parasit der Kohlblattlaus (Brevicoryne brassicae L.). Nachrichtenbl. Dtsch. Pflanzenschutzdienstes 1964, 18, 81–86. [Google Scholar]
- Askari, A.; Alishah, A. Courtship behavior and evidence for a sex pheromone in Diaeretiella rapae (Hymenoptera: Braconidae), the cabbage aphid primary parasitoid. Ann. Entomol. Soc. Am. 1979, 72, 749–750. [Google Scholar] [CrossRef]
- Blande, J.D.; Pickett, J.A.; Poppy, G.M. Attack rate and success of the parasitoid Diaeretiella rapae on specialist and generalist feeding aphids. J. Chem. Ecol. 2004, 30, 1781–1795. [Google Scholar] [CrossRef]
- Martínez, M.Á.; Duarte, L.; Ceballos, M. Biology and vertical life table of Diaeretiella rapae McIntosh under laboratory conditions. Rev. Prot. Veg. 2013, 28, 23–26. [Google Scholar]
- Gazmer, R.; Gupta, M.K.; Singh, M.D. Biology of Diaeretiella rapae (McIntosh) (Hymenoptera: Aphidiidae) on cabbage aphid (Brevicoryne brassicae Linnaeus) and influence of host age on the developmental duration. J. Biol. Control 2015, 29, 38–42. [Google Scholar] [CrossRef]
- McIntosh, C. The Book of the Garden, 2nd ed.; William Blackwood and Sons: Edinburgh, Scotland; London, UK, 1855; p. 194. [Google Scholar]
- Curtis, J. Farm Insects: Being the Natural History and Economy of the Insects Injurious to the Field Crops of Great Britain and Ireland, and also Those Which Infest Barns and Granaries. With Suggestions for Their Destruction, 1st ed.; Blackie and Son: Glasgow, Scotland; London, UK, 1860; pp. 37–74. [Google Scholar]
- Mackauer, M. Comments on the authorship, generic position, and synonymy of Diaeretiella rapae (M‘Intosh) (Hymenoptera: Aphidiidae). Contrib. Entomol. 1964, 14, 53–58. [Google Scholar]
- Gahan, A.B. Some synonymy and other notes on Aphidiinae. Proc. Entomol. Soc. Wash. 1910, 12, 179–189. [Google Scholar]
- Marshall, T.A. A Catalogue of British Hymenoptera: Chrysididae, Ichneumonidae, Braconidae and Evaniidae; Entomolical Society of London: London, UK, 1872; p. 136. [Google Scholar]
- Starý, P. The generic classification of the family Aphidiidae (Hymenoptera). Acta Soc. Entomol. Czechoslov. 1960, 57, 238–252. [Google Scholar]
- Mackauer, M. Die Typen der Unterfamilie Aphidiinae des Britischen Museums London (Hymenoptera: Braconidae). Contrib. Entomol. 1961, 11, 96–154. [Google Scholar]
- Mackauer, M. Die Gattungen der Familie Aphidiidae und ihreverwandtschaftliche Zuordnung (Hymenoptera: Ichneumonoidea). Contrib. Entomol. 1961, 11, 792–803. [Google Scholar]
- Akhtar, M.S.; Dey, D.; Usmani, M.K. Redescription of Diaeretiella rapae (M’Intosh) (Hymenoptera: Braconidae: Aphidiinae) with morphological variability of several populations from India. Munis Entomol. Zool. 2011, 6, 194–203. [Google Scholar]
- Pramanik, A.; Dey, D.; Kumar, A. Redescription of Diaeretiella rapae (McIntosh) (hymenoptera: Braconidae: Aphidiinae) with emphasis on morphometrics. J. Entomol. Res. 2012, 36, 77–82. [Google Scholar]
- Herrick, G.W.; Hungate, J.W. The Cabbage Aphis; Cornell University: New York, NY, USA, 1911; Volume 300, p. 734. [Google Scholar]
- Ashmead, W.H. Descriptions of new Braconidae in the collection of the US National Museum. Proc. U.S. Natl. Mus. 1889, 11, 611–671. [Google Scholar] [CrossRef]
- Marshall, T.A. A Monograph of British Braconidae. Part VIII. Trans. R. Entomol. Soc. Lond. 1899, 47, 1–79. [Google Scholar] [CrossRef]
- Dalla Torre, K.W. Braconidae. In Catalogus Hymenopterorum; Guilelmi Engelmann: Leipzig, Germany, 1898; Volume 4. [Google Scholar]
- Marshall, T.A. Les Braconides. In Species des Hyménoptères d’Europe et d’Algérie; André, E., Ed.; Gray: Beaune, Germany, 1891. [Google Scholar]
- Mackauer, M.; Starý, P. Hymenoptera: Ichneumonoidea, World Aphidiidae. In Index of Entomophagous Insects; Delucchi, V., Remaudière, G., Eds.; Le Fransois: Paris, France, 1967. [Google Scholar]
- Mackauer, M. A re-examination of CF Baker’s collection of aphid parasites (Hymenoptera: Aphidiidae). Can. Entomol. 1963, 95, 921–935. [Google Scholar] [CrossRef]
- Gahan, A.B. Aphidiinae of North America. Agric. Exp. Stn. Maryland College Park Bull. 1911, 152, 147–200. [Google Scholar]
