Abstract
The pollen stores of bumble bees host diverse microbiota that influence overall colony fitness. Yet, the taxonomic identity of these symbiotic microbes is relatively unknown. In this descriptive study, we characterized the microbial community of pollen provisions within captive-bred bumble bee hives obtained from two commercial suppliers located in North America. Findings from 16S rRNA and ITS gene-based analyses revealed that pollen provisions from the captive-bred hives shared several microbial taxa that have been previously detected among wild populations. While diverse microbes across phyla Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and Ascomycota were detected in all commercial hives, significant differences were detected at finer-scale taxonomic resolution based on the supplier source. The causative agent of chalkbrood disease in honey bees, Ascosphaera apis, was detected in all hives obtained from one supplier source, although none of the hives showed symptoms of infection. The shared core microbiota across both commercial supplier sources consisted of two ubiquitous bee-associated groups, Lactobacillus and Wickerhamiella/Starmerella clade yeasts that potentially contribute to the beneficial function of the microbiome of bumble bee pollen provisions.
1. Introduction
Organisms across the tree of life span a continuum of reliance on microbial symbionts [1,2], collectively referred to as their “microbiome” [3,4]. Host–microbiome interactions can range from mutualistic to pathogenic [2] depending on the taxonomic and functional composition of the microbiome. Non-pathogenic symbionts allow hosts to obtain nutrients from inaccessible substrates [5], utilize novel energy sources [6], and thrive in extreme habitats [7]. Hosts that derive nutritional benefits from their symbionts often demonstrate increased dependence on their microbial partners [8]. In some cases, extreme dependence on nutritional mutualists has led to major evolutionary transitions among several eukaryotic hosts [9]. For instance, it has been speculated that bees diversified from their predatory wasp ancestors by acquiring specialized pollen-digesting gut microbiota [10]. Indeed, the overall fitness of social bees depends on a sustained symbioses with their gut microbiome [3,11,12], which influences several aspects of host physiology, including pollen digestion, carbohydrate utilization, and immune function [13,14,15,16,17,18,19].
Microbes that are routinely isolated from healthy replicates are likely to have important functions within host environments, and are collectively referred to as the “core microbiota” [20]. Although the core gut microbiome of corbiculate bees, including honey bees [11,21,22,23,24] and bumble bees [12,13,14,22,25,26,27] share several related bacterial taxa, the manner in which these microbes are transferred through generations varies between bee groups. The inheritance of the gut microbiota is influenced by multiple factors including life history strategies [28], foraging preferences [29], and domestication practices [30]. Honey bee colonies are founded by a large number of workers and a queen, and microbial transmission across generations occurs through trophallaxis and other social interactions between nestmates [11,21,23]. In contrast, bumble bees live in annual colonies, founded by a single overwintered queen [31] who transmits her microbiota to the progeny. Therefore, the microbiome of newly initiated bumble bee colonies reflects that of its queen, and this generational inheritance through a single founding individual imposes a transmission bottleneck on the diversity of the gut microbiota [32]. For captive-bred bumble bees, which have fewer opportunities to acquire microbes during foraging, this may result in loss of overall microbiome diversity and/or of key bacterial strains [33,34]. The taxonomic reorganization of the microbial community can lead to subtle shifts in gut microbiome function, and can contribute to lower pathogen resistance and increased infection rates among captive-bred colonies [35]. Indeed, commercially-reared bumble bee colonies often harbor numerous bee pathogens, many of which have been transmitted to healthy populations through contact with contaminated pollen from infected hives [36,37,38,39]. Such pathogen spillover from commercial to wild colonies has caused dramatic population losses and range contractions among several bumble bee species across North America [40,41,42].
While the structure and function of the gut microbiome has been well-investigated [11], relatively little is known about the symbioses between bees and the external community of microbes associated with their pollen stores. Pollen-associated microbes play a central role in the development and nutrition of numerous social [43,44] and solitary bees [45]. Since the complex structure of pollen grains pose a digestive challenge for bees [46], raw pollen is initially mixed with nectar, regurgitated enzymes, and microbes, and fermented to varying degrees before being fed to the larvae. Past research in honey bees demonstrates that an external community of microbial symbionts facilitates this enzymatic process [44,47,48,49,50,51,52], enhancing the digestibility and nutrient content of the raw pollen substrate [50,53]. The resulting nutrient-dense pollen provision, (referred to as beebread in honey bees) consists of pre-digested pollen, nectar, and diverse microbes, and forms the sole source of nutrition for the larvae [22]. Nutrients derived from pollen [54] and pollen-associated microbiota [55] within beebread are assimilated into the biomass of overwintering honey bees and stored until spring. Furthermore, these external symbionts are involved in the production of vital macromolecules [50], long term preservation of stored pollen [56,57,58] and disease prevention [59].
Compared to honey bees, which store copious amounts of beebread [58], bumble bees do not overwinter and their colonies do not contain vast pollen reserves [60]. Instead, pollen brought back to the colony is quickly consumed by the residents [61], and unlike beebread, there may be less opportunity for microbial pre-digestion. Nevertheless, most pollen-storing species within the genus Bombus age their pollen to some degree before feeding it to their larvae [62,63]. In pollen-storing species, including B. impatiens and B. terrestris, bee-collected pollen is stored in specialized cells and periodically transferred to the developing larvae inside the brood chambers [63]. The microbe-rich environment of the storage cells suggests that these external symbionts may be involved in some way with larval nutrition. Indeed, empirical estimates reveal that pollen-borne microbes are a significant source of dietary proteins for bumble bees, their contributions often exceeding that of plant-based substrates [55]. Furthermore, direct evidence shows that exposure to fungicides can cause adverse shifts in the microbial community associated with bumble bee pollen provisions and can lead to severe mortality [45,64]. Taken together, growing evidence suggests that the sustained partnership between bumble bees and their external symbionts is integral to colony fitness.
In this study, we explored the bacterial and fungal diversity within the pollen provisions of commercially available bumble bee hives. Commercial hives were purchased from two supplier sources located within different bioregions of North America and reared indoors under semi-sterile conditions. The microbiome of pollen provisions was characterized using Illumina sequencing of the bacterial 16S rRNA and fungal ITS genes. The specific aims of this study were to: (1) characterize and compare the taxonomic diversity of bacteria and fungi within the pollen provisions of bumble bee hives obtained from the two supplier sources, (2) identify the “core microbiome” within the pollen provisions, and (3) use publicly available data to compare the bacterial community of bumble bee pollen provisions with that of the bumble bee gut and other insect–microbe symbioses.
2. Materials and Methods
2.1. Rearing and Sample Collection
Bombus impatiens hives were purchased from Koppert Biological Systems (Howell, Michigan, USA) in April 2016 (Source K, replicate hives K1, K2, and K3), and Biobest (Leamington, Ontario, Canada) in June 2016 (Source B, replicate hives B1, B2, B3, and B4). Prior to shipment, colonies from both suppliers were raised in captivity at the rearing facility and fed a mixture of commercially obtained pollen and nectar. Once in the laboratory, all hives were acclimatized with trap doors closed within a biosafety cabinet for 7 d. Next, bees were anesthetized by placing them in a −20 °C freezer for 10 min, and the initial number of workers recorded. The mother queen was removed from the hive using sterilized forceps and weighed using a microbalance that was previously sanitized with 90% ethanol. Prior to feeding to the bees, commercially purchased honey bee-collected pollen was sterilized to minimize the chances of introducing any environmentally acquired microbes. Pollen was sterilized by freeze-drying for 72 h, (Labconco, Freezone 2.5+, Kansas City, MO, USA), followed by soaking in 90% ethanol, and drying overnight in a hood under UV light. Sterility was verified by the absence of microbial growth by plating a subsample of the sterilized pollen on general-purpose agar media and incubating at 28 °C for 48 h. Based on previously published values, approximately 4.27 g d−1 of sterilized pollen was individually weighed into sterile petri dishes and introduced into the hive through the trap door for 28 d using autoclaved powder funnels [65,66]. At the end of the study, hives were chilled by placing them in a −20 °C freezer for 20 min to record the final worker number and queen weight. Using standard aseptic technique, hives were dissected to exhume pollen provisions from the brood cells. Pollen provisions were stored in PCR collection tubes at −80 °C and transported on ice for DNA analysis.
