A Multi-Agent Deep Reinforcement Learning Approach for Enhancement of COVID-19 CT Image Segmentation
Abstract
:1. Introduction
- Manual extraction and quality control take hours to process an image.
- Classical or manual labeling, at the pixel level, consumes a lot of time.
- The class discrepancy in the image datasets varies greatly, which may result in unwanted influences in the learned methods.
- Introducing an effective system to enhance semantic segmentation by modifying the traditional Deep-Q-Network to learn superior mask extraction compared to the most advanced methods.
- Proposing a multi-agent reinforcement learning (MARL) framework to extract COVID-19 masks automatically to improve the efforts of CT experts and enhance the potential of lung CT segmentation. We selected learning in the Markov decision process (MDP) to find optimal approaches in mask extraction for CT experts.
- Reducing the waiting time for experts to obtain masks manually, by introducing an alternative automatic approach.
2. Related Works
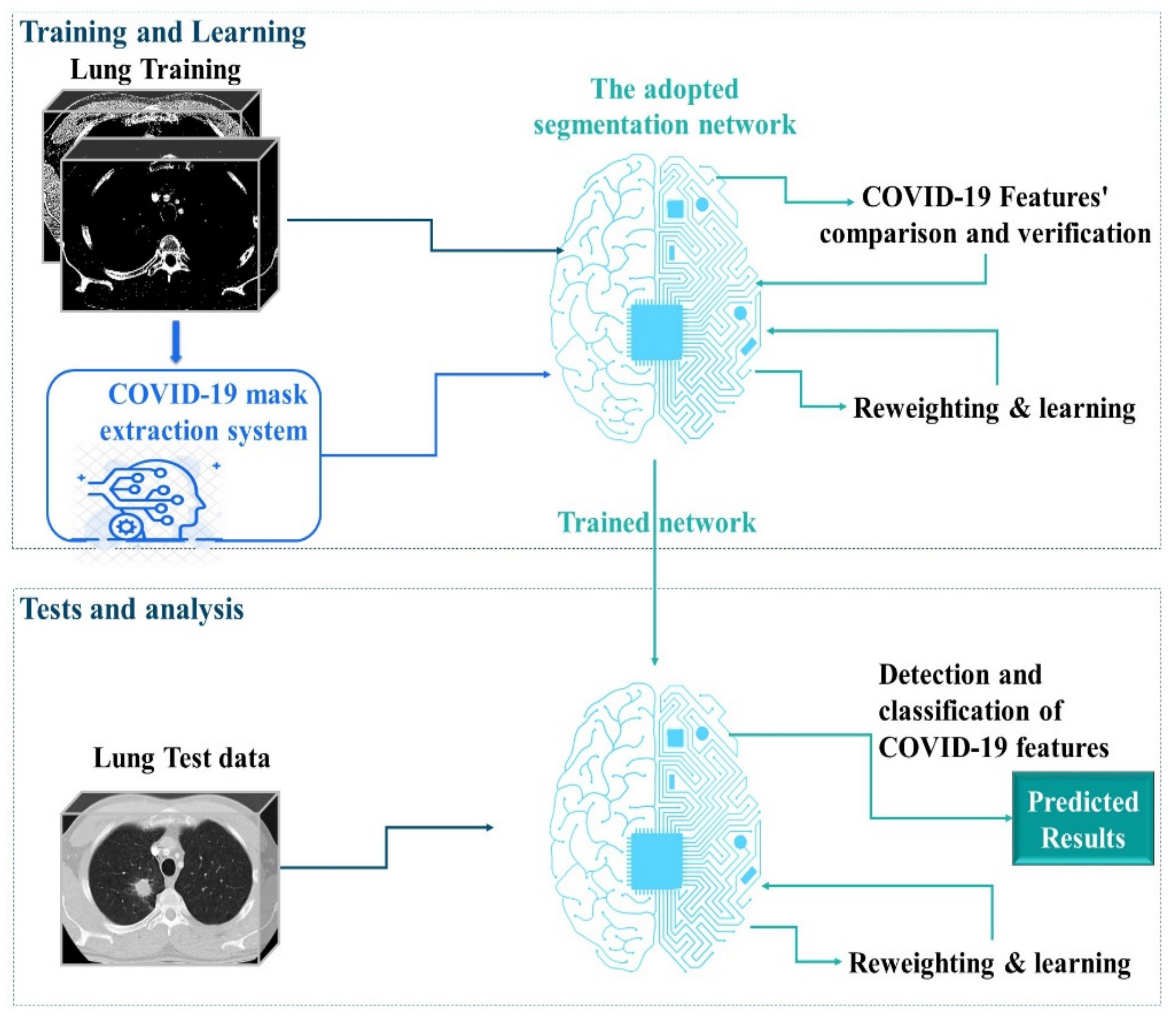
3. The Proposed Semantic Segmentation Environment
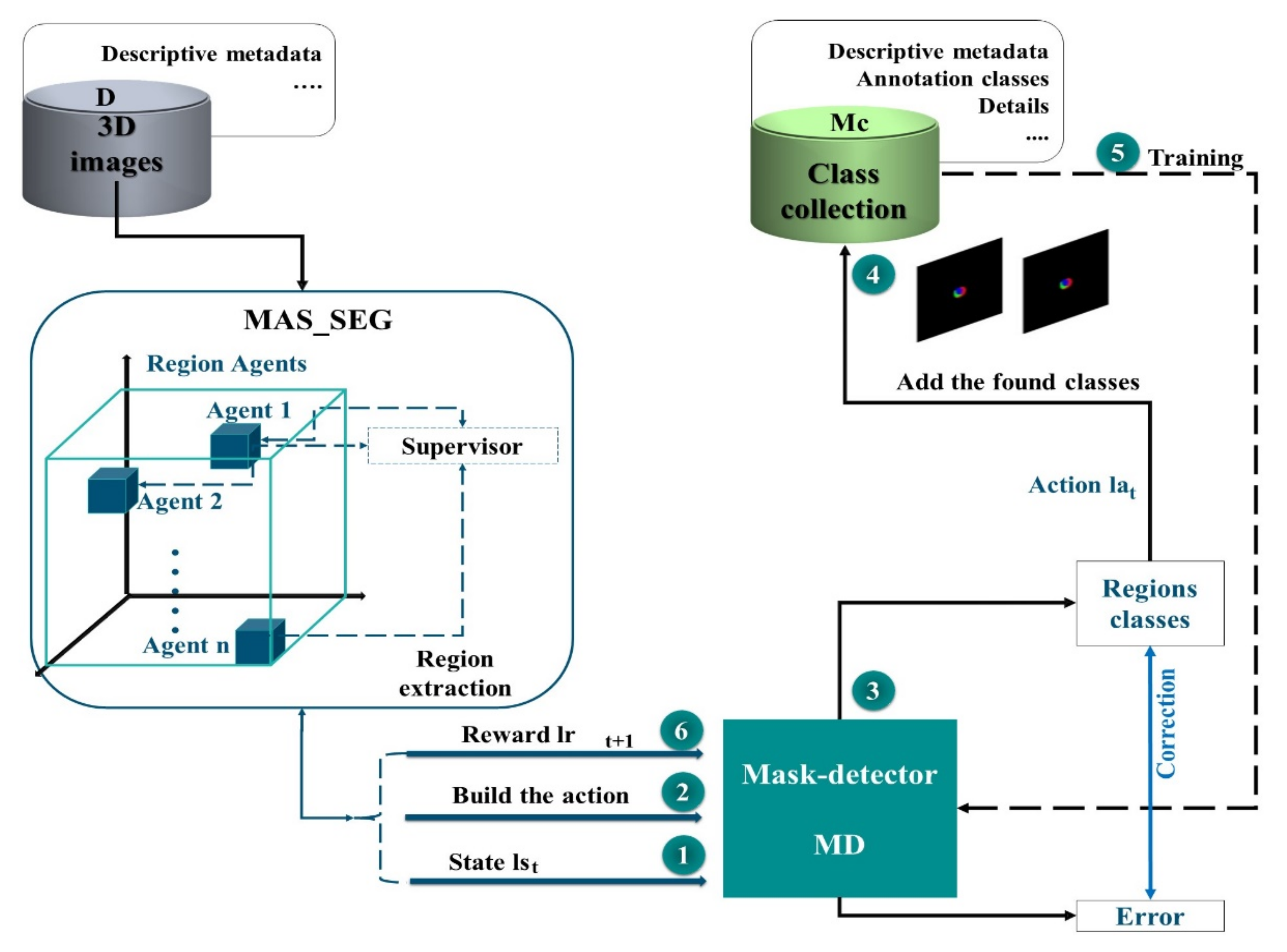
3.1. RL Architecture for Mask Extraction
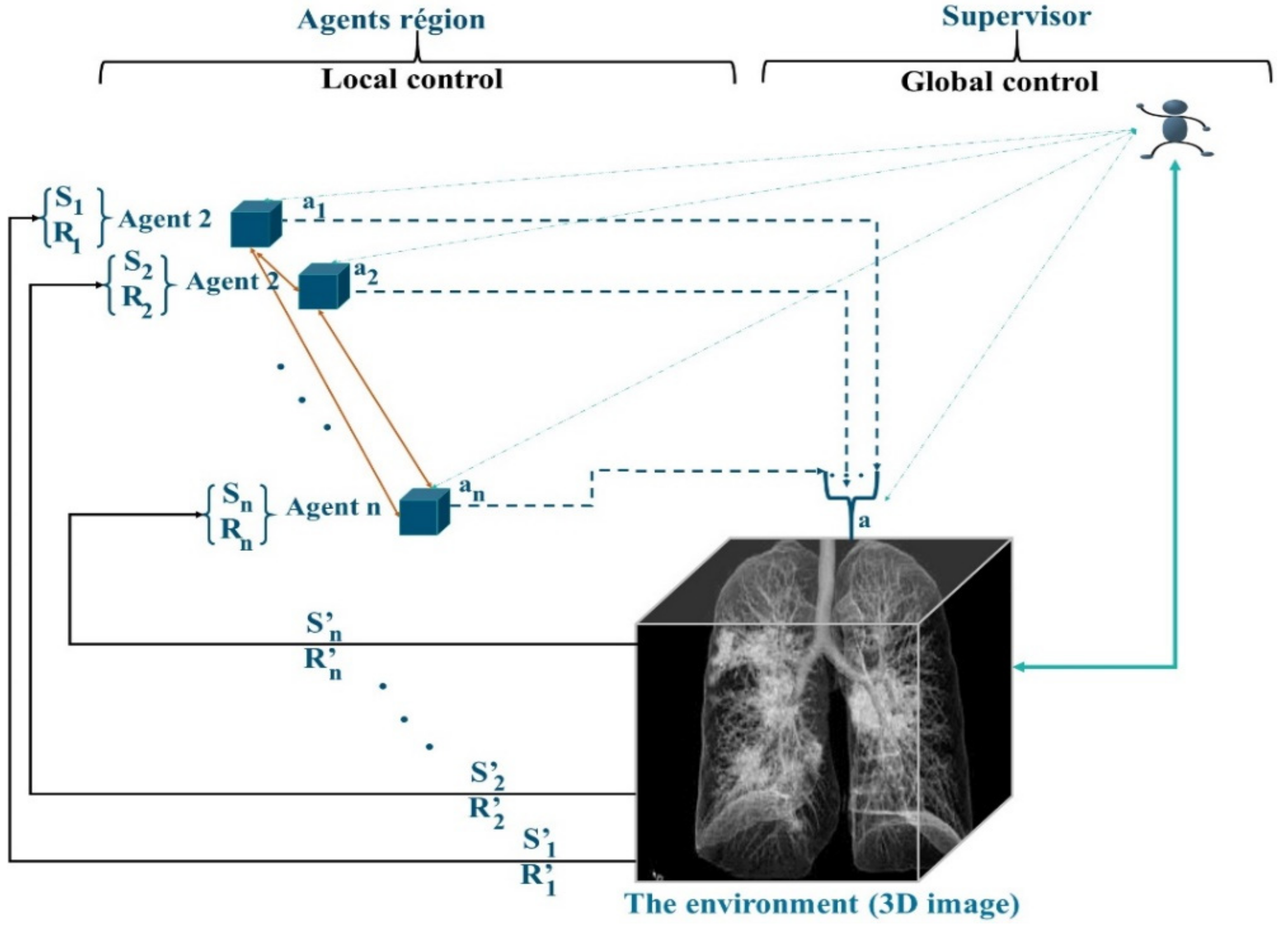
3.2. Structural Properties of MAS_SEG
3.3. The MAS Q-Learning Algorithm and the ε-Greedy Strategy
- , , , and represent respectively the state, action, reward, and Q-value on the Cth episode.
- is the learning factor of the Cth episode.
- is the discount factor.
- α indicates the weight the system gives to new rewards by discounting the long-term value.
- γ is the factor that reduces the contribution of max, which is the maximum Q-value expected in the new state.
3.4. Role of the Supervisor Agent
- Regional agents that cooperate.
- A supervisor agent, which incorporates a neural network to manage the interactions of the MAS agents and subsequently participates in the learning of the mask detector.
| Algorithm 1. QL_MAS |
| Initialize: The capacity Cap of the memory M, the values Q: ∀r, a|Q (r, a) = 0, The estimation weight LSTM-DQN θ = θ_0, the weight of the LSTM-DQN objectives θ′ For episode = 1 → ep do # ep represents the number of episodes Fix the initial positions of the agents according to the map from the RAG For i = 1 -> Reg do # Reg is the number of regions #N is the number of agents N = Reg Implement in each Reg i an agent Ai End for Do as long as t < Cap For j = 1 -> N do Calculation of the initial actions (2) Calculation of initial Q-value (1) Verification of the best adjacent neighbors satisfying the similarity criteria Negotiation to decide the optimal proposal End for Fusion Update of the map of the regions Update Reg N = Reg For j = 1 -> N do Calculation of the actions (2) Calculation of Q-value (1) Calculation of reward (4) Next state calculation (3) Save data d = {state(t), action(t), R(t), state(t + 1)} in memory M End for End Do Reset End for |
3.5. Region Generation Proposal
- The first output determines whether the anchor is in the foreground or background. If it is a foreground, it suggests there is an object in the rectangle.
