Tomato Chlorosis Virus Infection Facilitates Bemisia tabaci MED Reproduction by Elevating Vitellogenin Expression
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
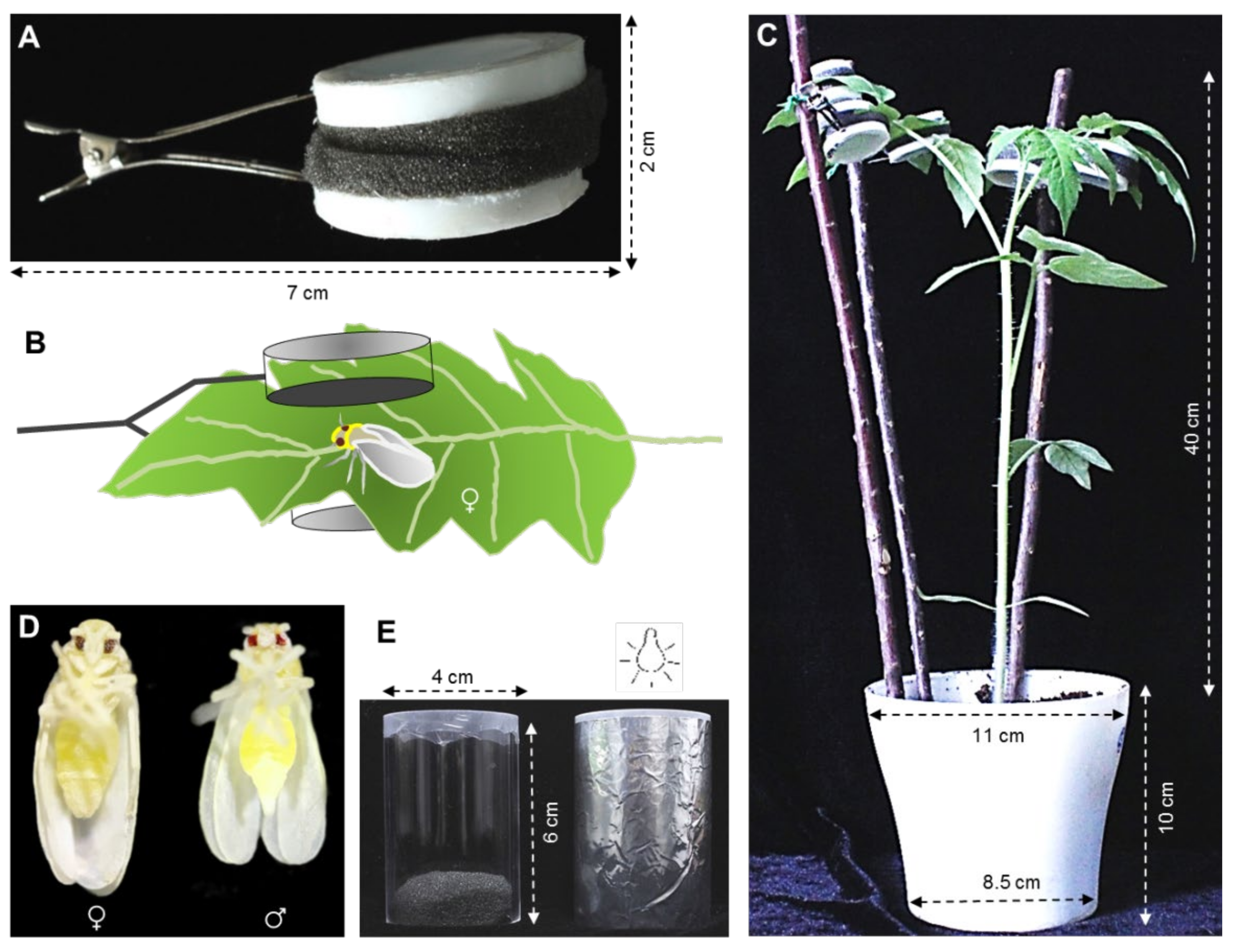
2.1. Host Plants and Whitefly Rearing
2.2. Whitefly Fecundity on ToCV-Infected and Uninfected Tomato Plants
2.3. Whitefly Vg Expression on ToCV-Infected and Uninfected Tomato Plants
2.4. The Role of Vg on Whitefly Reproduction in the Presence and Absence of ToCV
2.4.1. dsRNA Synthesis
2.4.2. Dietary RNAi on Whitefly Vg Expression
2.4.3. Dietary RNAi on the Development of Whitefly Ovaries
2.4.4. Dietary RNAi on Whitefly Fecundity
2.5. Data Analysis
3. Results
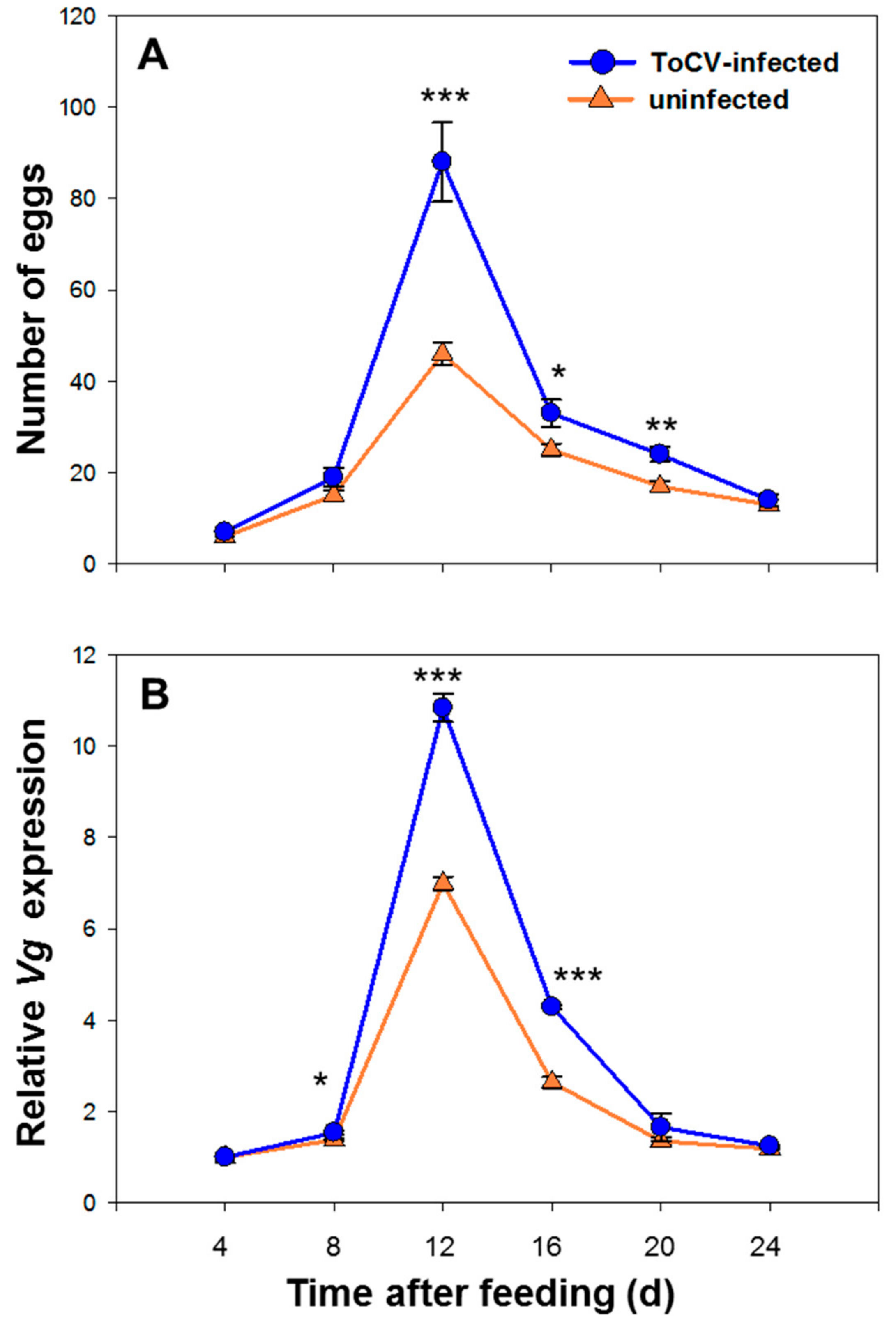
3.1. Whitefly Fecundity on ToCV-Infected and Uninfected Tomato Plants
3.2. Vg Relative Expression Level of Whitefly on ToCV-Infected and Uninfected Tomato Plants
3.3. The Role of Vg on Whitefly Reproduction in the Presence and Absence of ToCV
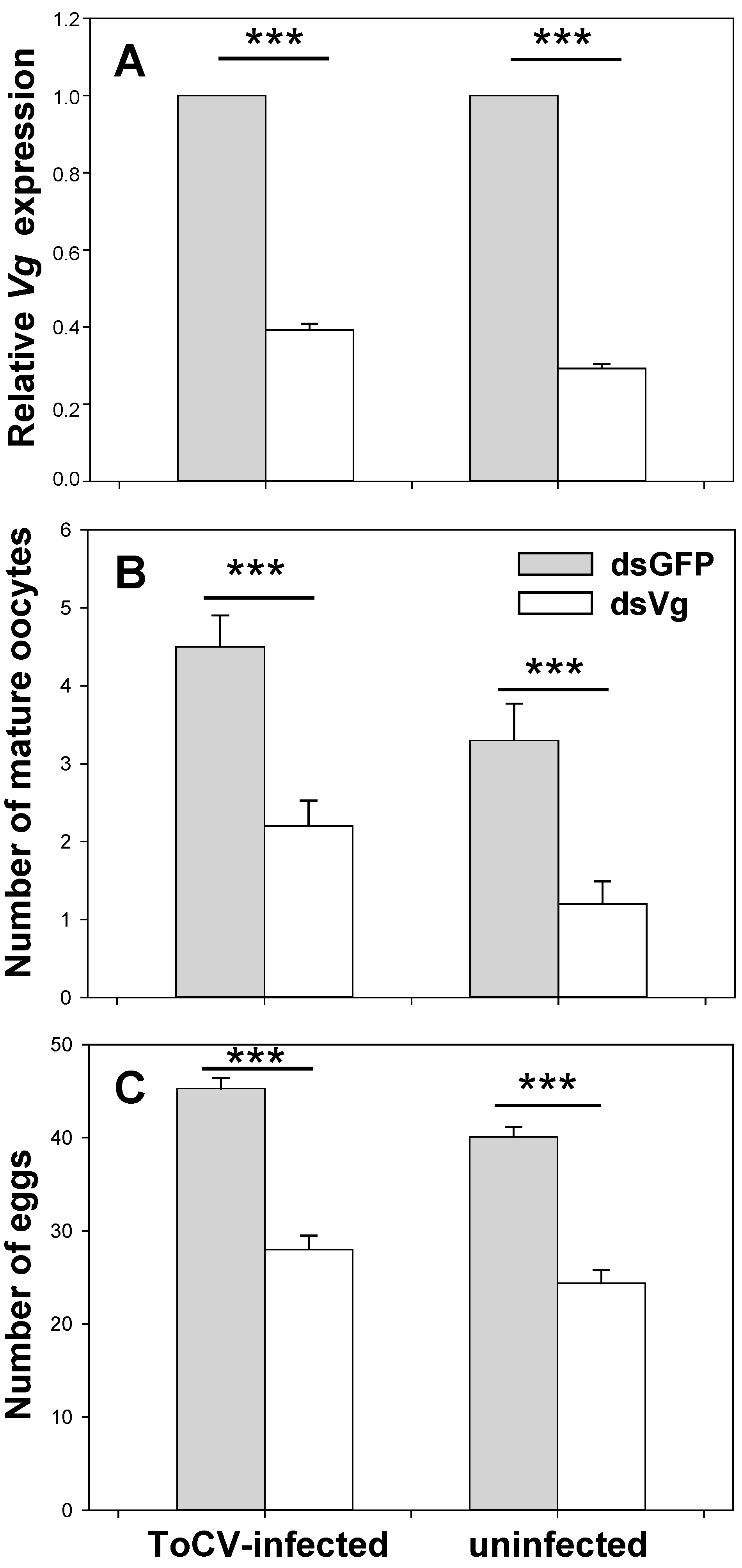
3.3.1. Vg Expression on Whiteflies with Dietary RNAi
3.3.2. Development of Whitefly Ovaries with Dietary RNAi
3.3.3. Whitefly Fecundity with Dietary RNAi
4. Discussion
4.1. ToCV Infection Facilitated the Whitefly Reproduction
4.2. ToCV Infection Facilitates Whitefly Reproduction through Elevating Vg Expression
4.3. RNAi on Vg as a New Approach in ToCV/Whitefly Control
5. Summary and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, K.B.G.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant. Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef] [PubMed]
- Elena, S.F.; Fraile, A.; García-Arenal, F. Evolution and emergence of plant viruses. Adv. Virus Res. 2014, 88, 161–191. [Google Scholar]
- Andret-Link, P.; Fuchs, M. Transmission specificity of plant viruses by vectors. J. Plant. Pathol. 2005, 87, 153–165. [Google Scholar]