2.2. DNA Isolation and Sequence Processing
One sample weighing approximately 0.3 g of pollen provision was collected from each hive, PCR-amplified, and sequenced at the UW Biotechnology Center. Briefly, DNA from pollen provisions was extracted using lysozyme solution, lysed, and then centrifuged at 10,000 G for 30 s at room temperature (Mo Bio Power Fecal® DNA isolation kit, Hilden, Germany) [45]. The supernatant was transferred to a collection tube and incubated with a protein precipitation solution. After adding an aqueous bind solution, the supernatant was transferred to a spin filter, centrifuged, and eluted in a clean collection tube. The isolated DNA was quantified and normalized to 2 ng μL−1 by fluorometric analysis. Reactions were prepared to analyze ITS (fungal), and 16S (bacterial) components of each pollen-provision sample [67]. A nested 2-step PCR protocol was developed using next-generation sequencing libraries targeting the 16S rRNA V3/V4 variable region and ITS/5.8s rRNA spacer region [68,69]. Region specific primers were modified to add sequencer-specific adapter overhang nucleotide sequences to the gene-specific sequences [70,71]. Following amplification, library size was verified and quantified by electrophoretic mobility, and cleaned using solid phase reversible immobilization beads. The 16S and ITS amplicons were pooled quantitatively to create a single amplicon pool for each sample. Sequencer specific adapters and sample specific dual indexes were added using the specific primers and PCR amplified [45]. The quality and quantity of the finished libraries was assessed using electrophoretic mobility, and fluorometry, respectively. All libraries were standardized and pooled prior to sequencing. All 16S rRNA and ITS sequence data were processed in Mothur v.1.38.0 [72]. After merging both PE libraries into single contigs, homopolymers, chimeric sequences, ambiguous sequences, and sequences larger than expected were removed. Unique filtered sequences from the 16S rRNA gene were aligned to the SILVA SSU database Version 123 [73]; the sequences from the ITS gene were aligned to the UNITE database [74]. A distance matrix between successfully aligned sequences was calculated for further OTU assignation using the average neighbor method used in Mothur. Finally, the abundance of each OTU was computed using the make.shared command with label = 0.03 (97% similarity). OTU tables for each gene were imported into R [75] for further statistical analysis (Supplementary Material, Table S1).
2.3. Community Analysis
Bacterial, fungal, and overall microbial community structure was visualized using nonmetric multidimensional scaling (NMDS) of the Bray-Curtis dissimilarity coefficient [76]. The generalized and variance-adjusted weighted Unifrac distances between each library were calculated using the R package GUNIFRAC [77], and the PermanovaG function of the same package was used to determine the differences in the fungal and bacterial communities between the two commercial sources. Shannon’s diversity and Pielou’s evenness metrics were computed and compared using the R package Vegan [78]. Microbial OTUs that showed > 50% prevalence and > 1% relative abundance across all replicates within each source were assigned as “core microbiota” [79]. The strength of association between the relative abundance of microbial taxa present in all hives was calculated by Pearson correlation analysis using the corrplot package in R [80]. The bacterial community of bumble bee pollen provisions reported in this study was compared to publicly available data from other insect–microbe symbioses that also amplified the V3/V4 regions of the 16S rRNA gene [34,81,82,83,84]. Clustering of bacterial communities from all selected studies was visualized by NMDS analysis based on the proportions table using the metaMDS function in the R package Vegan.
3. Results
Permutation multivariate analysis of variance PermanovaG indicated that the bacterial (F1,4 = 15.81, p = 0.035) and fungal (F1,4 = 5.54; p = 0.011) community of bumble bee pollen provisions varied significantly between supplier sources. The bacterial community of Source K had significantly lower diversity (H) (t5 = −6.24, p = 0.03) and species evenness (J) (t5 = −6.35, p < 0.01) compared to Source B. In contrast, the fungal community of Source B had significantly lower diversity (t5 = 2.87, p = 0.04) and species evenness (t5 = 2.86, p = 0.04) compared to Source K (Supplementary Material, Table S2). NMDS analysis revealed that the bacterial, fungal, and overall microbial communities clustered more closely in replicate hives from the same supplier source, compared to hives from different sources (Figure 1).
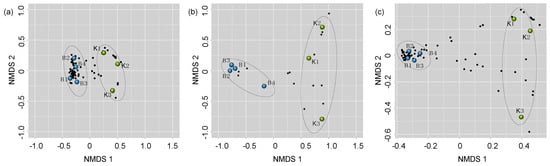
Figure 1.
Nonmetric multidimensional scaling of (a) bacterial, (b) fungal, and (c) overall microbial community compositions of pollen provisions from Source K (green circles), and Source B (blue circles). Points that are close together are more similar in diversity and abundance to one another than points that are far apart. Each black circle represents a unique microbial taxon.
Bacteria detected from pollen provisions included members of four phyla (Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria). Source K had higher abundance of Firmicutes, mainly belonging to the genus Lactobacillus. Actinobacteria included genera Streptomyces and Bifidobacterium, while there were minimal contributions from Proteobacteria and Bacteroidetes. In contrast, Source B had greater abundance of Proteobacteria (genera Ochrobactrum within Alphaproteobacteria, Comamonas and Delftia within Betaproteobacteria, and Stenotrophomonas and Pseudomonas within Gammaproteobacteria), and Bacteroidetes (Chryseobacterium and Sphingobacterium). While there was a lower abundance of Lactobacillus compared to Source K, Source B included other Firmicutes, including Enterococcus and an unclassified member within the Planococcaceae family (Figure 2a, Supplementary Material, Table S3).
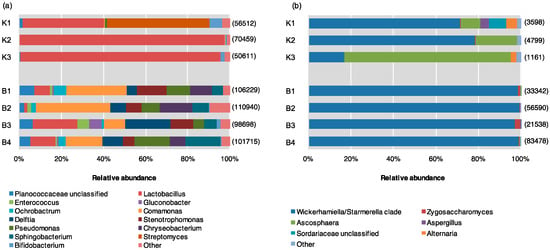
Figure 2.
Relative abundance of (a) bacterial and (b) fungal genera within pollen provisions of indoor–reared bumble bee colonies from Source K (N = 3), and Source B (N = 4). Sequences belonging to different microbial genera are shown in different colors. The total number of sequences isolated is displayed to the right of each library.
All sequences identified from ITS-based analyses belonged to the phylum Ascomycota, predominantly represented by classes Eurotiomycetes and Saccharomycetes (the only described class in the budding yeast subphylum Saccharomycotina). However, significant differences were noted at the genus rank between supplier sources; Source K had the greatest abundance from Saccharomycetes, dominated by yeasts of the Wickerhamiella/Starmerella (W/S) clade (subphylum Saccharomycotina) with minor contributions from Zygosaccharomyces. Source K hives also contained substantial amounts of members within genera Ascosphaera, including Ascosphaera apis, the causative agent of chalkbrood disease in honey bees. Minor contributions were also noted from Aspergillus, Alternaria, and an unclassified member of Sordariaceae. More than 99% of the fungal community of Source B hived consisted of members within Saccharomycetes, especially W/S clade yeasts, and Zygosaccharomyces at lower abundance. The remainder of the Source B fungal community included trivial contributions (<0.5%) from members within genus Ascosphaera and Aspergillus (Figure 2b).
Microbial OTUs that showed > 50% prevalence and > 1% relative abundance across all replicates within each source were assigned as “core microbiota” [79]. This cut off criterion was chosen based on previous work [79] and did not represent an established biological parameter. Based on this characterization, the core microbiota of Source K included two fungal and two bacterial taxa, whereas that of Source B consisted of one fungal and eight bacterial taxa. The shared core microbiota between the two sources consisted of only two groups, Lactobacillus and W/S clade yeasts (Figure 3). Pearson correlation analysis revealed significant associations between the relative abundance of multiple pairs of microbial taxa across all hives (Figure 4). Of these, the strongest positive correlations were detected between Enterococcaceae and Acetobacteraceae (r = 0.956, p < 0.001), Zygosaccharomyces and Acetobacteraceae (r = 0.959, p < 0.001), and Streptomycetaceae and Aspergillus (r = 0.995, p < 0.001). The strongest negative correlations were detected between W/S clade yeasts and Ascosphaera (r = −0.974, p < 0.001), and Lactobacillaceae and Comamonadaceae (r = −0.867, p < 0.01).
