Pharmacogenomics in Orofacial Clefts Care: Insights from Whole-Genome Sequencing of Case-Parents Trios
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Sample Collection and DNA Extraction
2.3. Whole-Genome Sequencing and Bioinformatics Analysis
2.4. Selection of Genes
2.5. Variant Classification and Structural Analysis
3. Results
3.1. Identification and Characterization of Pathogenic Variants in Drug-Metabolizing and Drug-Transporting Genes
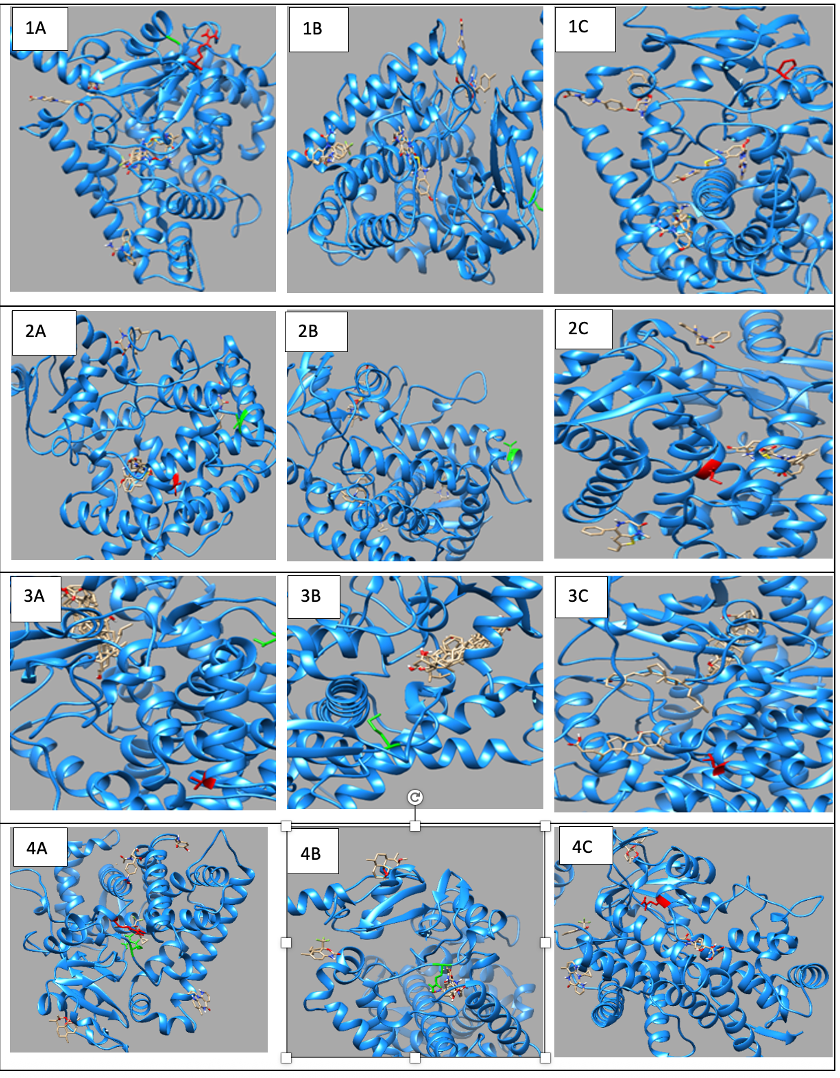
3.2. Structural and Functional Impact Analysis
3.2.1. CYP1A2 Variants
3.2.2. CYP2C18 Variants
3.2.3. CYP27A1 Variants
3.2.4. CYP2B6 Variants
3.3. Molecular Docking Analysis and Drug-Binding Implications
3.3.1. CYP1A2 Drug Interactions
3.3.2. CYP2C18 Substrate Binding
3.3.3. CYP27A1 Metabolic Effects
3.3.4. CYP2B6 Metabolic Effects
3.4. Drug Transport Gene Variants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADR | Adverse drug reaction |
| ACMG | American College of Medical Genetics and Genomics |
| CL | Cleft lip |
| CP | Cleft palate |
| GoF | Gain-of-function |
| LoF | Loss-of-function |
| OFC | Orofacial clefts |
| WGS | Whole-genome sequencing |
References
- Manlove, K.R.; Johnson, C.K.; Hayes, B.H. Molecular mechanisms of craniofacial development. Dev. Biol. 2020, 460, 132–145. [Google Scholar] [CrossRef]
- Khan, N.E.; Smith, M.H.; Johnson, D.R. Role of genetic factors in orofacial clefts. Gene 2020, 752, 144793. [Google Scholar] [CrossRef]
- Nasreddine, G.; El Hajj, J.; Ghassibe-Sabbagh, M. Orofacial clefts embryology, classification, epidemiology, and genetics. Mutat. Res. Rev. Mutat. Res. 2021, 787, 108373. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Chen, Y.; Wang, R. Syndromic forms of orofacial clefts. Clin. Genet. 2019, 96, 402–414. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, B.; Zhang, Q.; Wu, Y. Global, regional and national burden of orofacial clefts from 1990 to 2019: An analysis of the Global Burden of Disease Study 2019. Ann. Med. 2023, 55, 2215540. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Putri, F.A.; Pattamatta, M.; Anita, S.E.S.; Maulina, T. The Global Occurrences of Cleft Lip and Palate in Pediatric Patients and Their Association with Demographic Factors: A Narrative Review. Children 2024, 11, 322. [Google Scholar] [CrossRef]