- Hohn, T. Plant virus transmission from the insect point of view. Proc. Natl. Acad. Sci. USA 2007, 104, 17905–17906. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Ammar, E.-D.; Whitfield, A.E.; Redinbaugh, M.G. Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 2008, 46, 327–359. [Google Scholar] [CrossRef]
- Stout, M.J.; Thaler, J.S.; Thomma, B.P. Plant-mediated interactions between pathogenic microorganisms and herbivorous arthropods. Annu. Rev. Entomol. 2006, 51, 663–689. [Google Scholar] [CrossRef]
- Eigenbrode, S.D.; Bosque-Pérez, N.A.; Davis, T.S. Insect-borne plant pathogens and their vectors: Ecology, evolution, and complex interactions. Annu. Rev. Entomol. 2018, 63, 169–191. [Google Scholar] [CrossRef]
- Oliveira, M.R.V.; Henneberry, T.J.; Anderson, P. History, current status, and collaborative research projects for Bemisia tabaci. Crop. Prot. 2001, 20, 709–723. [Google Scholar] [CrossRef]
- Berlinger, M.J. Host plant resistance to Bemisia tabaci. Agric. Ecosyst. Environ. 1986, 17, 69–82. [Google Scholar] [CrossRef]
- Jones, D.R. Plant viruses transmitted by whiteflies. Eur. J. Plant. Pathol. 2003, 109, 195–219. [Google Scholar] [CrossRef]
- Bragard, C.; Caciagli, P.; Lemaire, O.; Lopez-Moya, J.J.; MacFarlane, S.; Peters, D.; Susi, P.; Torrance, L. Status and prospects of plant virus control through interference with vector transmission. Annu. Rev. Phytopathol. 2013, 51, 177–201. [Google Scholar] [CrossRef] [PubMed]
- Polston, J.E.; De Barro, P.J.; Boykin, L.M. Transmission specificities of plant viruses with the newly identified species of the Bemisia tabaci species complex. Pest. Manag. Sci. 2014, 70, 1547–1552. [Google Scholar] [CrossRef] [PubMed]
- Cathrin, P.B.; Ghanim, M. Chapter 4—Recent advances on interactions between the whitefly Bemisia tabaci and begomoviruses, with emphasis on Tomato yellow leaf curl virus. In Plant Virus–Host Interaction; Academic Press: Cambridge, MA, USA, 2014; pp. 79–103. [Google Scholar]