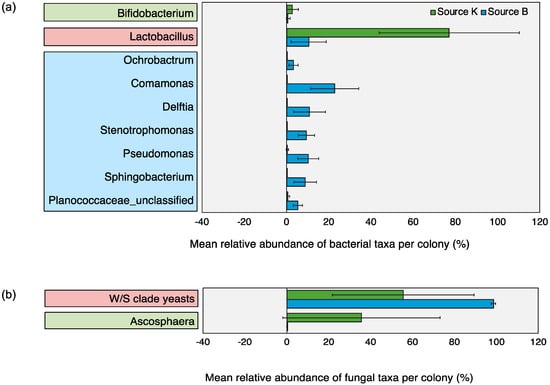
Figure 3.
Relative abundance of core (a) bacterial and (b) fungal taxa detected within pollen provisions of bumble bee hives from Source K (green bars) and Source B (blue bars) (Mean ± 1SD). Microbial OTUs that showed > 50% prevalence and >1% relative abundance across all replicates within each source were assigned as core microbiota (based on Graystock et al [79]). Core microbiota of Source K hives are enclosed within green boxes, that of Source B hives are enclosed within blue boxes, and the shared core microbiota across both sources are enclosed within red boxes.
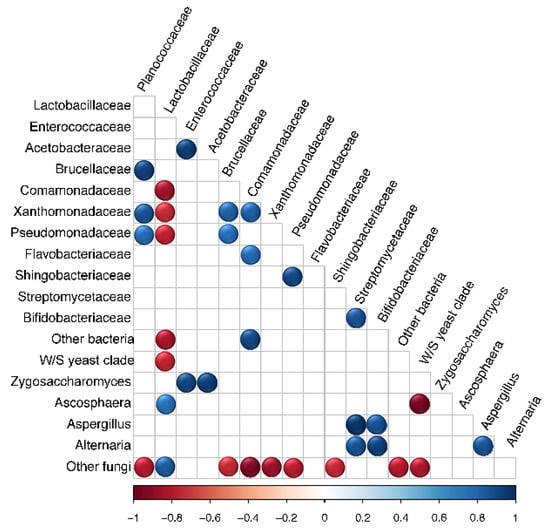
Figure 4.
Pearson correlation matrix showing the relative abundance of microbial taxa within bumble bee pollen provisions from both sources (N = 7). Significant positive and negative correlations are displayed in blue and red circles, respectively (p < 0.05). Color intensity is proportional to the correlation coefficients.
Hives from both sources showed a significant increase in the number of workers (Source K: t2 = −8.07, p = 0.02; Source B: t2 = −6.23, p < 0.01), although the percent increase was significantly higher in Source B compared to Source K (t5 = −3.26, p = 0.02). However, no statistical difference was noted between the initial and final weights of the mother queen for either source over time (Source K: t2 = 0.37, p = 0.75; Source B: t3 = −0.43, p = 0.69) (Supplementary Material, Table S4).
4. Discussion
Our findings revealed that the pollen stores of commercial bumble bee hives harbor several microbial taxa that are frequently associated with diverse bee species and their environment. Unlike the highly conserved bee gut microbiome, the taxonomic composition of bumble bee stored pollen varied significantly based on the commercial supplier sources. Both the bacterial and fungal communities were more similar among replicate hives obtained from the same source, compared to hives from different sources. Hives from both sources shared several members within phyla Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and Ascomycota. However, there were significant differences between the microbial community structure at lower ranks based on the commercial supplier. While Source K supported a significantly more diverse and even fungal community, Source B supported a significantly more diverse and even bacterial community. We speculate that the unique characteristics of each supplier, including the microbiome of the foundress queens and pollen diet used during captive breeding may have contributed to the differences between the microbial communities of bumble bee pollen stores.
Bumble bee colonies are founded by a single overwintered queen, and only the microbes colonizing the queen are expected to be transferred to the residents of the newly initiated colony. The single-queen generational inheritance can cause severe transmission bottlenecks, leading to a loss in microbial diversity and emergent function [30,85,86]. Commercial colonies, which have fewer opportunities to acquire microbes from their foraging environment, may be more vulnerable to such losses in microbial diversity. Indeed, past work has shown that commercial bumble bee hives, harbor a subset of the microbes associated with wild-caught colonies [33,34], and that subtle shifts in their gut microbiome may cause increased disease susceptibility among domesticated lineages [30]. Although there is limited evidence at this time, it is possible that captive-bred colonies experience similar alterations to the microbial community of their pollen stores, and that the loss of key symbiotic taxa could contribute to the increased pathogen loads observed within commercial bumble bee hives.
The microbial OTUs that showed > 50% prevalence and > 1% relative abundance across all replicate hives within each source were assigned as “core” groups. This cut off criterion was chosen based on previous work [79] and did not represent an established biological parameter. Based on this criterion, the shared core microbiota of the two commercial sources was restricted to only two putative pollen fermenters, Lactobacillus and yeasts of the Wickerhamiella/Starmerella (W/S) clade. Past research indicates that both groups perform unique beneficial functions for their hosts, including fermentation and long-term preservation of hive-stored pollen across managed and wild bee populations [24,44,49,50,57,76,87,88,89]. Lactobacilli are ubiquitously associated with multiple genera of bees, spanning a variety of locations, environmental conditions, and genetic traits [23,79,90]. Prior investigations into the gut and pollen microbiome of corbiculate bees indicate that Lactobacillus play a central role in bee metabolism, nutrition, and immune function, suggesting a similar functionality within bumble bee pollen provisions as well. Members of the genus Lactobacillus within the honey bee gut and hive environment mediate carbohydrate metabolism [11], defend against pathogens [44], produce essential secondary metabolites [43], and prevent spoilage of hive-stored food [56]. Culture-based studies with small number of isolates have previously hinted at the close association of W/S clade yeasts within diverse bee taxa [48,91,92]. Past work in honey bees [47], bumble bees [93], and stingless bees [94] has speculated that fermentative yeasts, including those belonging to the W/S clade [95], play an important role in bee nutrition. Our present study is the first to confirm and extend these findings using culture-independent sequencing techniques.
4.1. Bacterial Community
Hives from both sources harbored bacterial taxa that were consistent with known bee- and pollen-associated groups (e.g., Streptomycetaceae, Bifidobacteriaceae, Bacillales, Flavobacteriaceae, and Acetobacteraceae) [12,22,24,32]. Many of these groups are specific to beebread, and aid in maintaining general hygiene, inhibit parasites and pathogens, and prevent spoilage of stored pollen [34,52,76]. In addition to Lactobacillus, the core bacterial community of Source K included Bifidobacterium, which is speculated to be involved with pollen digestion in honey bees [22]. One library within Source K included a high abundance of Streptomyces, which has been previously detected in pollen stores of honey bees [76] and stingless bees [96], and is known to defend against pathogens including Paenibacillus larvae and Melisococcus plutonius that cause American foulbrood and European foulbrood disease, respectively [97,98]. The bacterial community of Source B was significantly more diverse compared to Source K, and contained higher abundances of Betaproteobacteria, Gammaproteobacteria, and Bacteroides within the Flavobacteriaceae and Sphingobacteriaceae family, which have been previously detected in beebread [43,88]. Source B hives also included Alphaproteobacteria belonging to the Acetobacteraceae family, a osmotolerant and acid resistant bacteria that serves as a source of inoculum for the early instar larval gut of honey bees [99]. The communities of pollen-associated bacteria identified in our study were distinct from those detected from other insect–microbe symbioses. This could suggest a high phylogenetic specificity of the bacterial community within bumble bee pollen provisions (Figure 5). Furthermore, the bacterial community of stored pollen from both commercial sources were more similar to each other than they were to the bumble bee gut microbiome [11,34]. This divergence between the microbiome of bumble bee gut and pollen provision could suggest that these two microhabitats support distinct, and well-adapted communities that likely perform varying roles within their respective environments [22,23,25,43,76].