- If the item is not properly located in the middle of the rectangle, the second output (error) is triggered to adjust the bounding box to best fit the detected object.
3.6. Markov Decision Process for Masking
- The MD is trained using a subset of the TMc mask class collection to guarantee the active learning set for several episodes and learn a good acquisition function that maximizes performance with a budget of regions .
- The MD network is tested on a separate subset.
- To calculate the reward, we employ a different subset.
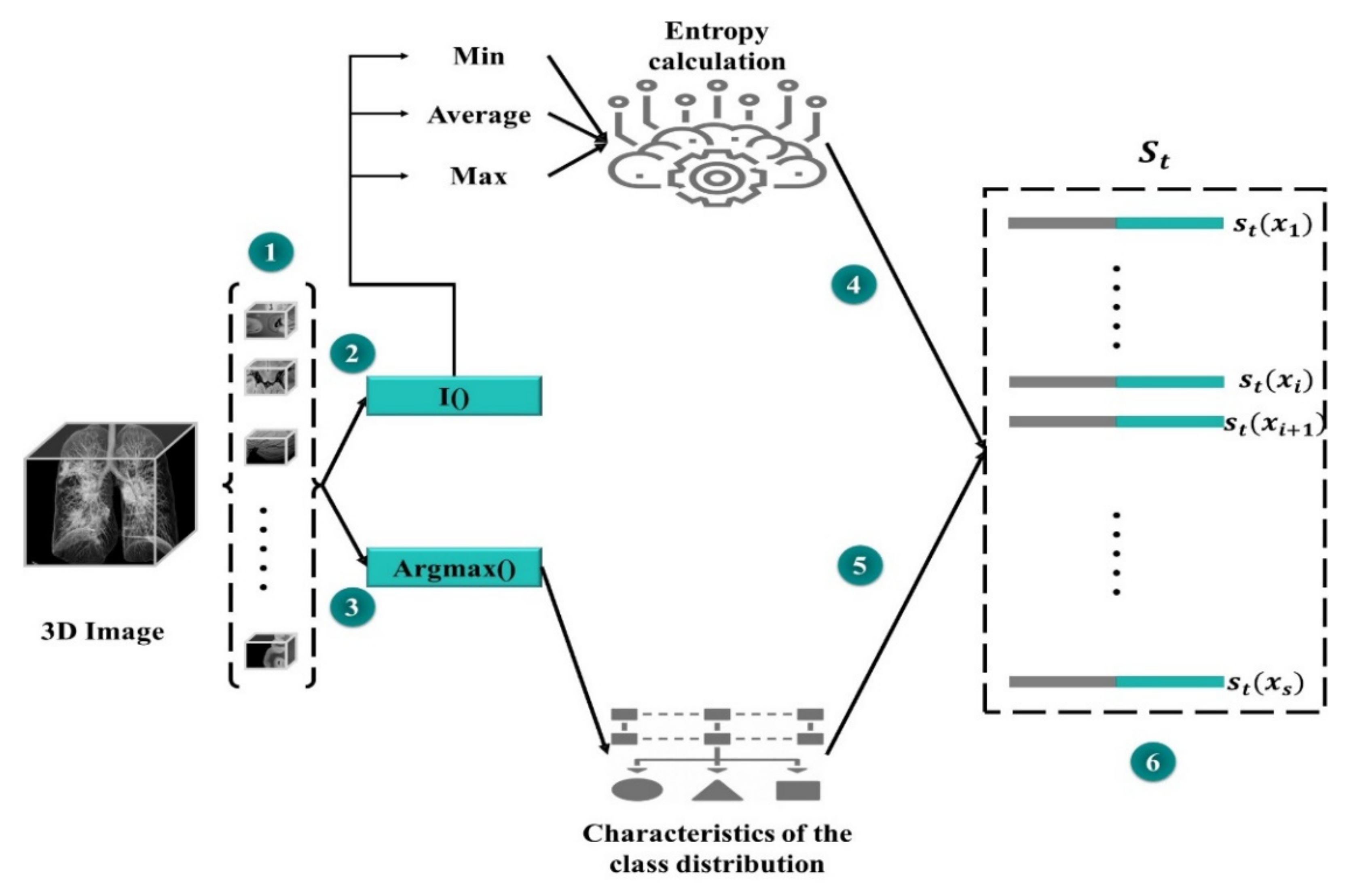
- The state representation is built using the set.
- The state lst is calculated according to the value of the function F iteration t ‘’ and .
- A controlled action space is constructed with group with regions, sampled uniformly from the processed set . For every single region within each cluster, we calculate its sub-actions representation .
- The MD, RL agent, selects K sub-actions using ε-greedy (greedy policy). Respectively, each single sub-action is defined as the selection of a region (on ) to be processed from a set .