- Ellsworth, P.C. Whitefly Management in Arizona: Looking at the Whole System; University of Arizona: Tucson, AZ, USA, 1998; pp. 65–68. [Google Scholar]
- Shi, X.B.; Tang, X.; Zhang, X.; Zhang, D.Y.; Li, F.; Yan, F.; Zhang, Y.J.; Zhou, X.G.; Liu, Y. Transmission efficiency, preference and behavior of Bemisia tabaci MEAM1 and MED under the influence of Tomato Chlorosis Virus. Front. Plant. Sci. 2018, 8, 2271. [Google Scholar] [CrossRef] [PubMed]
- Tzanetakis, I.E.; Martin, R.R.; Wintermantel, W.M. Epidemiology of criniviruses: An emerging problem in world agriculture. Front. Microbiol. 2013, 4, 119. [Google Scholar] [CrossRef] [PubMed]
- Lozano, G.; Moriones, E.; Navas-Castillo, J. Complete nucleotide sequence of the RNA2 of the crinivirus Tomato chlorosis virus. Arch. Virol. 2006, 151, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olive, E.; Navas-Castillo, J. Tomato chlorosis virus, an emergent plant virus still expanding its geographical and host ranges. Mol. Plant. Pathol. 2019, 20, 1307–1320. [Google Scholar] [CrossRef]
- Kaur, N.; Chen, W.B.; Zheng, Y.; Hasegawa, D.K.; Ling, K.S.; Fei, Z.J.; Wintermantel, W.M. Transcriptome analysis of the whitefly, Bemisia tabaci MEAM1 during feeding on tomato infected with the crinivirus, Tomato chlorosis virus, identifies a temporal shift in gene expression and differential regulation of novel orphan genes. BMC Genom. 2017, 18, 370. [Google Scholar] [CrossRef]
- Snigirevskaya, E.S.; Raikhel, A.S. Receptor-mediated endocytosis of yolk proteins in insect oocytes. In Reproductive Biology of Invertebrates: Progress in Vitellogenesis; CRC Press: Boca Raton, FL, USA, 2005; Volume 12, pp. 199–228. [Google Scholar]
- Cho, K.H.; Cheon, H.M.; Kokoza, V.; Raikhel, A.S. Regulatory region of the vitellogenin receptor gene sufficient for high-level, germ line cell-specific ovarian expression in transgenic Aedes aegypti mosquitoes. Insect Biochem. Mol. Biol. 2006, 36, 273–281. [Google Scholar] [CrossRef]