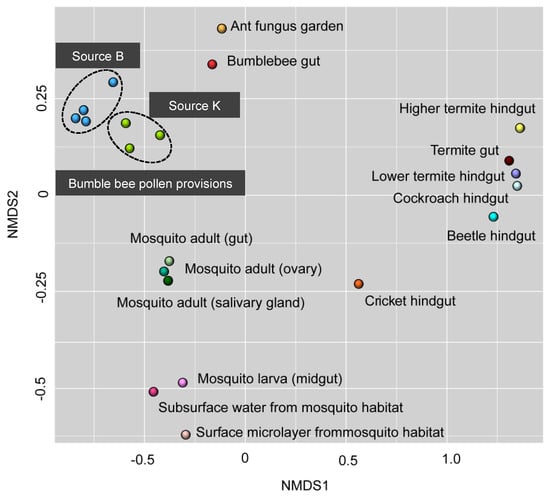
Figure 5.
Comparative analysis of nonmetric multidimensional scaling using 16S sequencing data obtained from past studies of insect–microbe symbiosis. Points that are close together are more similar in diversity and abundance to one another than points that are far apart. Each symbol represents the bacterial community detected from a unique insect–microbe symbiosis. See text for references.
4.2. Fungal Community
The core fungal microbiota within pollen provisions from both sources were dominated by W/S clade yeasts. Budding yeasts are ubiquitous within pollen provisions consumed by healthy larvae of social and solitary bees, and appear to be vital for bee nutrition [47,48,91]. All hives also contained other common bee-associated fungi, including representatives from Eurotiomycetes, Sordariomycetes, and Dothideomycetes at lower abundances [45,48,93]. However, there were significant differences in the fungal communities between commercial sources. Hives from Source K showed a high relative abundance of Ascosphaeraceae, including the honey bee pathogen, Ascosphaera apis, which causes symptoms similar to that of chalkbrood disease in bumble bees [100] (Supplementary Material, Table S5). Although none of the Source K hives showed signs of chalkbrood, it is interesting to note that they had a significantly slower growth rate compared to Source B hives. Since previous work has detected chalkbrood disease within living, asymptomatic larval honey bees [101,102], it is possible that the decreased colony growth rate of Source K hives was indicative of a subclinical A. apis infection.
In contrast to Source K, the fungal community of Source B hives had a higher abundance of other non-pathogenic fungal taxa, with W/S clade yeasts contributing to over 95% of the overall community. Past research in honey bees indicates that beebread from chalkbrood-resistant colonies have a higher abundance of non-pathogenic yeasts and molds, many of which produce inhibitory antimycotics against A. apis [103]. Our data from pairwise comparisons of microbial abundance within bumble bee pollen provisions was consistent with this earlier finding. Across all significant associations identified in this study, the strongest negative correlation was observed between the relative abundance of non-pathogenic W/S clade yeasts and Ascosphaera sp. (Figure 4). Although W/S clade yeasts may not directly contribute to increased resistance to chalkbrood disease, it is possible that their interactions with other microbial taxa can indirectly affect the abundance of A. apis [103]. Members within the genus Bacillus, Bifidobacterium, and Streptomyces present within the gut and hive environments of social bees produce antibiotics, antifungals and secondary metabolites, which prevent fungal infections in bees [16,17,24,76,104,105]. Based on these past studies, it is possible that greater diversity in the overall and core bacterial community could explain the lower loads of pathogenic Ascosphaera observed within Source B hives.
4.3. Implications for Bumble Bee Health
There are several documented cases of pathogen transmission from honey bees to bumble bees in Europe and North America [106,107,108,109,110,111]. The honey bee pathogen A. apis has been detected within the colonies of many captive-reared bumble bees species, including B. griseocollis, B. nevadensis, B. vosnesenskii, and B. terrestris [112]. A recent study reveals that infected colonies of B. terrestris showed symptoms similar to that of chalkbrood disease in honey bees [100]. However, this study is the first to report the appearance of A. apis within the hive-stored pollen of commercially reared B. impatiens, which adds to the growing concern about pathogen-driven bumble bee decline [40]. Our study shows that while all hives were reared under similar laboratory conditions using sterilized pollen diets, the abundance of A. apis was substantially higher in Source K hives. On average, A. apis contributed to nearly 30% of the fungal community in hives obtained from Source K, compared to 0.1% in those from Source B. This suggests that A. apis contamination could likely be linked to the supplier of Source K hives, and that these hives may have been exposed to the pathogen at the rearing facility during captive breeding, prior to commercial distribution.
One potential route of infection among commercially reared bumble bees is the use of infected foundress queens. Captive colonies are often founded by wild-caught queen bees that may have been exposed to various pathogens within their natural environment before capture [113,114,115,116]. When such infected queens are used for commercial rearing, there is a high risk of pathogen transmission into the newly initiated colonies [117]. Another potential route of exposure is the use of infected honey bee-collected pollen as diet for the developing colonies reared in captivity [118]. Contaminated honey bee pollen, which often carries bumble bee pathogens, has been reported to cause widespread infections at commercial rearing facilities and can lead to rapid declines in bumble bee populations [112,118,119,120,121]. The risk of pathogen spillover from the release of such infected colonies to native bee populations poses a significant threat to the health of wild bees [35,38,41,118,120,122,123].
Colony initiation in bumble bees occurs anew every year through a single overwintered founding queen, whereas honey bee colonies are formed by large interacting groups of overwintering bees. It is hypothesized that while trophallaxis and other social interactions promote recurrent gut colonization among honey bees [124], the inheritance of microbes through a single founding queen contributes to the high heterogeneity of the bumble bee gut microbiome [32,125]. Each generation of bumble bees can acquire a unique microbiome from the new founding queen, and these colony-level differences can be further amplified during the extreme isolation experienced within artificial rearing facilities. Since foragers of captive-bred colonies have less opportunity to acquire microbes from the environment, the microbial community within the hives used in this study likely differed from that of wild populations. Given that the microbial communities varied significantly between supplier sources, it is possible that our results could have stemmed from the inheritance of unique microbiota specific to the founding queens, and/or microbes within distinct honey bee-collected pollen diets used at the two rearing facilities. Indeed, the commercial suppliers used in this study were located in different countries within North America and may be subject to different policies and regulations pertaining to local land management, agricultural practices, and pesticide regulations within their respective landscapes. Such unique characteristics of each breeding facility based on supplier location could be important in explaining the source-specific differences reported in our study. Another factor that may have influenced our results involves the controlled laboratory conditions of the experiment. The natural variability experienced by wild bees is likely to promote a high diversity of environmentally acquired microbes. In contrast, the hives used in this study were reared indoors under semi-sterile conditions and fed sterilized pollen, leaving minimal opportunity for the influx of environmental- and diet-derived microbes. This could have potentially led to an underestimation of the microbial diversity associated with the pollen provisions of natural bee populations.
5. Conclusions
Our findings indicated that while the microbial community of bumble bee pollen provisions were quite variable across commercial sources, the shared core microbiota across both suppliers was restricted to only two groups: Lactobacillus and W/S clade yeasts. Although the exact composition of the “healthy” microbiome remains unclear, the reshuffling of key taxa within the community of these external symbionts can have unpredictable impacts on commercial bee hives. Future investigations comparing pollen stores of captive and wild bumble bee colonies could reveal critical links between microbiome structure and colony fitness, and help establish a predictive framework to conserve bee populations.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/11/4/250/s1. Table S1: Unique OTUs, taxonomy, and sequence read number. Each row is a unique OTU, and each column is a pollen provision library. Table S2: Shannon’s diversity (H’) and Pielou’s evenness (J’) of bacterial and fungal community; Table S3: Distribution of sequencing reads of bacterial and fungal taxa at the family and order rank respectively; Table S4: Colony demography and mother queen weights of hives; Table S5: Top blast hit (Mothur) of genus Ascosphaera isolated in this study. All sequences are deposited in GenBank under accession number PRJNA414625.