- Eslami, N.; Bardideh, E.; Tatari, P.; Dehghani, L. Orofacial dysfunction in cleft lip and palate patients using the nordic orofacial test-screening. J. World Fed. Orthod. 2024, 13, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Van Dyck, J.; Cadenas de Llano-Pérula, M.; Willems, G.; Verdonck, A. Dental development in cleft lip and palate patients: A systematic review. Forensic Sci Int. 2019, 300, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Mingzhe, H.; Johnson, K.L.; Roberts, M.P. Pharmacokinetics and pharmacodynamics in clinical practice. Adv. Drug Deliv. Rev. 2021, 175, 113–128. [Google Scholar] [CrossRef]
- Debon, A.; Siirola, E.; Snajdrova, R. Enzymatic Bioconjugation: A Perspective from the Pharmaceutical Industry. JACS Au. 2023, 3, 1267–1283. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Reed, L.; Arlt, V.M.; Phillips, D.H. The role of cytochrome P450 enzymes in carcinogen activation and detoxication: An in vivo-in vitro paradox. Carcinogenesis 2018, 39, 851–859. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Manikandan, P.; Nagini, S. Cytochrome P450 Structure, Function and Clinical Significance: A Review. Curr. Drug Targets 2018, 19, 38–54. [Google Scholar] [CrossRef] [PubMed]
- McDermott, J.H.; Sharma, V.; Keen, J.; Newman, W.G. Embedding Pharmacogenetics Into Clinical Practice to Improve Patient Outcomes. Ann. Hum. Genet. 2025, 89, 398–405. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, M.; Ma, J.; Li, M.; Zhang, Y.; Jiang, B.; Zhao, X.; Huai, C.; Shen, L.; Zhang, N.; He, L.; et al. Cytochrome P450 Enzymes and Drug Metabolism in Humans. Int. J. Mol. Sci. 2021, 22, 12808. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Obebi Cliff-Eribo, K.; Sammons, H.; Star, K.; Ralph Edwards, I.; Osakwe, A.; Choonara, I. Adverse drug reactions in Nigerian children: A retrospective review of reports submitted to the Nigerian Pharmacovigilance Centre from 2005 to 2012. Paediatr. Int. Child Health 2016, 36, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Awotoye, W.; Mossey, P.A.; Hetmanski, J.B.; Gowans, L.J.J.; Eshete, M.A.; Adeyemo, W.L.; Alade, A.; Zeng, E.; Adamson, O.; Naicker, T.; et al. Whole-genome sequencing reveals de-novo mutations associated with nonsyndromic cleft lip/palate. Sci. Rep. 2022, 12, 11743. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Anderson, D.; Lassmann, T. An expanded phenotype centric benchmark of variant prioritization tools. Hum. Mutat. 2022, 43, 539–546. [Google Scholar] [CrossRef] [PubMed]
- PharmGKB. Very Important Pharmacogenes (VIPs). PharmGKB Knowledgebase. Available online: https://www.pharmgkb.org (accessed on 8 February 2024).