- Tufail, M.; Takeda, M. Molecular characteristics of insect vitellogenins. J. Insect Physiol. 2008, 54, 1447–1458. [Google Scholar] [CrossRef]
- Yang, X.; Xie, W.; Li, R.M.; Zhou, X.M.; Wang, S.L.; Wu, Q.J.; Yang, N.N.; Xia, J.X.; Yang, Z.Z.; Guo, L.T.; et al. RNA interference-mediated knockdown of the hydroxyacid-oxoacid transhydrogenase gene decreases thiamethoxam resistance in adults of the whitefly Bemisia tabaci. Sci. Rep. 2017, 7, 41201. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Wan, F.H.; Zhang, Y.J.; Brown, J.K. Change in the biotype composition of Bemisia tabaci in Shandong province of China from 2005 to 2008. Environ. Entomol. 2010, 39, 1028–1036. [Google Scholar] [CrossRef] [PubMed]
- Khasdan, V.; Levin, I.; Rosner, A.; Morin, S.; Kontsedalov, S.; Maslenin, L.; Horowitz, A.R. DNA markers for identifying biotypes B and Q of Bemisia tabaci (Hemiptera: Aleyrodidae) and studying population dynamics. Bull. Entomol. Res. 2005, 95, 605–613. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Li, R.M.; Xie, W.; Wang, S.L.; Wu, Q.J.; Yang, N.N.; Yang, X.; Pan, H.P.; Zhou, X.M.; Bai, L.Y.; Xu, B.Y.; et al. Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS ONE 2013, 8, e53006. [Google Scholar] [CrossRef]
- Wei, J.; He, Y.Z.; Guo, Q.; Liu, Y.Q.; Zhou, X.P.; Liu, S.S.; Wang, X.W. Vector development and vitellogenin determine the transovarial transmission of begomoviruses. Proc. Natl. Acad. Sci. USA 2017, 114, 6746–6751. [Google Scholar] [CrossRef]
- Guo, J.Y.; Ye, G.Y.; Dong, S.Z.; Liu, S.S. An invasive whitefly feeding on a virus-infected plant increased its egg production and realized fecundity. PLoS ONE 2010, 5, e11713. [Google Scholar] [CrossRef]
- Shi, X.B.; Pan, H.P.; Xie, W.; Wu, Q.J.; Wang, S.L.; Liu, Y.; Fang, Y.; Chen, G.; Gao, X.W.; Zhang, Y.J. Plant virus differentially alters the plant’s defense response to its closely related vectors. PLoS ONE 2013, 8, e83520. [Google Scholar] [CrossRef]