Author Contributions
P.S.D. and S.A.S. conceived and performed the experiment and wrote the manuscript; P.S.D. and L.D.-G. analyzed the data; M.A.B.H., C.R.C., C.T.H., and J.Z. provided technical assistance and contributed to the manuscript. All authors approved the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by USDA-ARS appropriated funds (CRIS #3655-21220-001, awarded to SAS and JZ), Wisconsin Department of Agriculture, Trade, and Consumer Protection (#197199, awarded to SAS), NSF (DEB-1442148, awarded to CTH), the DOE Great Lakes Bioenergy Research Center (BER DE-FC02-07ER64494 and DE-SC0018409, awarded to CRC and CTH), and the USDA NIFA (Hatch projects 1003258 and 1020204, awarded to CTH). CTH is a Pew Scholar in the Biomedical Sciences and H. I. Romnes Faculty Fellow, supported by the Pew Charitable Trusts and Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation (WARF), respectively.
Acknowledgments
The author(s) thank the UW-Biotechnology Center DNA Sequencing Facility for providing amplification and sequencing services; Caitlin Carlson, Jennifer Knack, and Jake Otto for assistance with molecular analysis.
Conflicts of Interest
The authors declare that they have no potential conflict of interest in relation to the study in this paper.
References
- Hammer, T.J.; Sanders, J.G.; Fierer, N. Not all animals need a microbiome. FEMS Microbiol. Lett. 2019, 366. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.M.; Henry, L.M.; Cornwallis, C.K.; Kiers, E.T.; West, S.A. The evolution of host-symbiont dependence. Nat. Commun. 2017, 8, 15973. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Kwong, W.K.; McFrederick, Q.; Anderson, K.E.; Barribeau, S.M.; Chandler, J.A.; Cornman, R.S.; Dainat, J.; De Miranda, J.R.; Doublet, V.; et al. The bee microbiome: Impact on bee health and model for evolution and ecology of host-microbe interactions. MBio 2016, 7, e02164-15. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Raaijmakers, J.M. Cross-kingdom similarities in microbiome functions. ISME J. 2015, 9, 1905–1907. [Google Scholar] [CrossRef]
- Moran, N.A.; Plague, G.R.; Sandström, J.P.; Wilcox, J.L. A genomic perspective on nutrient provisioning by bacterial symbionts of insects. Proc. Natl. Acad. Sci. USA 2003, 14543–14548. [Google Scholar] [CrossRef]
- Kleiner, M.; Wentrup, C.; Lott, C.; Teeling, H.; Wetzel, S.; Young, J.; Chang, Y.J.; Shah, M.; VerBerkmoes, N.C.; Zarzycki, J.; et al. Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use. Proc. Natl. Acad. Sci. USA 2012, 109, E1173–E1182. [Google Scholar] [CrossRef]
- Bang, C.; Dagan, T.; Deines, P.; Dubilier, N.; Duschl, W.J.; Fraune, S.; Hentschel, U.; Hirt, H.; Hülter, N.; Lachnit, T.; et al. Metaorganisms in extreme environments: Do microbes play a role in organismal adaptation? Zoology 2018, 127, 1–19. [Google Scholar] [CrossRef]
- Gibson, C.M.; Hunter, M.S.; Cara, M.G.; Martha, S.H. Extraordinarily widespread and fantastically complex: Comparative biology of endosymbiotic bacterial and fungal mutualists of insects. Ecol. Lett. 2009, 13, 223–234. [Google Scholar] [CrossRef]
- Sachs, J.L.; Skophammer, R.G.; Regus, J.U. Evolutionary transitions in bacterial symbiosis. Proc. Natl. Acad. Sci. USA 2011, 10800–10807. [Google Scholar] [CrossRef]
- Cardinal, S.; Danforth, B.N. Bees diversified in the age of eudicots. Proc. R. Soc. B Biol. Sci. 2013, 280, 20122686. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Abrol, D.P.; Li, J.; Schmid-Hempel, P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol. Ecol. 2013, 27, 2028–2044. [Google Scholar] [CrossRef]
- Koch, H.; Schmid-Hempel, P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. USA 2011, 108, 19288–19292. [Google Scholar] [CrossRef] [PubMed]
- Cariveau, D.P.; Elijah Powell, J.; Koch, H.; Winfree, R.; Moran, N.A. Variation in gut microbial communities and its association with pathogen infection in wild bumble bees (Bombus). ISME J. 2014, 8, 2369–2379. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Cisarovsky, G.; Schmid-Hempel, P. Ecological effects on gut bacterial communities in wild bumblebee colonies. J. Anim. Ecol. 2012, 81, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Vásquez, A.; Forsgren, E.; Fries, I.; Paxton, R.J.; Flaberg, E.; Szekely, L.; Olofsson, T.C. Symbionts as major modulators of insect health: Lactic acid bacteria and honeybees. PLoS ONE 2012, 7, e33188. [Google Scholar]
- Killer, J.; Dubná, S.; Sedláček, I.; Švec, P. Lactobacillus apis sp. nov., from the stomach of honeybees (Apis mellifera), having an in vitro inhibitory effect on the causative agents of American and European foulbrood. Int. J. Syst. Evol. Microbiol. 2014, 64, 152–157. [Google Scholar] [CrossRef]
- Palmer-Young, E.C.; Raffel, T.R.; McFrederick, Q.S. pH-mediated inhibition of a bumble bee parasite by an intestinal symbiont. Parasitology 2018, 146, 380–388. [Google Scholar] [CrossRef]
- Ribière, C.; Hegarty, C.; Stephenson, H.; Whelan, P.; O’Toole, P.W. Gut and Whole-Body Microbiota of the Honey Bee Separate Thriving and Non-thriving Hives. Microb. Ecol. 2019, 78, 195–205. [Google Scholar] [CrossRef]
- Shade, A.; Handelsman, J. Beyond the Venn diagram: The hunt for a core microbiome. Environ. Microbiol. 2012, 14, 4–12. [Google Scholar] [CrossRef]
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Martinson, V.G.; Danforth, B.N.; Minckley, R.L.; Rueppell, O.; Tingek, S.; Moran, N.A.; TIingek, S.; Moran, N.A. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 2011, 20, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A.; Hansen, A.K.; Powell, J.E.; Sabree, Z.L. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE 2012, 7, e36393. [Google Scholar] [CrossRef]
- Corby-Harris, V.; Maes, P.; Anderson, K.E. The bacterial communities associated with honey bee (Apis mellifera) foragers. PLoS ONE 2014, 9, e95056. [Google Scholar] [CrossRef]
- Koch, H.; Schmid-Hempel, P. Bacterial communities in Central European bumblebees: Low diversity and high specificity. Microb. Ecol. 2011, 62, 121–133. [Google Scholar] [CrossRef]
- Li, J.; Powell, J.E.; Guo, J.; Evans, J.D.; Wu, J.; Williams, P.; Lin, Q.; Moran, N.A.; Zhang, Z. Two gut community enterotypes recur in diverse bumblebee species. Curr. Biol. 2015, 25, R652–R653. [Google Scholar] [CrossRef]
- Powell, E.; Ratnayeke, N.; Moran, N.A. Strain diversity and host specificity in a specialized gut symbiont of honeybees and bumblebees. Mol. Ecol. 2016, 25, 4461–4471. [Google Scholar] [CrossRef]
- Michener, C.D. The Social Behavior of the Bees: A Comparative Study; Harvard University Press: Cambridge, MA, USA, 1974; Volume 36. [Google Scholar]
- Leonhardt, S.D.; Blüthgen, N. The same, but different: Pollen foraging in honeybee and bumblebee colonies. Apidologie 2012, 43, 449–464. [Google Scholar] [CrossRef]
- Mockler, B.K.; Kwong, W.K.; Moran, N.A.; Koch, H. Microbiome structure influences infection by the parasite Crithidia bombi in bumble bees. Appl. Environ. Microbiol. 2018, 84, e02335-17. [Google Scholar] [CrossRef]
- Michener, C.D. The Bees of the World, 2nd ed.; Hopkins, J., Ed.; Academic Press: Baltimore, MD, USA, 2007. [Google Scholar]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.W.; Soh, E.J.Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic microbiome evolution in social bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef]
