Gene Flow Patterns among Aedes aegypti (Diptera: Culicidae) Populations in Sri Lanka
Abstract
1. Introduction
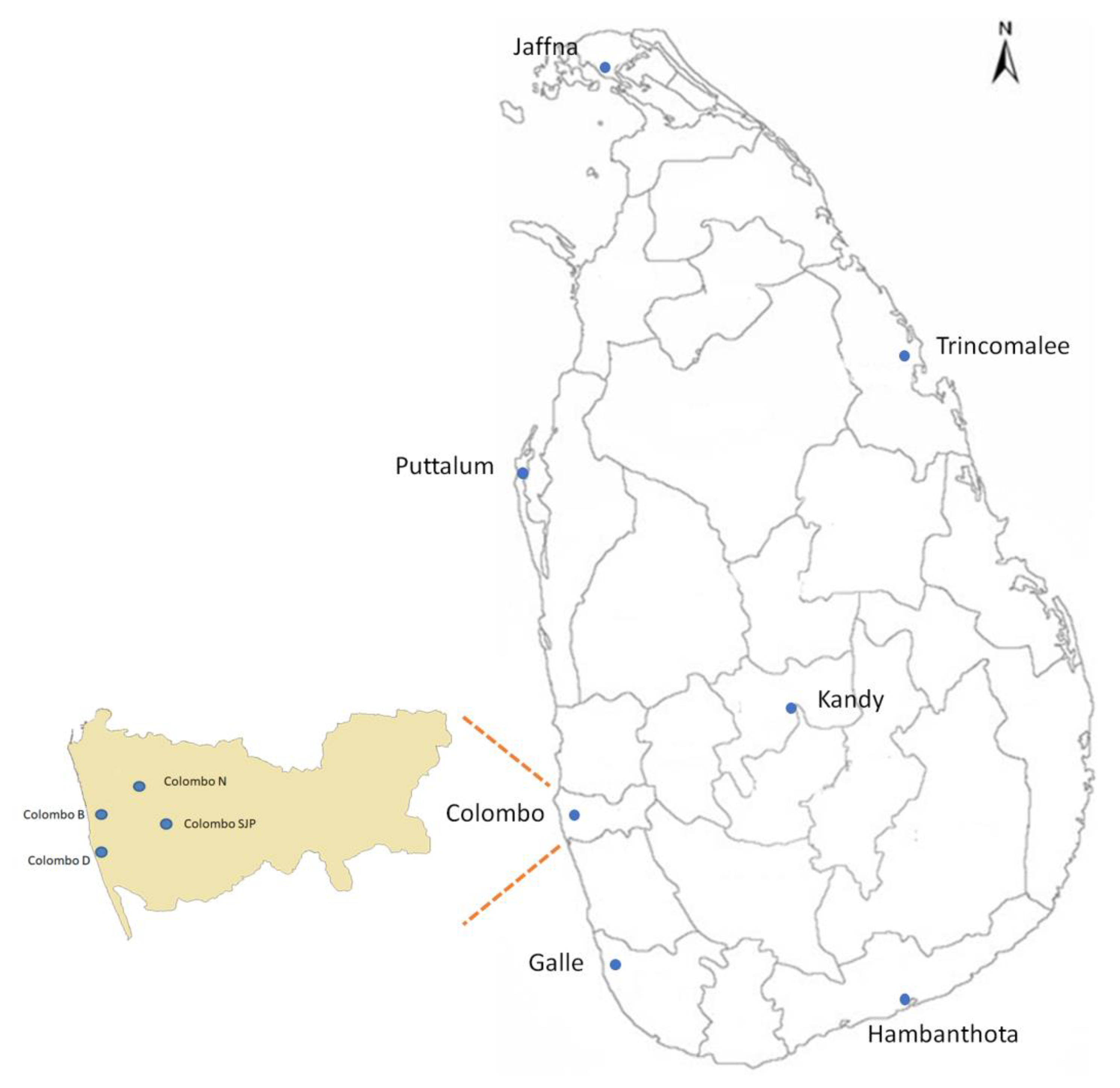
2. Materials and Methods
2.1. Mosquito Collection and Genomic DNA Isolation
2.2. Microsatellite Genotyping
2.3. Population Structure Analyses
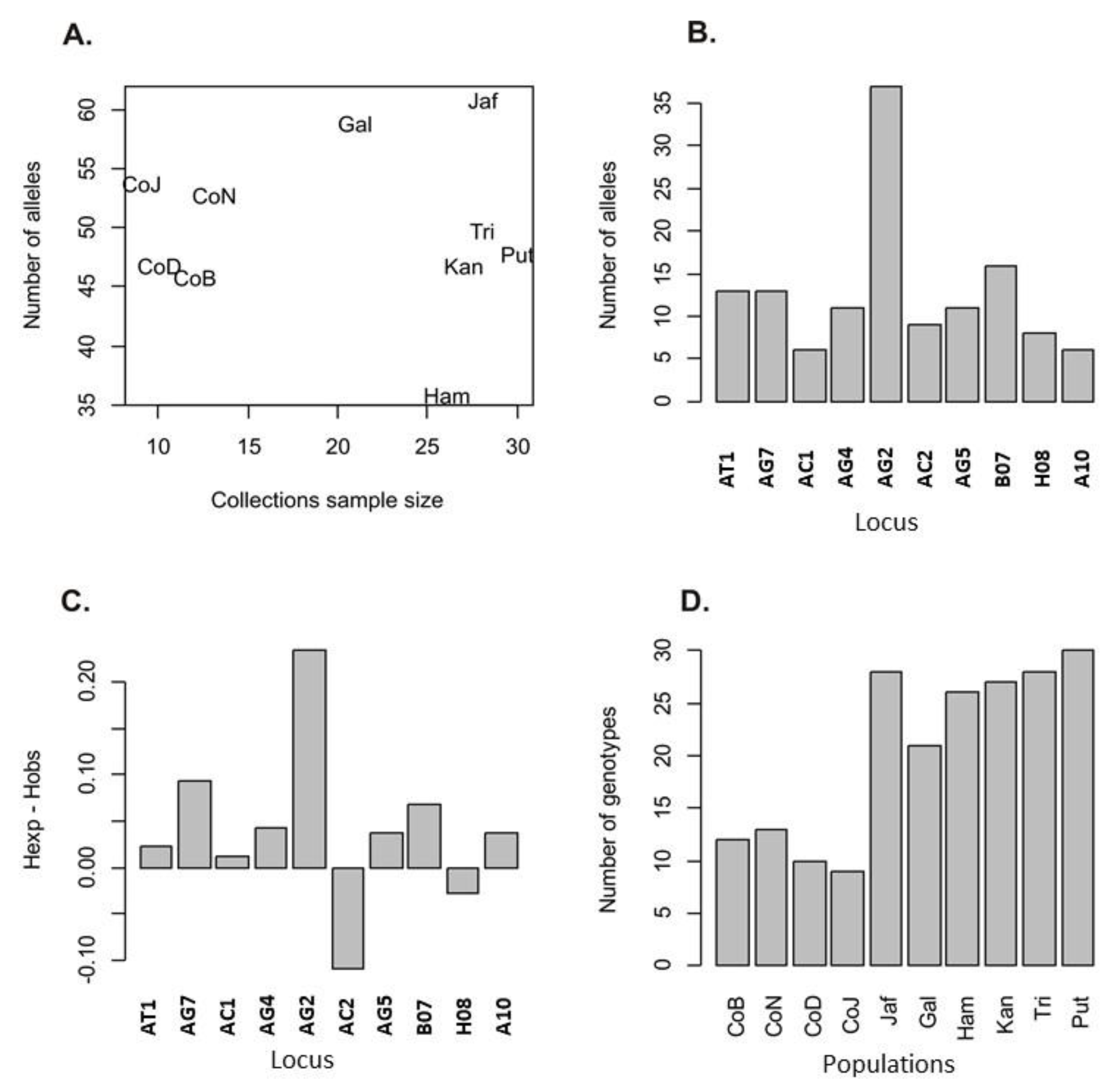
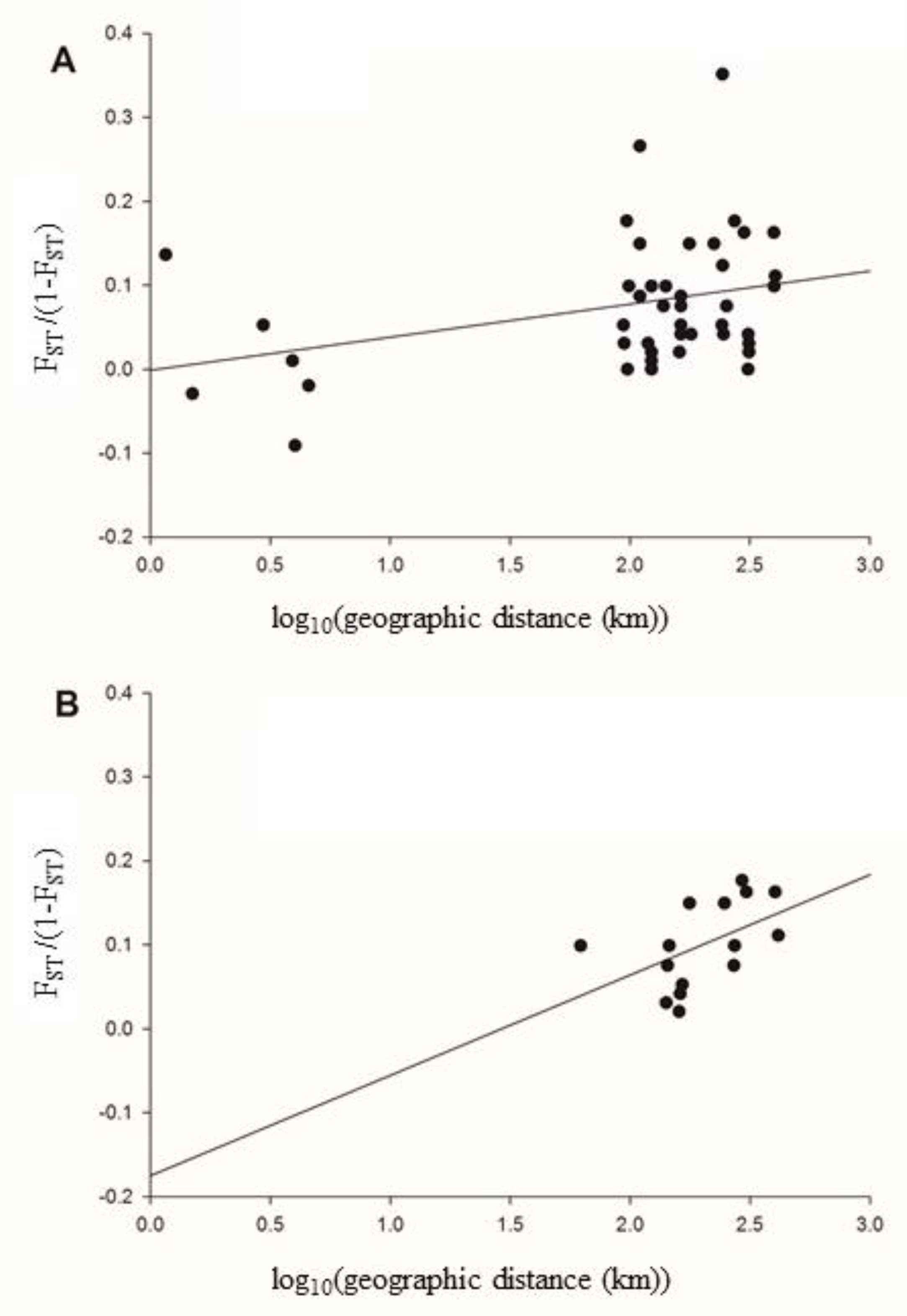
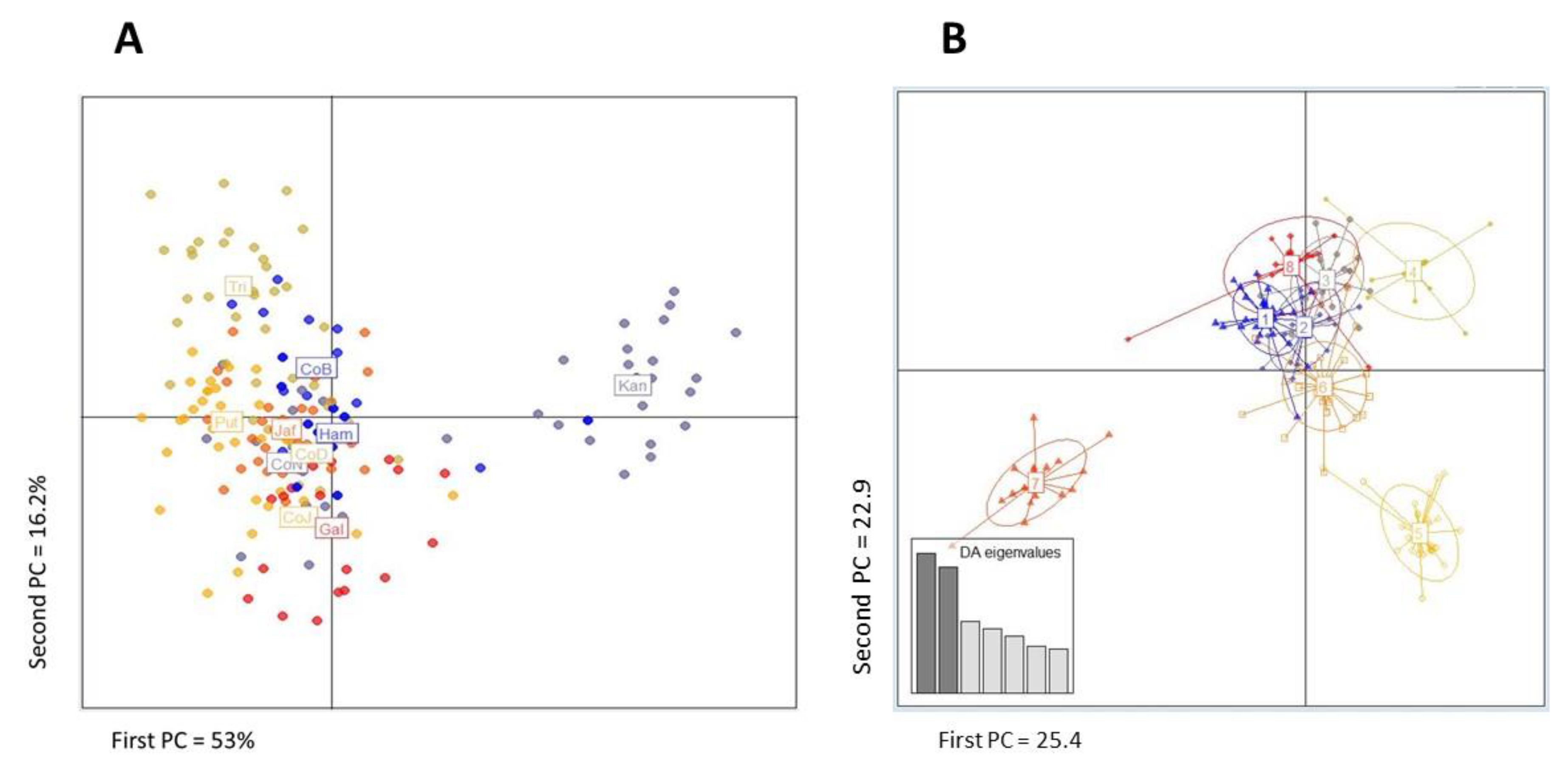