- Jiu, M.; Zhou, X.P.; Tong, L.; Xu, J.; Yang, X.; Wang, F.H.; Liu, S.S. Vector-virus mutualism accelerates population increase of an invasive whitefly. PLoS ONE 2007, 2, e182. [Google Scholar] [CrossRef]
- Guo, J.Y.; Dong, S.Z.; Yang, X.L.; Cheng, L.; Wan, F.H.; Liu, S.S.; Zhou, X.P.; Ye, G.Y. Enhanced vitellogenesis in a whitefly via feeding on a begomovirus-infected plant. PLoS ONE 2012, 7, e43567. [Google Scholar] [CrossRef]
- Fereres, A.; Peñaflor, M.F.G.V.; Favaro, C.F.; Azevedo, K.E.X.; Landi, C.H.; Maluta, N.K.P.; Bento, J.M.S.; Lopes, J.R.S. Tomato infection by whitefly-transmitted circulative and non-circulative viruses induce contrasting changes in plant volatiles and vector behaviour. Viruses 2016, 8, 225. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, L.F.M.; Bello, V.H.; Marchi, B.R.D.; Sartori, M.M.P.; Pavan, M.A.; Krause-Sakate, R. Performance of Bemisia tabaci MEAM1 and Trialeurodes vaporariorum on Tomato chlorosis virus (ToCV) infected plants. J. Appl. Entomol. 2018, 142, 1008–1015. [Google Scholar] [CrossRef]
- Li, J.; Ding, T.B.; Chi, H.; Chu, D. Effects of Tomato chlorosis virus on the performance of its key vector, Bemisia tabaci, in China. J. Appl. Entomol. 2018, 142, 296–304. [Google Scholar] [CrossRef]
- Maluta, N.; Fereres, A.; Lopes, J.R.S. Plant-mediated indirect effects of two viruses with different transmission modes on Bemisia tabaci feeding behavior and fitness. J. Pest. Sci. 2019, 92, 405–416. [Google Scholar] [CrossRef]
- Su, Q.; Preisser, E.L.; Zhou, X.M.; Xie, W.; Liu, B.M.; Wang, S.L.; Wu, Q.J.; Zhang, Y.J. Manipulation of host quality and defense by a plant virus improves performance of whitefly vectors. J. Econ. Entomol. 2015, 108, 11–19. [Google Scholar] [CrossRef]