- A designation of the region masks is made, then the sets are updated: (adding the new mask images in Mc) and (removing these regions from set D).
- The MD agent is trained through one iteration on the newly added regions .
- RL receives the award rt+1 as the variation in performance between and on .
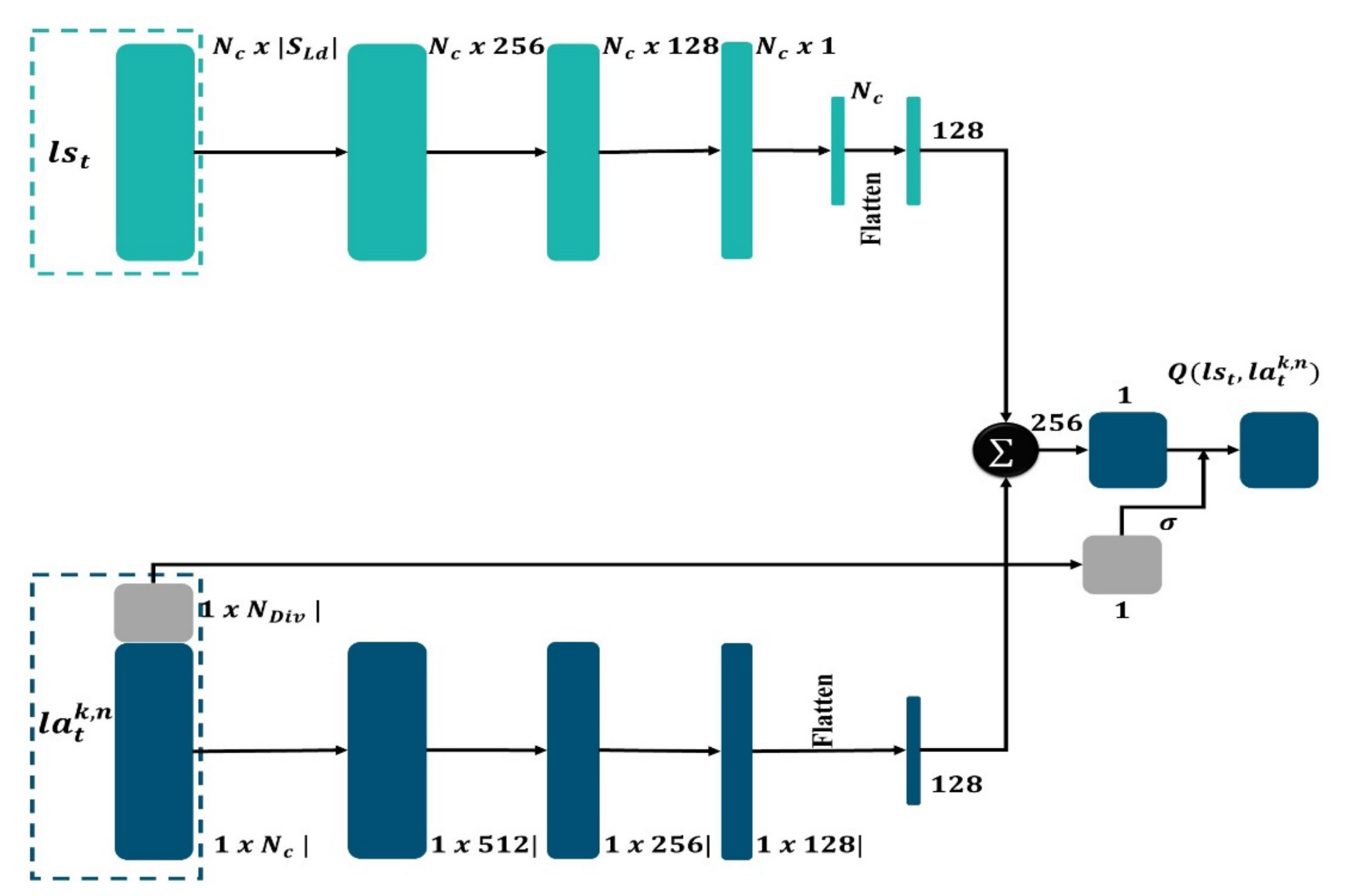
3.7. State Representation
3.8. Action Representation
3.9. Mask-Detector (MD)
4. Results and Discussion
4.1. Dataset Collection
- COVID-19-A: the public TCIA dataset [61] containing many CT lung scans (for non-COVID-19 subjects). These data are used for the training phase. The mask detector studies these images to learn the thoracic characteristics.
- COVID-19-B: The public dataset of Ma et al. [62], which consists of annotated volumes of CT images (COVID-19), for network training. The scans were obtained via the Coronacases initiative and the Radiopaedia database.
- COVID-19-C: publicly available database with thoracic CT images [63]. (These data are used for the test phase.)