- Rodrigues-Soares, F.; Peñas-Lledó, E.M.; Tarazona-Santos, E.; Sosa-Macías, M.; Terán, E.; López-López, M.; Rodeiro, I.; Moya, G.E.; Calzadilla, L.R.; Ramírez-Roa, R.; et al. Genomic Ancestry, CYP2D6, CYP2C9, and CYP2C19 Among Latin Americans. Clin. Pharmacol. Ther. 2020, 107, 257–268. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information (NCBI). Gene Summaries for CYP450 Enzymes and Transporters. Available online: https://www.ncbi.nlm.nih.gov (accessed on 8 February 2024).
- Parsons, M.T.; de la Hoya, M.; Richardson, M.E.; Tudini, E.; Anderson, M.; Berkofsky-Fessler, W.; Caputo, S.M.; Chan, R.C.; Cline, M.S.; Feng, B.J.; et al. Evidence-based recommendations for gene-specific ACMG/AMP variant classification from the ClinGen ENIGMA BRCA1 and BRCA2 Variant Curation Expert Panel. Am. J. Hum. Genet. 2024, 111, 2044–2058. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Venselaar, H.; Te Beek, T.A.; Kuipers, R.K.; Hekkelman, M.L.; Vriend, G. Protein structure analysis of mutations causing inheritable diseases: An e-Science approach with life scientist friendly interfaces. BMC Bioinform. 2010, 11, 548. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Land, H.; Humble, M.S. YASARA: A Tool to Obtain Structural Guidance in Biocatalytic Investigations. Methods Mol. Biol. 2018, 1685, 43–67. [Google Scholar] [CrossRef] [PubMed]
- Vriend, G. WHAT IF: A molecular Modeling and Drug Design Program. Available online: https://swift.cmbi.umcn.nl/gv/whatif/ (accessed on 8 February 2024).
- Bordoli, L.; Kiefer, F.; Arnold, K.; Benkert, P.; Battey, J.; Schwede, T. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 2009, 4, 1–13. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jo, S.; Cheng, X.; Lee, J.; Kim, S.; Park, S.J.; Patel, D.S.; Beaven, A.H.; Lee, K.I.; Rui, H.; Park, S.; et al. CHARMM-GUI 10 years for biomolecular modeling and simulation. J. Comput. Chem. 2017, 38, 1114–1124. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shao, Y.; Molnar, L.F.; Jung, Y.; Kussmann, J.; Ochsenfeld, C.; Brown, S.T.; Gilbert, A.T.B.; Slipchenko, L.V.; Levchenko, S.V.; O’Neill, D.P.; et al. Advances in methods and algorithms in a modern quantum chemistry program package. Phys. Chem. Chem. Phys. 2006, 8, 3172–3191. [Google Scholar] [CrossRef]
- Deenadayalan, A.; Vijayalakshmi, S.; Janaki, C.S.; Jayaraman, S. Molecular docking analysis of stevioside with Akt and PPARγ. Bioinformation 2021, 17, 283–288. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Luzum, J.A.; Petry, N.; Taylor, A.K.; Van Driest, S.L.; Dunnenberger, H.M.; Cavallari, L.H. Moving Pharmacogenetics Into Practice: It’s All About the Evidence! Clin. Pharmacol. Ther. 2021, 110, 649–661. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2018, 138, 103–141. [Google Scholar] [CrossRef]