- Starý, P. A revision of the genus Diaeretiella Starý (Hymenoptera: Aphidiidae). Acta Entomol.Musei Natl. Pragae. 1961, 34, 383–397. [Google Scholar]
- Mackauer, M. Hymenopterorum Catalogus. Pars 3. Aphidiidae; Junk: Gravenhage, Netherlands, 1968. [Google Scholar]
- Starý, P. Aphid parasites of Czechoslovakia. A review of the Czechoslovak Aphidiidae (Hymenoptera); Springer: Hague, The Netherlands, 1966; p. 242. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Tomanović, Ž.; Žikić, V.; Petrović, A. Aphidius Nees (Hymenoptera, Braconidae, Aphidiinae) in Serbia: Key to species identification including parasitoid–aphid host list. Acta Entomol. Serbica 2023, 28, 61–76. [Google Scholar]
- Sharkey, M.J.; Wharton, R.A. Morphology and terminology. In Manual of the New World Genera of the Family Braconidae; Wharton, R.A., Marsh, P.M., Sharkey, M.J., Eds.; International Society of Hymenopterists: Washington, DC, USA, 1997. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol 1994, 3, 294–299. [Google Scholar]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial DNA in Humans and Chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-Del Barrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsis, S.E.; Sanchez-Gracia, A. DnaSP v6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, syy032. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. 2018. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef]
- Xia, X.; Xie, Z.; Salemi, M.; Chen, L.; Wang, Y. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 2003, 26, 1–7. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree, version 1.4.4; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, Scotland, 2009. [Google Scholar]
- Tomanović, Ž.; Rakhshani, E.; Starý, P.; Kavallieratos, N.G.; Stanisavljević, L.Ž.; Žikić, V.; Athanassiou, C.G. Phylogenetic relationships between the genera Aphidius and Lysaphidus (Hymenoptera: Braconidae: Aphidiinae) with description of Aphidius iranicus sp. nov. The Canadian Entomologist 2007, 139, 297–307. [Google Scholar] [CrossRef]
- Ferreri, M.; Qu, W.; Han, B.O. Phylogenetic networks: A tool to display character conflict and demographic history. Afr. J. Biotechnol. 2011, 10, 12799–12803. [Google Scholar]
- Elliott, N.C.; Burd, J.D.; Armstrong, J.; Walker, C.B.; Reed, D.K.; Peairs, F.B. Release and recovery of imported parasitoids of the Russian wheat aphid in eastern Colorado. Southwest. Entomol. 1995, 20, 125–129. [Google Scholar]
- Brewer, M.J.; Nelson, D.J.; Ahern, R.G.; Donahue, J.D.; Prokrym, D.R. Recovery and range expansion of parasitoids (Hymenoptera: Aphelinidae and Braconidae) released for biological control of Diuraphis noxia (Homoptera: Aphididae) in Wyoming. Environ. Entomol. 2001, 30, 578–588. [Google Scholar] [CrossRef]
- Tanigoshi, L.K.; Pike, K.S.; Miller, R.H.; Miller, T.D.; Allison, D. Search for, and release of, parasitoids for the biological control of Russian wheat aphid in Washington State (USA). Agric. Ecosyst. Environ. 1995, 52, 25–30. [Google Scholar] [CrossRef]
- Carver, M.; Starý, P. A preliminary review of the Aphidiidae (Hymenoptera: Ichneumonoidea) of Australia and New Zealand. Aust. J. Entomol. 1974, 13, 235–240. [Google Scholar] [CrossRef]
- Teulon, D.A.J.; Drayton, G.M.; Scott, I.A.W. Exotic introductions of primary parasitoids of aphids in New Zealand: The good and the bad. In Proceedings of the Third International Symposium on Biological Control of Arthropods, Christchurch, New Zealand, 8–13 February 2008; pp. 421–430. [Google Scholar]
- Žikić, V.; Stanković, S.S.; Petrović, A.; Ilić Milošević, M.; Tomanović, Ž.; Klingenberg, C.P.; Ivanović, A. Evolutionary relationships of wing venation and wing size and shape in Aphidiinae (Hymenoptera: Braconidae). Org. Divers. Evol. 2017, 17, 607–617. [Google Scholar] [CrossRef]
- Peters, R.S.; Krogmann, L.; Mayer, C.; Donath, A.; Gunkel, S.; Meusemann, K.; Kolozov, A.; Podsiadlowski, L.; Petersen, M.; Lanfear, R.; et al. Evolutionary history of the Hymenoptera. Curr. Biol. 2017, 27, 1013–1018. [Google Scholar] [CrossRef]
- Polilov, A.A. Small is beautiful: Features of the smallest insects and limits to miniaturization. Annu. Rev. Entomol. 2015, 60, 103–121. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).