- COVID-19-D: A collection of 3D CT images of 10 confirmed COVID-19 cases, which are made available online by the Coronacases initiative [64]. (These data are used for the test phase).
4.2. Evaluation Metrics
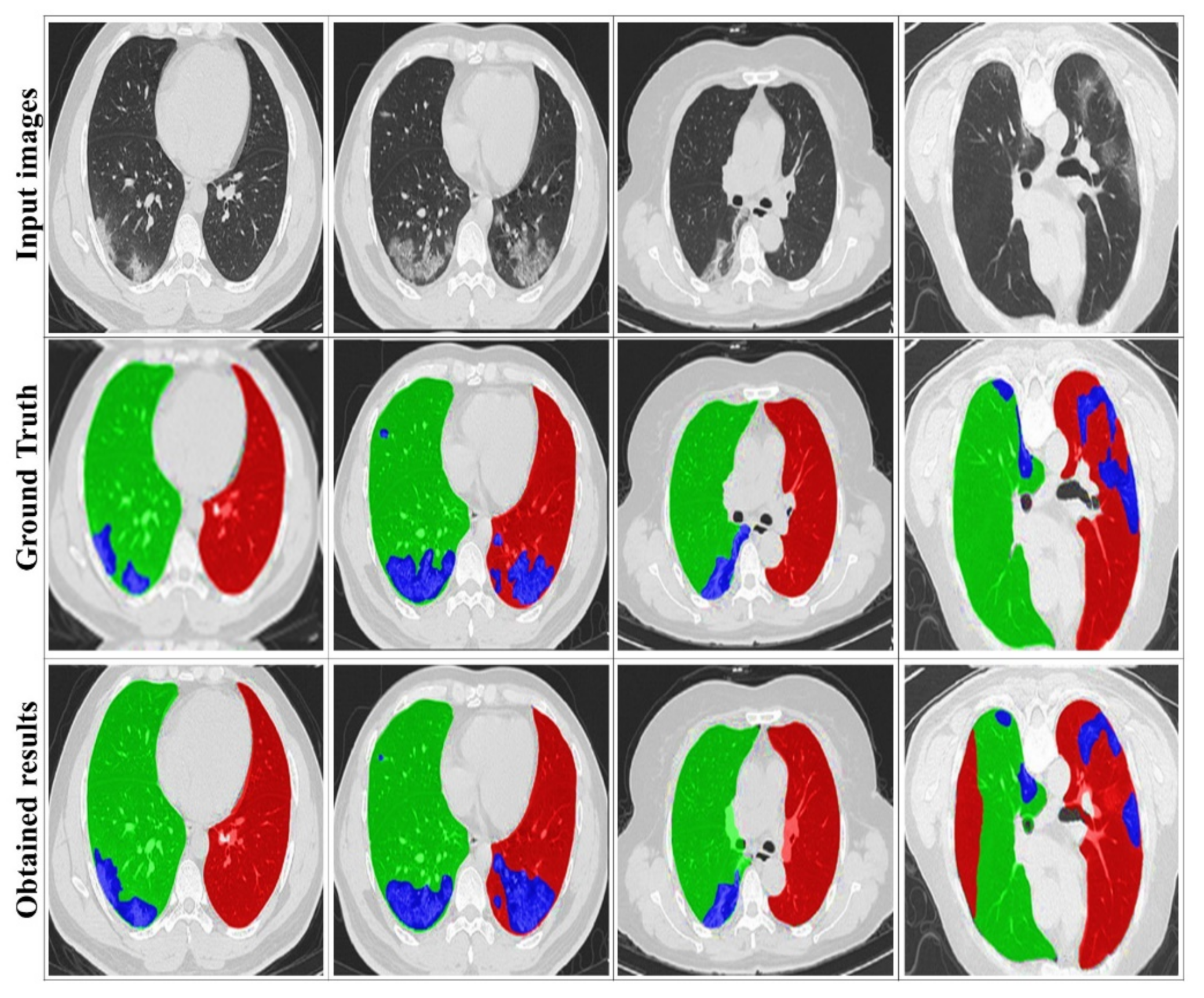
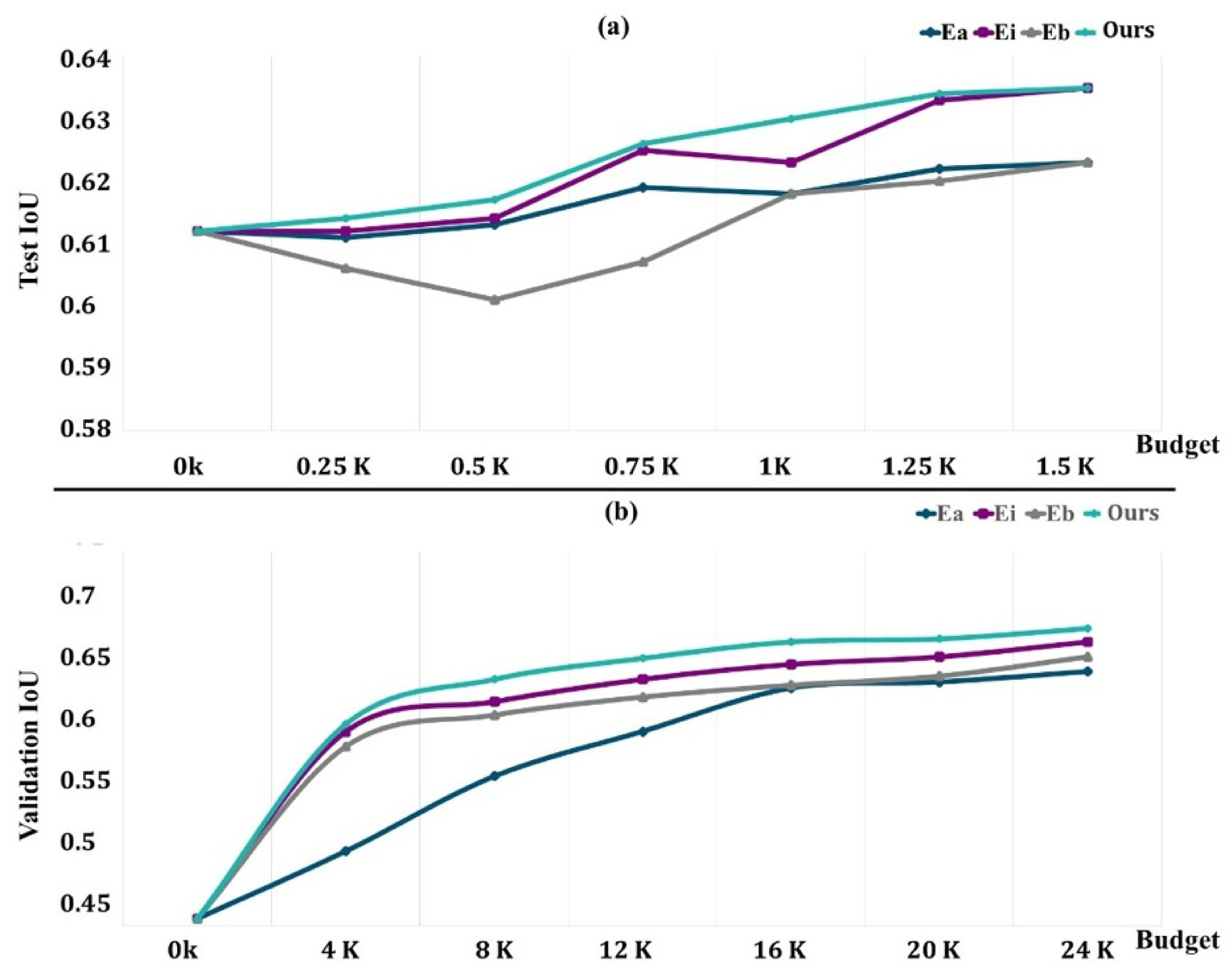
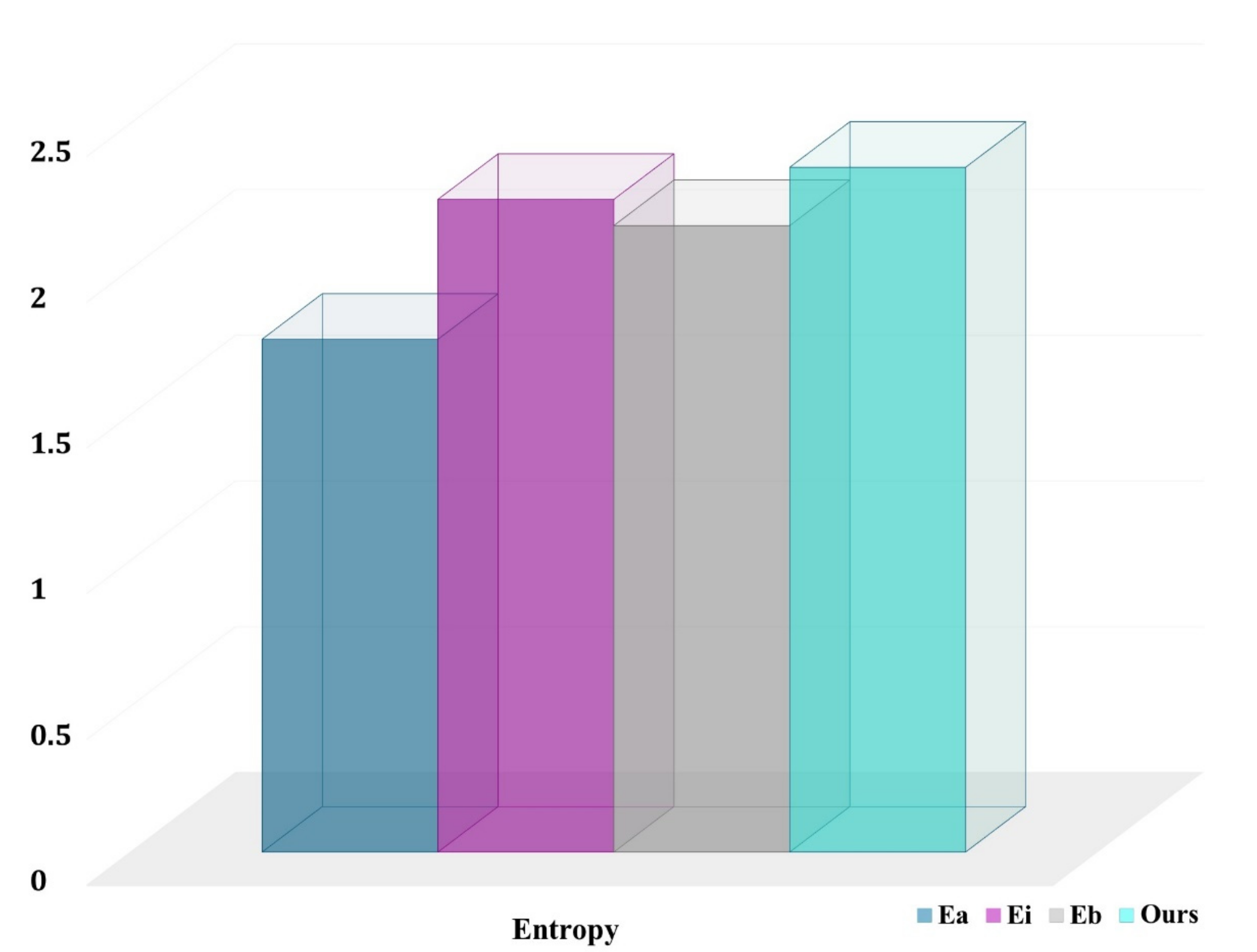
4.3. Evaluation of the DRL System
- is the uniform random selection of areas to be labeled from all potential regions in the dataset at each phase;
- is an uncertainty sampling approach that picks voxel-level areas with the highest cumulative Shannon entropy;
- chooses locations with the highest cumulative BALD measure at the pixel level [65].
4.4. Comparison with State-of-the-Art Methods
5. Conclusions and Future Works
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haque, I.R.I.; Neubert, J. Deep learning approaches to biomedical image segmentation. Inform. Med. Unlocked 2020, 18, 100297. [Google Scholar] [CrossRef]