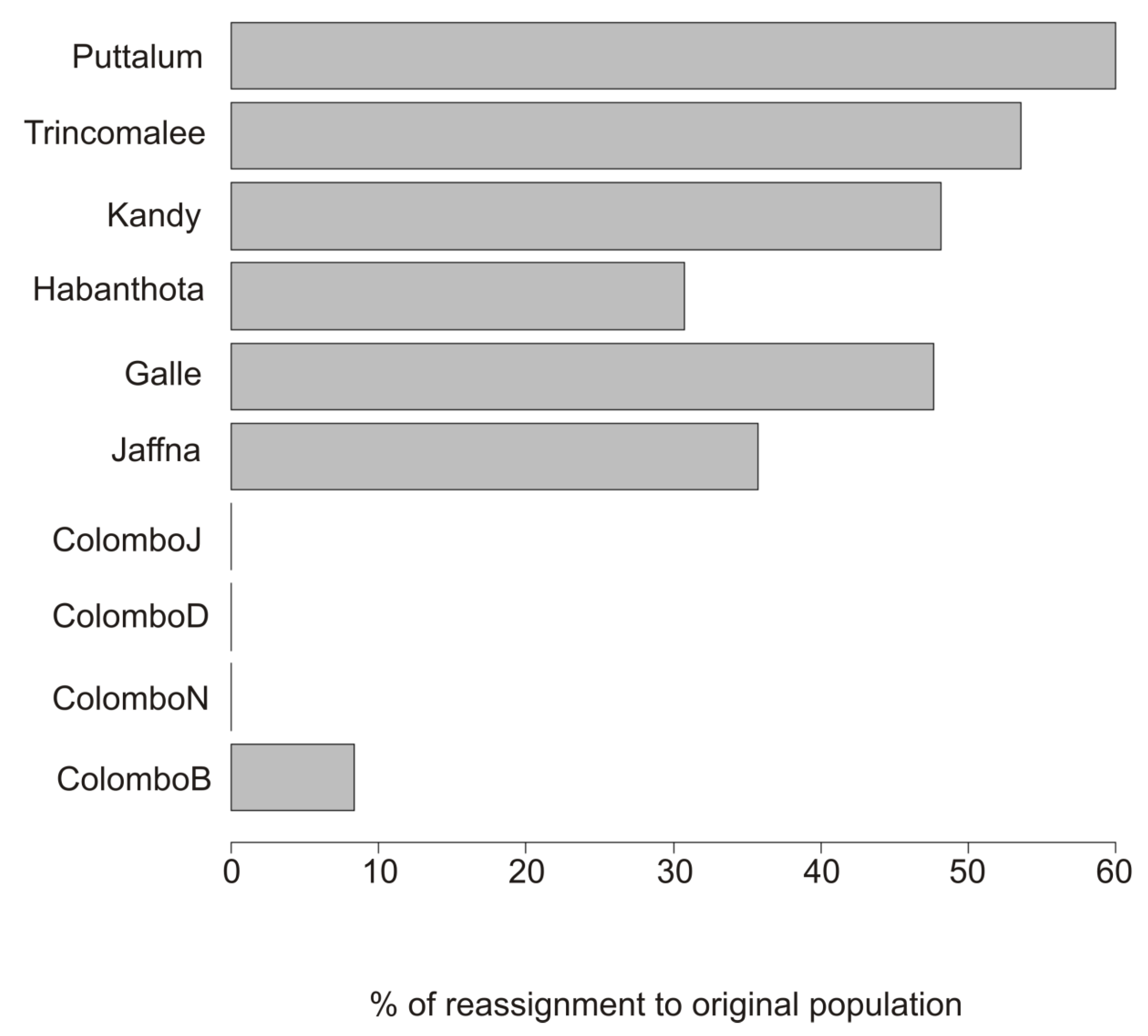
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Association of dengue symptoms with haematological parameters: A retrospective study of 10 hospitals in India. In Global Strategy for Dengue Prevention and Control 2012–2020; WHO: Geneva, Switzerland, 2012. [Google Scholar]
- World Health Organization. Dengue and Dengue Haemorrhagic Fever Fact Sheet. Available