- Pikuleva, I.A.; Cartier, N. Cholesterol Hydroxylating Cytochrome P450 46A1: From Mechanisms of Action to Clinical Applications. Front Aging Neurosci. 2021, 13, 696778. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- van de Steeg, E.; Wagenaar, E.; van der Kruijssen, C.M. Role of multidrug resistance protein 3 (MRP3/ABCC3) in hepatobiliary and enteric transport of bile salts. Mol. Pharmacol. 2009, 75, 154–160. [Google Scholar] [CrossRef]
- Adedapo, A.D.A.; Adedeji, W.A.; Adedapo, I.A.; Adedapo, K.S. Cohort study on adverse drug reactions in adults admitted to the medical wards of a tertiary hospital in Nigeria: Prevalence, incidence, risk factors, and fatality. Br. J. Clin. Pharmacol. 2021, 87, 1878–1889. [Google Scholar] [CrossRef]
- Almazroo, O.A.; Miah, M.K.; Venkataramanan, R. Drug Metabolism in the Liver. Clin. Liver Dis. 2017, 21, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, Y.C.; Demir, O. Use of Phenytoin, Phenobarbital Carbamazepine, Levetiracetam Lamotrigine and Valproate in Pregnancy and Breastfeeding: Risk of Major Malformations, Dose-dependency, Monotherapy vs Polytherapy, Pharmacokinetics and Clinical Implications. Curr. Neuropharmacol. 2021, 19, 1805–1824. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, P.F.; Neiner, A.; Kharasch, E.D. Efavirenz Metabolism: Influence of Polymorphic CYP2B6 Variants and Stereochemistry. Drug Metab. Dispos. 2019, 47, 1195–1205, Erratum in: Drug Metab. Dispos. 2019, 47, 1231. https://doi.org/10.1124/dmd.119.086348err. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Weinshilboum, R.M.; Wang, L. Pharmacogenomics: Precision medicine and drug response. Mayo Clin. Proc. 2017, 92, 1711–1722. [Google Scholar] [CrossRef]

| GENE | Genomic Coordinate | Genotype of Proband | HGVSc | HGVSp | Variant in Father? (Genotype) | Variant in Mother? (Genotype) | Number of Tools That Predicted Pathogenicity | Prevalence |
|---|---|---|---|---|---|---|---|---|
| Families (Individuals) | ||||||||
| CYP1A2 | 15:74749955 (rs45565238) | G/A | NM_000761.5:c.217G>A | p.Gly73Arg | Yes (G/A) | None | 8 | 2 (4) |
| 15:74750007 (Novel) | G/C | NM_000761.5:c.269G>C | p.Arg90Pro | Yes (G/C) | None | 6 | 1 (2) | |
| CYP2C18 | 10:94724372 (rs59636573) | G/T | NM_000772.3:c.988G>T | p.Val330Leu | None | Yes (G/T) | 6 | 2 (4) |
| 10:94720472 (rs60181876) | C/T | NM_000772.3:c.896C>T | p.Thr299Ile | Yes (C/T) | None | 6 | 2 (4) | |
| CYP27A1 | 2:218814105 (rs145722193) | G/T | NM_000784.4:c.1102G>T | p.Val368Leu | None | Yes (G/T) | 6 | 1 (2) |
| 2:218814998 (rs151117761) | G/A | NM_000784.4:c.1564G>A | p.Val522Met | Yes (G/A) | None | 6 | 2 (4) | |
| CYP2B6 | 19:41016652 (rs764288403) | G/A | NM_000767.5:c.1301G>A | p.Arg434Gln | None | Yes (G/A) | 8 | 1 (2) |
| 19:41004122 (rs372295360) | G/A | NM_000767.5:c.293G>A | p.Arg98Gln | Yes (G/A) | None | 9 | 1 (2) | |
| SLC6A2 | 16:55691982 (rs45564432) | C/G | NM_001043.3:c.848C>G | p.Thr283Arg | Yes (C/G) | None | 6 | 2 (4) |
| ABCC3 | 17:50683692 (rs11568591) | G/A | NM_003786.4:c.3890G>A | p.Arg1297His | Yes (G/A) | None | 6 | 3 (6) |