- Parmentier, L.; Meeus, I.; Mosallanejad, H.; de Graaf, D.C.; Smagghe, G. Plasticity in the gut microbial community and uptake of Enterobacteriaceae (Gammaproteobacteria) in Bombus terrestris bumblebees’ nests when reared indoors and moved to an outdoor environment. Apidologie 2016, 47, 237–250. [Google Scholar] [CrossRef]
- Meeus, I.; Parmentier, L.; Billiet, A.; Maebe, K.; Van Nieuwerburgh, F.; Deforce, D.; Wäckers, F.; Vandamme, P.; Smagghe, G. 16S rRNA amplicon sequencing demonstrates that indoor-reared bumblebees (Bombus terrestris) harbor a core subset of bacteria normally associated with the wild host. PLoS ONE 2015, 10, e0125152. [Google Scholar] [CrossRef] [PubMed]
- Graystock, P.; Yates, K.; Evison, S.E.F.; Darvill, B.; Goulson, D.; Hughes, W.O.H. The Trojan hives: Pollinator pathogens, imported and distributed in bumblebee colonies. J. Appl. Ecol. 2013, 50, 1207–1215. [Google Scholar] [CrossRef]
- Otterstatter, M.C.; Thomson, J.D. Does pathogen spillover from commercially reared bumble bees threaten wild pollinators? PLoS ONE 2008, 3, e2771. [Google Scholar] [CrossRef]
- Cameron, S.A.; Lim, H.C.; Lozier, J.D.; Duennes, M.A.; Thorp, R. Test of the invasive pathogen hypothesis of bumble bee decline in North America. Proc. Natl. Acad. Sci. USA 2016, 113, 4386–4391. [Google Scholar] [CrossRef]
- Sachman-Ruiz, B.; Narváez-Padilla, V.; Reynaud, E. Commercial Bombus impatiens as reservoirs of emerging infectious diseases in central México. Biol. Invasions 2015, 17, 2043–2053. [Google Scholar] [CrossRef]
- Meeus, I.; Brown, M.J.F.F.; De Graaf, D.C.; Smagghe, G. Effects of Invasive Parasites on Bumble Bee Declines. Conserv. Biol. 2011, 25, 662–671. [Google Scholar] [CrossRef]
- Cameron, S.A.; Lozier, J.D.; Strange, J.P.; Koch, J.B.; Cordes, N.; Solter, L.F.; Griswold, T.L.; Robinson, G.E. Patterns of widespread decline in North American bumble bees. Proc. Natl. Acad. Sci. USA 2011, 108, 662–667. [Google Scholar] [CrossRef]
- Szabo, N.D.; Colla, S.R.; Wagner, D.L.; Gall, L.F.; Kerr, J.T. Do pathogen spillover, pesticide use, or habitat loss explain recent North American bumblebee declines? Conserv. Lett. 2012, 5, 232–239. [Google Scholar] [CrossRef]
- Thorp, R.W.; Strickler, K.; Cane, J.H. Bumble bees (Hymenoptera: Apidae): Commercial use and environmental concerns. In Thomas Say Publications in Entomology; Proceedings: Lanham, MD, USA, 2003. [Google Scholar]
- Saraiva, M.A.; Zemolin, A.P.P.; Franco, J.L.; Boldo, J.T.; Stefenon, V.M.; Triplett, E.W.; de Oliveira Camargo, F.A.; Roesch, L.F.W. Relationship between honeybee nutrition and their microbial communities. Antonie van Leeuwenhoek 2015, 107, 921–933. [Google Scholar] [CrossRef]
- Vásquez, A.; Olofsson, T.C. The lactic acid bacteria involved in the production of bee pollen and bee bread. J. Apic. Res. 2009, 48, 189–195. [Google Scholar] [CrossRef]
- Steffan, S.A.; Dharampal, P.S.; Diaz-Garcia, L.A.; Currie, C.R.; Zalapa, J.E.; Hittinger, C.T. Empirical, metagenomic, and computational techniques illuminate the mechanisms by which fungicides compromise bee health. JoVE 2017, 54631, e54631. [Google Scholar] [CrossRef] [PubMed]
- Roulston, T.H.; Cane, J.H. Pollen nutritional content and digestibility for animals. Plant Syst. Evol. 2000, 222, 187–209. [Google Scholar] [CrossRef]
- Pain, J.A.; Mugenet, J. Biochemical and physiological research on pollen stored by bees. Ann. Abeille 1966, 9, 209–236. [Google Scholar] [CrossRef][Green Version]
- Gilliam, M. Microbiology of pollen and bee bread: The yeasts. Apidologie 1979, 10, 43–53. [Google Scholar] [CrossRef]
- Gilliam, M. Microbiology of pollen and bee bread: The genus Bacillus. Apidologie 1979, 10, 269–274. [Google Scholar] [CrossRef]
- Gilliam, M. Identification and roles of non-pathogenic microflora associated with honey bees. FEMS Microbiol. Lett. 1997, 155, 1–10. [Google Scholar] [CrossRef]
- Gilliam, M.; Prest, D.B.; Lorenz, B.J. Microbiology of pollen and bee bread: Taxonomy and enzymology of molds. Apidology 1989, 20, 53–68. [Google Scholar] [CrossRef]
- Gilliam, M.; Roubik, D.; Lorenz, B. Microorganisms associated with pollen, honey, and brood provisions in the nest of a stingless bee, Melipona fasciata. Apidologie 1990, 21, 89–97. [Google Scholar] [CrossRef]
- Nicolson, S.W.; Da Silva Das Neves, S.; Human, H.; Pirk, C.W.W. Digestibility and nutritional value of fresh and stored pollen for honey bees (Apis mellifera scutellata). J. Insect Physiol. 2018, 107, 302–308. [Google Scholar] [CrossRef]
- Degrandi-Hoffman, G.; Eckholm, B.; Huang, M. Methods for comparing nutrients in beebread made by Africanized and European honey bees and the effects on hemolymph protein titers. J. Vis. Exp. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Steffan, S.A.; Dharampal, P.S.; Danforth, B.N.; Gaines-Day, H.R.; Takizawa, Y.; Chikaraishi, Y. Omnivory in bees: Elevated trophic positions among all major bee families. Am. Nat. 2019, 194, 704281. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Carroll, M.J.; Sheehan, T.; Mott, B.M.; Maes, P.; Corby-Harris, V. Hive-stored pollen of honey bees: Many lines of evidence are consistent with pollen preservation, not nutrient conversion. Mol. Ecol. 2014, 23, 5904–5917. [Google Scholar] [CrossRef]
- Anderson, K.E.; Sheehan, T.H.; Eckholm, B.J.; Mott, B.M.; DeGrandi-Hoffman, G. An emerging paradigm of colony health: Microbial balance of the honey bee and hive (Apis mellifera). Insectes Sociaux 2011, 58, 431–444. [Google Scholar] [CrossRef]
- Carroll, M.J.; Brown, N.; Goodall, C.; Downs, A.M.; Sheenan, T.H.; Anderson, K.E. Honey bees preferentially consume freshlystored Pollen. PLoS ONE 2017, 12, e0175933. [Google Scholar] [CrossRef] [PubMed]
- Pettis, J.S.; Lichtenberg, E.M.; Andree, M.; Stitzinger, J.; Rose, R.; Vanengelsdorp, D. Crop pollination exposes honey bees to pesticides which alters their susceptibility to the gut pathogen Nosema ceranae. PLoS ONE 2013, 8, e70182. [Google Scholar] [CrossRef]
- Goulson, D. Bumblebees: Behaviour, ecology, and conservation; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Stabler, D.; Paoli, P.P.; Nicolson, S.W.; Wright, G.A. Nutrient balancing of the adult worker bumblebee (Bombus terrestris) depends on the dietary source of essential amino acids. J. Exp. Biol. 2015, 218, 793–802. [Google Scholar] [CrossRef]
- Goulson, D. Bumblebees: Their behaviour and ecology; Oxford University Press: Oxford, UK, 2003; ISBN 0198526075. [Google Scholar]
- Evans, E.; Burns, I.; Spivak, M. Befriending bumble bees: A practical guide to raising local bumble bees; University of Minnesota Extension Service: St. Paul, MN, USA, 2007. [Google Scholar]
- Bernauer, O.M.; Gaines-Day, H.R.; Steffan, S.A. Colonies of bumble bees (Bombus impatiens) produce fewer workers, less bee biomass, and have smaller mother queens following fungicide exposure. Insects 2015, 6, 478–488. [Google Scholar] [CrossRef]
- Goulson, D.; Peat, J.; Stout, J.C.; Tucker, J.; Darvill, B.; Derwent, L.C.; Hughes, W.O.H. Can alloethism in workers of the bumblebee, Bombus terrestris, be explained in terms of foraging efficiency? Anim. Behav. 2002, 64, 123–130. [Google Scholar] [CrossRef]
- Evans, E.C.; Spivak, M. Effects of honey Bee (Hymenoptera: Apidae) and bumble bee (Hymenoptera: Apidae) presence on cranberry (Ericales: Ericaceae) pollination. J. Econ. Entomol. 2006, 99, 614–620. [Google Scholar] [CrossRef]
- Scientific, T. ThermoFisher Scientific User Guide: Qubit dsDNA HS Assay Kits. Available online: https://tools.thermofisher.com/content/sfs/manuals/Qubit_dsDNA_HS_Assay_UG.pdf (accessed on 5 June 2017).