online: https://www.who.int/neglected_diseases/integrated_media/integrated_media_dengue/en/ (accessed on 25 January 2017).
- Gloria-Soria, A.; Ayala, D.; Bheecarry, A.; Calderon-Arguedas, O.; Chadee, D.D.; Chiappero, M.; Coetzee, M.; Elahee, K.B.; Fernandez-Salas, I.; Kamal, H.A.; et al. Global genetic diversity of Aedesaegypti. Mol. Ecol. 2016, 25, 5377–5395. [Google Scholar] [CrossRef] [PubMed]
- Kumara, Y.G.; Weerakoon, K.G. Review of Port City as a Place; Port City in Hambantota, Sri Lanka. Eur. Acad. Res. 2014, 2, 2350–2370. [Google Scholar]
- Sun, W.; Xue, L.; Xie, X.X. Spatial-temporal distribution of dengue and climate characteristics for two clusters in Sri Lanka from 2012 to 2016. Sci. Rep-UK 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Louis, V.R.; Montenegro Quinonez, C.A.; Kusumawathie, P.; Palihawadana, P.; Janaki, S.; Tozan, Y.; Wijemuni, R.; Wilder-Smith, A.; Tissera, H.A. Characteristics of and factors associated with dengue vector breeding sites in the City of Colombo, Sri Lanka. Pathog. Glob. Health 2016, 110, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Epidemiology Unit MoH: Dengue Update. Epidemiology Unit. Available online: http://www.epid.gov.lk/ (accessed on 1 December 2017).