- Su, Q.; Mescher, M.C.; Wang, S.L.; Chen, G.; Xie, W.; Wu, Q.J.; Wang, W.K.; Zhang, Y.J. Tomato yellow leaf curl virus differentially influences plant defence responses to a vector and a non-vector herbivore. Plant. Cell Environ. 2016, 39, 597–607. [Google Scholar] [CrossRef]
- Fiebig, M.; Poehling, H.M.; Borgemeister, C. Barley yellow dwarf virus, wheat, and Sitobion avenae: A case of trilateral interactions. Entomol. Exp. Appl. 2004, 110, 11–21. [Google Scholar] [CrossRef]
- Mauck, K.E.; De Moraes, C.M.; Mescher, M.C. Biochemical and physiological mechanisms underlying effects of Cucumber mosaic virus on host-plant traits that mediate transmission by aphid vectors. Plant. Cell Environ. 2014, 37, 1427–1439. [Google Scholar] [CrossRef]
- Shrestha, A.; Srinivasan, R.; Riley, D.G.; Culbreath, A.K. Direct and indirect effects of a thrips-transmitted Tospovirus on the preference and fitness of its vector, Frankliniella fusca. Entomol. Exp. Appl. 2012, 145, 260–271. [Google Scholar] [CrossRef]
- Zotti, M.J.; Smagghe, G. RNAi technology for insect management and protection of beneficial insects from diseases: Lessons, challenges and risk assessments. Neotrop. Entomol. 2015, 44, 197–213. [Google Scholar] [CrossRef]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: What we know so far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, J.G.; Duan, J.J. RNAi-based insecticidal crops: Potential effects on non-target species. Bioscience 2013, 63, 657–665. [Google Scholar] [CrossRef]
| Gene | Primer Sequence (5′-3′) * |
|---|---|
| Actin | F: CGCTGCCTCCACCTCATT R: ACCGCAAGATTCCATACCC |
| EF-1α | F: TAGCCTTGTGCCAATTTCCG R: CCTTCAGCATTACCGTCC |
| qPCR Vg | F: CTTCTCCGCTGCTTTCTT R: TGTTGGCGTATTTGTTGG |
| dsGFP | F: TAATACGACTCACTATAGGGTTCAGTGGAGAGGGTGAAGGT R: TAATACGACTCACTATAGGGTGTGTGGACAGGTAATGGTTG |
| dsVg | F: ATTCTCTAGAAGCTTAATACGACTCACTATAGGGTCTGTGATGCCTTAGTT R: ATTCTCTAGAAGCTTAATACGACTCACTATAGGGCTCTCTTGAGGTTTTGT |