| 17:50677861 (rs34620384) | C/T | NM_003786.4:c.3496C>T | p.Arg1166Cys | None | Yes (C/T) | 6 | 2 (4) |
| Gene | Variants | 2D Depiction of Amino Acid Change | 3D Depiction of Amino Acid Change |
|---|---|---|---|
| CYP1A2 | 15:74749955 (rs45565238) | 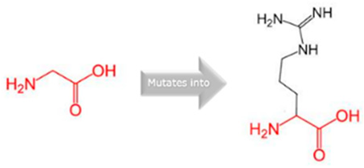 Gly into an Arg at position 73 |  Green: Wildtype (Glycine) Red: Mutant (Arginine) |
| 15:74750007 (Novel) |  Arg into a Pro at position 90 |  Green: Wildtype (Arginine) Red: Mutant (Proline) | |
| CYP2C18 | 10:94724372 (rs59636573) | 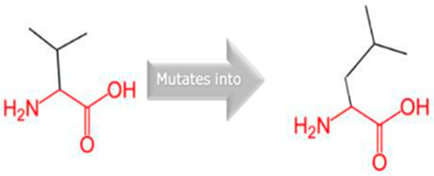 Val into a Leu at position 330 |  Green: Wildtype (Valine) Red: Mutant (Leucine) |
| 10:94720472 (rs60181876) | 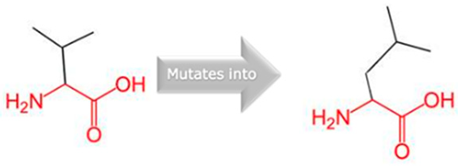 Thre into an Iso at position 299 |  Green: Wildtype (Threonine) Red: Mutant (Isoleucine) | |
| CYP27A1 | 2:218814105 (rs145722193) | 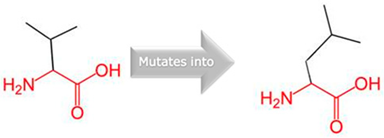 Val into a Leu at position 368 | 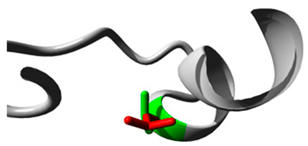 Green: Wildtype (Valine) Red: Mutant (Leucine) |
| 2:218814998 (rs151117761) | NA | NA | |
| CYP2B6 | 19:41016652 (rs764288403) | 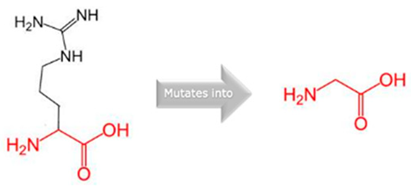 Arg into a Gln at position 434 | 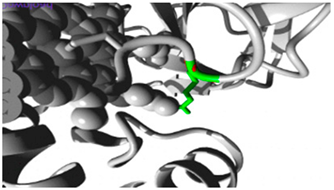 Green: Wildtype (Arginine) Red: Mutant (Glutamine) |
| 19:41004122 (rs372295360) |  Arg into a Gln at position 98 | 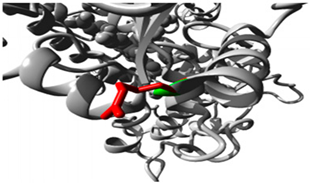 Green: Wildtype (Arginine) Red: Mutant (Glutamine) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poku-Adusei, E.; Mensah, G.O.; Asamoah, C.O.; Tsri, B.; Akeeya, H.; Danbaki, A.S.; Obiri-Yeboah, S.; Busch, T.D.; Borquaye, L.S.; Donkor, P.; et al. Pharmacogenomics in Orofacial Clefts Care: Insights from Whole-Genome Sequencing of Case-Parents Trios. J. Pers. Med. 2025, 15, 456. https://doi.org/10.3390/jpm15100456
Poku-Adusei E, Mensah GO, Asamoah CO, Tsri B, Akeeya H, Danbaki AS, Obiri-Yeboah S, Busch TD, Borquaye LS, Donkor P, et al. Pharmacogenomics in Orofacial Clefts Care: Insights from Whole-Genome Sequencing of Case-Parents Trios. Journal of Personalized Medicine. 2025; 15(10):456. https://doi.org/10.3390/jpm15100456
Chicago/Turabian StylePoku-Adusei, Elvis, Gideon Okyere Mensah, Christian Opoku Asamoah, Bruce Tsri, Hafsa Akeeya, Abass Shaibu Danbaki, Solomon Obiri-Yeboah, Tamara D. Busch, Lawrence Sheringham Borquaye, Peter Donkor, and et al. 2025. "Pharmacogenomics in Orofacial Clefts Care: Insights from Whole-Genome Sequencing of Case-Parents Trios" Journal of Personalized Medicine 15, no. 10: 456. https://doi.org/10.3390/jpm15100456
APA StylePoku-Adusei, E., Mensah, G. O., Asamoah, C. O., Tsri, B., Akeeya, H., Danbaki, A. S., Obiri-Yeboah, S., Busch, T. D., Borquaye, L. S., Donkor, P., Butali, A., & Gowans, L. J. J. (2025). Pharmacogenomics in Orofacial Clefts Care: Insights from Whole-Genome Sequencing of Case-Parents Trios. Journal of Personalized Medicine, 15(10), 456. https://doi.org/10.3390/jpm15100456