- Sirisena, P.D.N.N.; Noordeen, F. Evolution of dengue in Sri Lanka-changes in the virus, vector, and climate. Int. J. Infect Dis. 2014, 19, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Dickson, L.B.; Campbell, C.L.; Juneja, P.; Jiggins, F.M.; Sylla, M.; Black, W.C. Exon-Enriched Libraries Reveal Large Genic Differences Between Aedesaegypti from Senegal, West Africa, and Populations Outside Africa. G3-Genes Genom. Genet. 2017, 7, 571–582. [Google Scholar] [CrossRef]
- Rathnayake, J.; Wijeratne, A.W. Second container port in Sri Lanka; Hambanthota or Trincomalee: An analysis using the game theory. Int. J. Logist. Syst. Manag. 2012, 13, 358–378. [Google Scholar] [CrossRef]
- Belkin, J.N. The Mosquitoes of the South Pacific (Diptera, Culicidae); University of California Press: Berkeley/Los Angeles, CA, USA, 1962. [Google Scholar]
- Ballinger-Crabtree, M.E.; Black, W.C.; Miller, B.R. Use of Genetic Polymorphisms Detected by the Random-Amplified Polymorphic DNA-Polymerase Chain-Reaction (Rapd-Pcr) for Differentiation and Identification of Aedesaegypti Subspecies and Populations. Am. J. Trop. Med. Hyg. 1992, 47, 893–901. [Google Scholar] [CrossRef]
- Slotman, M.A.; Kelly, N.B.; Harrington, L.C.; Kitthawee, S.; Jones, J.W.; Scott, T.W.; Caccone, A.; Powell, J.R. Polymorphic microsatellite markers for studies of Aedesaegypti (Diptera: Culicidae), the vector of dengue and yellow fever. Mol. Ecol. Notes 2007, 7, 168–171. [Google Scholar] [CrossRef]
- Chambers, E.W.; Meece, J.K.; McGowan, J.A.; Lovin, D.D.; Hemme, R.R.; Chadee, D.D.; McAbee, K.; Brown, S.E.; Knudson, D.L.; Severson, D.W. Microsatellite isolation and linkage group identification in the yellow fever mosquito Aedes aegypti. J. Hered. 2007, 98, 202–210. [Google Scholar] [CrossRef]
- Garza, J.C.; Williamson, E.G. Detection of reduction in population size using data from microsatellite loci. Mol. Ecol. 2001, 10, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Ahmed, I. Adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Collins, C. A tutorial for discriminant analysis of principal components (DAPC) using adegenet 2.0.0. Imp Coll London-MRC Cent Outbreak. Anal. Model. 2015, 43, 1–43. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Available online: http://www.R-project.org (accessed on 8 May 2018).
- Piry, S.; Luikart, G.; Cornuet, J.M. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar]
- Luikart, G.; Cornuet, J.M. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv. Biol. 1998, 12, 228–237. [Google Scholar] [CrossRef]
- Beerli, P. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 2006, 22, 341–345. [Google Scholar] [CrossRef]
- Barraud, C.P. The fauna of British India including Ceylon and Burma; Taylor and Francis, Red Lion Court: London, UK, 1934. [Google Scholar]
- Yang, H.M.; Macoris, M.D.; Galvani, K.C.; Andrighetti, M.T.; Wanderley, D.M. Assessing the effects of temperature on the population of Aedesaegypti, the vector of dengue. Epidemiol. Infect 2009, 137, 1188–1202. [Google Scholar] [CrossRef]
- Marinho, R.A.; Beserra, E.B.; Bezerra-Gusmão, M.A.; Porto, V.D.; Olinda, R.A.; dos Santos, C.A. Effects of temperature on the life cycle, expansion, and dispersion of Aedesaegypti (Diptera: Culicidae) in three cities in Paraiba, Brazil. J. Vector Ecol. 2016, 41, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Trpis, M.; Hausermann, W. Dispersal and other population parameters of Aedesaegypti in an African village and their possible significance in epidemiology of vector-borne diseases. Am. J. Trop. Med. Hyg. 1986, 356, 1263–1279. [Google Scholar] [CrossRef] [PubMed]
- Reiter, P.; Amador, M.A.; Anderson, R.A.; Clark, G.G. Short report: Dispersal of Aedesaegypti in an urban area after blood feeding as demonstrated by rubidium-marked eggs. Am. J. Trop. Med. Hyg. 1995, 52, 177–179. [Google Scholar] [CrossRef] [PubMed]