| Treatments | Time | |||||
|---|---|---|---|---|---|---|
| 4 d | 8 d | 12 d | 16 d | 20 d | 24 d | |
| ToCV-infected | 7.0 ± 8.668 | 19.0 ± 1.988 | 88.0 ± 8.668 | 33.0 ± 3.016 | 24.0 ± 1.627 | 14.0 ± 1.314 |
| Uninfected | 6.0 ± 0.931 | 15.0 ± 1.192 | 46.0 ± 2.441 | 25.0 ± 1.315 | 17.0 ± 1.181 | 13.0 ± 0.965 |
| Treatments | Time | |||||
|---|---|---|---|---|---|---|
| 4 d | 8 d | 12 d | 16 d | 20 d | 24 d | |
| ToCV-infected | 1.000 ± 0.000 | 1.541 ± 0.093 | 10.840 ± 0.537 | 4.290 ± 0.037 | 1.656 ± 0.303 | 1.248 ± 0.002 |
| Uninfected | 1.000 ± 0.000 | 1.380 ± 0.030 | 6.995 ± 0.253 | 2.640 ± 0.117 | 1.353 ± 0.089 | 1.184 ± 0.003 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, L.; Shi, X.; Shi, J.; Zhang, Z.; Fang, Y.; Zhang, Z.; Pan, Q.; Zheng, L.; Gao, Y.; Zhang, D.; et al. Tomato Chlorosis Virus Infection Facilitates Bemisia tabaci MED Reproduction by Elevating Vitellogenin Expression. Insects 2021, 12, 101. https://doi.org/10.3390/insects12020101
Huang L, Shi X, Shi J, Zhang Z, Fang Y, Zhang Z, Pan Q, Zheng L, Gao Y, Zhang D, et al. Tomato Chlorosis Virus Infection Facilitates Bemisia tabaci MED Reproduction by Elevating Vitellogenin Expression. Insects. 2021; 12(2):101. https://doi.org/10.3390/insects12020101
Chicago/Turabian StyleHuang, Liping, Xiaobin Shi, Jizhe Shi, Zhuo Zhang, Yong Fang, Zhanhong Zhang, Qiuyi Pan, Limin Zheng, Yang Gao, Deyong Zhang, and et al. 2021. "Tomato Chlorosis Virus Infection Facilitates Bemisia tabaci MED Reproduction by Elevating Vitellogenin Expression" Insects 12, no. 2: 101. https://doi.org/10.3390/insects12020101
APA StyleHuang, L., Shi, X., Shi, J., Zhang, Z., Fang, Y., Zhang, Z., Pan, Q., Zheng, L., Gao, Y., Zhang, D., Tan, X., Liu, Y., & Zhou, X. (2021). Tomato Chlorosis Virus Infection Facilitates Bemisia tabaci MED Reproduction by Elevating Vitellogenin Expression. Insects, 12(2), 101. https://doi.org/10.3390/insects12020101