- Khadempour, L.; LeMay, V.; Jack, D.; Bohlmann, J.; Breuil, C. The Relative Abundance of Mountain Pine Beetle Fungal Associates Through the Beetle Life Cycle in Pine Trees. Microb. Ecol. 2012, 64, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Dorn-In, S.; Hölzel, C.S.; Janke, T.; Schwaiger, K.; Balsliemke, J.; Bauer, J. PCR-SSCP-based reconstruction of the original fungal flora of heat-processed meat products. Int. J. Food Microbiol. 2013, 162, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef] [PubMed]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. a Guid. to methods Appl. 1990, 18, 315–322. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.; Weber, C. Introducing mothur: Open source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef]
- Koljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2014, 22, 5271–5277. [Google Scholar] [CrossRef]
- R-Team Computers. R: A language and environment for statistical computing [Computer software]; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Anderson, K.E.; Sheehan, T.H.; Mott, B.M.; Maes, P.; Snyder, L.; Schwan, M.R.; Walton, A.; Jones, B.M.; Corby-Harris, V. Microbial ecology of the hive and pollination landscape: Bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS ONE 2013, 8, e83125. [Google Scholar] [CrossRef]
- Chen, J.; Bittinger, K.; Charlson, E.S.; Hoffmann, C.; Lewis, J.; Wu, G.D.; Collman, R.G.; Bushman, F.D.; Li, H. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 2012, 28, 2106–2113. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M. The vegan package. Community Ecol. Packag. 2007, 10, 631–637. [Google Scholar]
- Graystock, P.; Rehan, S.M.; McFrederick, Q.S. Hunting for healthy microbiomes: Determining the core microbiomes of Ceratina, Megalopta, and Apis bees and how they associate with microbes in bee collected pollen. Conserv. Genet. 2017, 18, 701–711. [Google Scholar] [CrossRef]
- An Introduction to corrplot package Introduction. Available online: https://cran.r-project.org/web/packages/corrplot/vignettes/corrplot-intro.html (accessed on 6 April 2020).
- Liberti, J.; Sapountzis, P.; Hansen, L.H.; Sørensen, S.J.; Adams, R.M.M.M.; Boomsma, J.J. Bacterial symbiont sharing in Megalomyrmex social parasites and their fungus-growing ant hosts. Mol. Ecol. 2015, 24, 3151–3169. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, C.; Köhler, T.; Brune, A. The cockroach origin of the termite gut microbiota: Patterns in bacterial community structure reflect major evolutionary events. Appl. Environ. Microbiol. 2014, 80, 2261–2269. [Google Scholar] [CrossRef] [PubMed]
- Otani, S.; Mikaelyan, A.; Nobre, T.; Hansen, L.H.; Koné, N.A.; Sørensen, S.J.; Aanen, D.K.; Boomsma, J.J.; Brune, A.; Poulsen, M. Identifying the core microbial community in the gut of fungus-growing termites. Mol. Ecol. 2014, 23, 4631–4644. [Google Scholar] [CrossRef] [PubMed]
- Gimonneau, G.; Tchioffo, M.T.; Abate, L.; Boissière, A.; Awono-Ambéné, P.H.; Nsango, S.E.; Christen, R.; Morlais, I. Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infect. Genet. Evol. 2014, 28, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Engel, P.; Koch, H.; Moran, N.A. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc. Natl. Acad. Sci. USA 2014, 111, 11509–11514. [Google Scholar] [CrossRef] [PubMed]
- Ellegaard, K.M.; Brochet, S.; Bonilla-Rosso, G.; Emery, O.; Glover, N.; Hadadi, N.; Jaron, K.S.; van der Meer, J.R.; Robinson-Rechavi, M.; Sentchilo, V.; et al. Genomic changes underlying host specialization in the bee gut symbiont Lactobacillus Firm5. Mol. Ecol. 2019, 28, 2224–2237. [Google Scholar] [CrossRef]
- Voulgari-Kokota, A.; Grimmer, G.; Steffan-Dewenter, I.; Keller, A. Bacterial community structure and succession in nests of two megachilid bee genera. FEMS Microbiol. Ecol. 2018, 95, fiy218. [Google Scholar] [CrossRef]
- Donkersley, P.; Rhodes, G.; Pickup, R.W.; Jones, K.C.; Wilson, K. Bacterial communities associated with honeybee food stores are correlated with land use. Ecol. Evol. 2018, 8, 4743–4756. [Google Scholar] [CrossRef]
- Gilliam, M.; Buchmann, S.L.S.; Lorenz, B.B.J. Microbial flora of the larval provisions of the solitary bees, Centris pallida and Anthophora sp. Apidologie 1984, 15, 1–10. [Google Scholar] [CrossRef]
- Ahn, J.H.; Hong, I.P.; Bok, J.I.; Kim, B.Y.; Song, J.; Weon, H.Y. Pyrosequencing analysis of the bacterial communities in the guts of honey bees Apis cerana and Apis mellifera in Korea. J. Microbiol. 2012, 50, 735–745. [Google Scholar] [CrossRef]
- Rosa, C.A.; Lachance, M.A.; Silva, J.O.C.; Teixeira, A.C.P.; Marini, M.M.; Antonini, Y.; Martins, R.P. Yeast communities associated with stingless bees. FEMS Yeast Res. 2003, 4, 271–275. [Google Scholar] [CrossRef]
- Lachance, M.A.; Starmer, W.T.; Rosa, C.A.; Bowles, J.M.; Barker, J.S.S.; Janzen, D.H. Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res. 2001, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Brysch-Herzberg, M. Ecology of yeasts in plant-bumblebee mutualism in Central Europe. FEMS Microbiol. Ecol. 2004, 50, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.C.P.; Marini, M.M.; Nicoli, J.R.; Antonini, Y.; Martins, R.P.; Lachance, M.A.; Rosa, C.A. Starmerella meliponinorum sp. nov., a novel ascomycetous yeast species associated with stingless bees. Int. J. Syst. Evol. Microbiol. 2003, 53, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Brysch-Herzberg, M.; Lachance, M.A. Candida bombiphila sp. nov., a new asexual yeast species in the Wickerhamiella clade. Int. J. Syst. Evol. Microbiol. 2004, 54, 1857–1859. [Google Scholar] [CrossRef] [PubMed]
- Cambronero-Heinrichs, J.C.; Matarrita-Carranza, B.; Murillo-Cruz, C.; Araya-Valverde, E.; Chavarría, M.; Pinto-Tomás, A.A. Phylogenetic analyses of antibiotic-producing Streptomyces sp. isolates obtained from the stingless-bee Tetragonisca angustula (Apidae: Meliponini). Microbiology (United Kingdom) 2019, 165, 292–301. [Google Scholar] [CrossRef]
- Promnuan, Y.; Kudo, T.; Chantawannakul, P. Actinomycetes isolated from beehives in Thailand. World J. Microbiol. Biotechnol. 2009. [Google Scholar] [CrossRef]