- Huber, K.; Le Loan, L.; Hoang, T.H.; Tien, T.K.; Rodhain, F.; Failloux, A.B. Temporal genetic variation in Aedesaegypti populations in ho chi Minh City (Vietnam). Heredity 2002, 89, 7–14. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Costa-Ribeiro, M.C.; Lourenço-de-Oliveira, R.; Failloux, A.B. Higher genetic variation estimated by microsatellites compared to isoenzyme markers in Aedesaegypti from Rio de Janeiro. Mem. Inst. Oswaldo Cruz. 2006, 101, 917–921. [Google Scholar] [CrossRef]
- Hlaing, T.; Tun-Lin, W.; Somboon, P.; Socheat, D.; Setha, T.; Min, S.; Thaung, S.; Anyaele, O.; De Silva, B.; Chang, M.S.; et al. Spatial genetic structure of Aedesaegypti mosquitoes in mainland Southeast Asia. Evol. Appl. 2010, 3, 319–339. [Google Scholar] [CrossRef]
- Rasheed, S.B.; Boots, M.; Frantz, A.C.; Butlin, R.K. Population structure of the mosquito Aedesaegypti (Stegomyiaaegypti) in Pakistan. Med. Vet. Entomol. 2013, 27, 430–440. [Google Scholar] [CrossRef]
- Huber, K.; Le Loan, L.; Chantha, N.; Failloux, A.B. Human transportation influences Aedesaegypti gene flow in Southeast Asia. Acta Trop. 2004, 90, 23–29. [Google Scholar] [CrossRef]
- Huber, K.; Le Loan, L.; Hoang, T.H.; Ravel, S.; Rodhain, F.; Failloux, A.B. Genetic differentiation of the dengue vector, Aedesaegypti (Ho Chi Minh City, Vietnam) using microsatellite markers. Mol. Ecol. 2002, 11, 1629–1635. [Google Scholar] [CrossRef]
- Endersby, N.M.; Hoffmann, A.A.; White, V.L.; Lowenstein, S.; Ritchie, S.; Johnson, P.H.; Rapley, L.P.; Ryan, P.A.; Nam, V.S.; Yen, N.T.; et al. Genetic structure of Aedesaegypti in Australia and Vietnam revealed by microsatellite and exon primed intron crossing markers suggests feasibility of local control options. J. Med. Entomol. 2009, 46, 1074–1083. [Google Scholar] [CrossRef]
- Hemme, R.R.; Thomas, C.L.; Chadee, D.D.; Severson, D.W. Influence of urban landscapes on population dynamics in a short-distance migrant mosquito: Evidence for the dengue vector Aedesaegypti. PLoS Negl. Trop. Dis. 2010, 4, e634. [Google Scholar] [CrossRef] [PubMed]
- Dutta, P.; Khan, S.A.; Sharma, C.K.; Doloi, P.; Hazarika, N.C.; Mahanta, J. Distribution of potential dengue vectors in major townships along the national highways and trunk roads of northeast India. Southeast Asian J. Trop. Med. Public Health 1998, 29, 173–176. [Google Scholar] [PubMed]
- Edman, J.D.; Scott, T.W.; Costero, A.; Morrison, A.C.; Harrington, L.C.; Clark, G.G. Aedesaegypti (Diptera: Culicidae) movement influenced by availability of oviposition sites. J. Med. Entomol. 1998, 35, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Suwonkerd, W.; Mongkalangoon, P.; Parbaripai, A.; Grieco, J.; Achee, N.; Roberts, D.; Chareonviriyaphap, T. The effect of host type on movement patterns of Aedesaegypti (Diptera: Culicidae) into and out of experimental huts in Thailand. J. Vector Ecol. 2006, 31, 311–318. [Google Scholar] [CrossRef]
- Paupy, C.; Chantha, N.; Huber, K.; Lecoz, N.; Reynes, J.M.; Rodhain, F.; Failloux, A.B. Influence of breeding sites features on genetic differentiation of Aedesaegypti populations analyzed on a local scale in Phnom Penh Municipality of Cambodia. Am. J. Trop. Med. Hyg. 2004, 71, 73–81. [Google Scholar] [CrossRef]
- Getis, A.; Morrison, A.C.; Gray, K.; Scott, T.W. Characteristics of the spatial pattern of the dengue vector, Aedesaegypti, in Iquitos, Peru. Am. J. Trop. Med. Hyg. 2003, 69, 494–505. [Google Scholar] [CrossRef]
- Guagliardo, S.A.; Barboza, J.L.; Morrison, A.C.; Astete, H.; Vazquez-Prokopec, G.; Kitron, U. Patterns of geographic expansion of Aedesaegypti in the Peruvian Amazon. PLoS Negl. Trop. Dis. 2014, 8, e3033. [Google Scholar] [CrossRef]
- Suwonkerd, W.; Suwannachote, N.; Boonti, T.; Charoenviriyaphap, T. Biting pattern of a dengue vectors, Aedesaegypti and Aedesalbopictus in urban and rural gradient in Chiang Mai province, Northern Thailand. Am. J. Trop. Med. Hyg. 2006, 75, 72–73. [Google Scholar]
- Beaty, B.J.; Black, W.C.; Eisen, L.; Flores, A.E.; García-Rejón, J.E.; Loroño-Pino, M.; Saavedra-Rodriguez, K. The intensifying storm: Domestication of Aedesaegypti, urbanization of arboviruses, and emerging insecticide resistance. In Global Health Im-pacts of Vector-Borne Diseases: Workshop Summary; National Academies Press: Washington, DC, USA, 2016. [Google Scholar]
- Schmidt, T.L.; Filipovic, I.; Hoffmann, A.A.; Rasic, G. Fine-scale landscape genomics of Aedesaegypti reveals loss of Wolbachiatransinfection, dispersal barrier and potential for occasional long distance movement. bioRxiv 2017, 103598. [Google Scholar]
- Saavedra-Rodriguez, K.; Beaty, M.; Lozano-Fuentes, S.; Denham, S.; Garcia-Rejon, J.; Reyes-Solis, G.; Machain-Williams, C.; Loroño-Pino, M.A.; Flores-Suarez, A.; Ponce-Garcia, G.; et al. Local evolution of pyrethroid resistance offsets gene flow among Aedesaegypti collections in Yucatan State, Mexico. Am. J. Trop. Med. Hyg. 2015, 92, 201–209. [Google Scholar] [CrossRef]

| Source of Variation | d.f. | Sum of Squares | Variance Components | F | % Variation |
|---|---|---|---|---|---|
| Among cities | 6 | 142.63 | 0.035 | 0.048 *** | 4.75 |
| Among collections in cities | 3 | 6.46 | 0.030 | 0.043 *** | 4.08 |
| Within populations | 2630 | 1749.07 | 0.665 | 0.088 * | 91.17 |
| Total | 2639 | 1898.15 | 0.729 |
| Locus# | ColomboB | ColomboN | ColomboD | Colombo J | Jaffna | Galle | Hambanthota | Kandy | Trincomalee | Puttalum |
|---|---|---|---|---|---|---|---|---|---|---|
| AT1 | 0.750 | 0.857 | 0.600 | 0.800 | 0.875 | 1.000 | 1.000 | 0.556 | 0.857 | 0.667 |
| AG7 | 0.600 | 0.571 | 0.800 | 0.667 | 0.750 | 1.000 | 0.600 | 0.800 | 0.400 | 0.571 |
| AC1 | 0.375 | 0.375 | 0.375 | 0.375 | 0.333 | 0.208 | 0.375 | 0.375 | 0.375 | 0.375 |
| AG4 | 1.000 | 0.417 | 1.000 | 0.333 | 0.308 | 0.417 | 0.364 | 0.462 | 0.417 | 0.115 |
| AG2 | 0.429 | 0.500 | 0.306 | 0.261 | 0.600 | 0.278 | 0.189 | 1.000 | 0.432 | 0.647 |
| AC2 | 1.000 | 1.000 | 1.000 | 0.318 | 1.000 | 0.417 | 0.667 | 1.000 | 0.049 | 1.000 |
| AG5 | 0.625 | 0.857 | 0.833 | 0.833 | 0.200 | 0.700 | 0.571 | 0.857 | 0.833 | 0.833 |
| B07 | 0.615 | 0.750 | 0.778 | 0.727 | 0.636 | 0.875 | 0.500 | 0.727 | 0.500 | 0.600 |
| H08 | 1.000 | 0.600 | 1.000 | 0.235 | 1.000 | 0.571 | 0.600 | 0.714 | 1.000 | 1.000 |
| A10 | 1.000 | 0.600 | 1.000 | 0.800 | 0.571 | 0.600 | 0.500 | 1.000 | 0.750 | 0.500 |
| Mean | 0.710 | 0.653 | 0.769 | 0.535 | 0.627 | 0.607 | 0.537 | 0.749 | 0.561 | 0.631 |
| s.d. | 0.243 | 0.206 | 0.262 | 0.250 | 0.285 | 0.285 | 0.216 | 0.227 | 0.289 | 0.272 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernando, H.S.D.; Hapugoda, M.; Perera, R.; Black IV, W.C.; De Silva, B.G.D.N.K. Gene Flow Patterns among Aedes aegypti (Diptera: Culicidae) Populations in Sri Lanka. Insects 2020, 11, 169. https://doi.org/10.3390/insects11030169
Fernando HSD, Hapugoda M, Perera R, Black IV WC, De Silva BGDNK. Gene Flow Patterns among Aedes aegypti (Diptera: Culicidae) Populations in Sri Lanka. Insects. 2020; 11(3):169. https://doi.org/10.3390/insects11030169
Chicago/Turabian StyleFernando, H.S.D., Menaka Hapugoda, Rushika Perera, William C. Black IV, and B.G.D.N.K. De Silva. 2020. "Gene Flow Patterns among Aedes aegypti (Diptera: Culicidae) Populations in Sri Lanka" Insects 11, no. 3: 169. https://doi.org/10.3390/insects11030169
APA StyleFernando, H. S. D., Hapugoda, M., Perera, R., Black IV, W. C., & De Silva, B. G. D. N. K. (2020). Gene Flow Patterns among Aedes aegypti (Diptera: Culicidae) Populations in Sri Lanka. Insects, 11(3), 169. https://doi.org/10.3390/insects11030169