- Kim, D.R.; Cho, G.; Jeon, C.W.; Weller, D.M.; Thomashow, L.S.; Paulitz, T.C.; Kwak, Y.S. A mutualistic interaction between Streptomyces bacteria, strawberry plants and pollinating bees. Nat. Commun. 2019, 10, 4802. [Google Scholar] [CrossRef]
- Vojvodic, S.; Rehan, S.M.; Anderson, K.E. Microbial gut diversity of Africanized and European honey bee larval instars. PLoS ONE 2013, 8, e72106. [Google Scholar] [CrossRef]
- de Sousa Pereira, K.; Meeus, I.; Smagghe, G. Honey bee-collected pollen is a potential source of Ascosphaera apis infection in managed bumble bees. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Bailón, E.; Higes, M.; Martínez-Salvador, A.; Antúnez, K.; Botías, C.; Meana, A.; Prieto, L.; Martín-Hernández, R. The prevalence of the honeybee brood pathogens Ascosphaera apis, Paenibacillus larvae and Melissococcus plutonius in Spanish apiaries determined with a new multiplex PCR assay. Microb. Biotechnol. 2013, 6, 731–739. [Google Scholar] [PubMed]
- James, R.R.; Skinner, J.S. PCR diagnostic methods for Ascosphaera infections in bees. J. Invertebr. Pathol. 2005, 90, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Gilliam, M.; Taber, S.; Lorenz, B.J.; Prest, D.B. Factors affecting development of chalkbrood disease in colonies of honey bees, Apis mellifera, fed pollen contaminated with Ascosphaera apis. J. Invertebr. Pathol. 1988, 52, 314–325. [Google Scholar] [CrossRef]
- Reynaldi, F.J.; De Giusti, M.R.; Alippi, A.M. Inhibition of the growth of Ascosphaera apis by Bacillus and Paenibacillus strains isolated from honey. Rev. Argent. Microbiol. 2004, 36, 52–55. [Google Scholar]
- Sabaté, D.C.; Carrillo, L.; Carina Audisio, M.; Audisio, M.C. Inhibition of Paenibacillus larvae and Ascosphaera apis by Bacillus subtilis isolated from honeybee gut and honey samples. Res. Microbiol. 2009, 160, 193–199. [Google Scholar] [CrossRef]
- Fürst, M.A.; McMahon, D.P.; Osborne, J.L.; Paxton, R.J.; Brown, M.J.F. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 2014, 506, 364–366. [Google Scholar] [CrossRef]
- Meeus, I.; de Miranda, J.R.; de Graaf, D.C.; Wäckers, F.; Smagghe, G. Effect of oral infection with Kashmir bee virus and Israeli acute paralysis virus on bumblebee (Bombus terrestris) reproductive success. J. Invertebr. Pathol. 2014, 121, 64–69. [Google Scholar] [CrossRef]
- Graystock, P.; Yates, K.; Darvill, B.; Goulson, D.; Hughes, W.O.H. Emerging dangers: Deadly effects of an emergent parasite in a new pollinator host. J. Invertebr. Pathol. 2013, 114, 114–119. [Google Scholar] [CrossRef]
- Evison, S.E.F.; Roberts, K.E.; Laurenson, L.; Pietravalle, S.; Hui, J.; Biesmeijer, J.C.; Smith, J.E.; Budge, G.; Hughes, W.O.H. Pervasiveness of parasites in pollinators. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Manley, R.; Boots, M.; Wilfert, L. Emerging viral disease risk to pollinating insects: Ecological, evolutionary and anthropogenic factors. J. Appl. Ecol. 2015, 52, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Mcmahon, D.P.; Fürst, M.A.; Caspar, J.; Theodorou, P.; Brown, M.J.F.; Paxton, R.J. A sting in the spit: Widespread cross-infection of multiple RNA viruses across wild and managed bees. J. Anim. Ecol. 2015, 84, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Maxfield-Taylor, S.A.; Mujic, A.B.; Rao, S. First detection of the larval chalkbrood disease pathogen Ascosphaera apis (Ascomycota: Eurotiomycetes: Ascosphaerales) in adult bumble bees. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.M.; Brown, M.J.F. Parasites and genetic diversity in an invasive bumblebee. J. Anim. Ecol. 2014, 83, 1428–1440. [Google Scholar] [CrossRef]
- Colgan, T.J.; Carolan, J.C.; Sumner, S.; Blaxter, M.L.; Brown, M.J.F. Infection by the castrating parasitic nematode Sphaerularia bombi changes gene expression in Bombus terrestris bumblebee queens. Insect Mol. Biol. 2019, 29, 170–182. [Google Scholar] [CrossRef]
- Çankaya, N.E.; Kaftanoglu, O. An investigation on some diseases and parasites of bumblebee queens (Bombus terrestris L.) in Turkey. Pakistan J. Biol. Sci. 2006, 9, 1282–1286. [Google Scholar]
- Mullins, J.L.; Strange, J.P.; Tripodi, A.D. Why Are Queens Broodless? Failed Nest Initiation Not Linked to Parasites, Mating Status, or Ovary Development in Two Bumble Bee Species of Pyrobombus (Hymenoptera: Apidae: Bombus). J. Econ. Entomol. 2019, 113, 575–581. [Google Scholar] [CrossRef]
- Brown, M.J.F.; Schmid-Hempel, R.; Schmid-Hempel, P. Strong context-dependent virulence in a host-parasite system: Reconciling genetic evidence with theory. J. Anim. Ecol. 2003, 72, 994–1002. [Google Scholar] [CrossRef]
- Goulson, D.; Hughes, W.O.H. Mitigating the anthropogenic spread of bee parasites to protect wild pollinators. Biol. Conserv. 2015, 191, 10–19. [Google Scholar] [CrossRef]
- Singh, R.; Levitt, A.L.; Rajotte, E.G.; Holmes, E.C.; Ostiguy, N.; Vanengelsdorp, D.; Lipkin, W.I.; Depamphilis, C.W.; Toth, A.L.; Cox-Foster, D.L. RNA viruses in hymenopteran pollinators: Evidence of inter-taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS ONE 2010, 5, e14357. [Google Scholar] [CrossRef]
- Graystock, P.; Jones, J.C.; Pamminger, T.; Parkinson, J.F.; Norman, V.; Blane, E.J.; Rothstein, L.; Wäckers, F.; Goulson, D.; Hughes, W.O.H. Hygienic food to reduce pathogen risk to bumblebees. J. Invertebr. Pathol. 2016, 136, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Stephen, W.P. Bombus (Bombus) occidentalis (Hymenoptera: Apiformes): In decline or recovery. Pan-Pac. Entomol. 2007, 83, 360–362. [Google Scholar] [CrossRef]
- Murray, T.E.; Coffey, M.M.F.; Kehoe, E.; Horgan, F.G.F. Pathogen prevalence in commercially reared bumble bees and evidence of spillover in conspecific populations. Biol. Conserv. 2013, 159, 269–276. [Google Scholar] [CrossRef]
- Arbetman, M.P.; Meeus, I.; Morales, C.L.; Aizen, M.A.; Smagghe, G. Alien parasite hitchhikes to Patagonia on invasive bumblebee. Biol. Invasions 2013, 15, 489–494. [Google Scholar] [CrossRef]
- Engel, P.; Martinson, V.G.; Moran, N.A. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. USA 2012, 109, 11002–11007. [Google Scholar] [CrossRef]
- Koch, H.; Schmid-Hempel, P. Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol. Lett. 2012, 15, 1095–1103. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).