- Hasoon, J.N.; Fadel, A.H.; Hameed, R.S.; Mostafa, S.A.; Khalaf, B.A.; Mohammed, M.A.; Nedoma, J. COVID-19 anomaly detection and classification method based on supervised machine learning of chest X-ray images. Results Phys. 2021, 31, 105045. [Google Scholar] [CrossRef] [PubMed]
- Muzammil, S.; Maqsood, S.; Haider, S.; Damaševičius, R. CSID: A Novel Multimodal Image Fusion Algorithm for Enhanced Clinical Diagnosis. Diagnostics 2020, 10, 904. [Google Scholar] [CrossRef] [PubMed]
- Alyasseri, Z.A.A.; Al-Betar, M.A.; Abu Doush, I.; Awadallah, M.A.; Abasi, A.K.; Makhadmeh, S.N.; Alomari, O.A.; Abdulkareem, K.H.; Adam, A.; Damasevicius, R.; et al. Review on COVID-19 diagnosis models based on machine learning and deep learning approaches. Expert Syst. 2021, e12759. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Singh, D.; Kaur, M.; Damaševičius, R. Overview of current state of research on the application of artificial intelligence techniques for COVID-19. PeerJ Comput. Sci. 2021, 7, e564. [Google Scholar] [CrossRef] [PubMed]
- Lawin, F.J.; Danelljan, M.; Tosteberg, P.; Bhat, G.; Khan, F.S.; Felsberg, M. Deep Projective 3D Semantic Segmentation. In Proceedings of the Computer Analysis of Images and Patterns: 17th International Conference, Ystad, Sweden, 22–24 August 2017; pp. 95–107. [Google Scholar] [CrossRef] [Green Version]
- Irfan, R.; Almazroi, A.; Rauf, H.; Damaševičius, R.; Nasr, E.; Abdelgawad, A. Dilated Semantic Segmentation for Breast Ultrasonic Lesion Detection Using Parallel Feature Fusion. Diagnostics 2021, 11, 1212. [Google Scholar] [CrossRef] [PubMed]
- Lateef, F.; Ruichek, Y. Survey on semantic segmentation using deep learning techniques. Neurocomputing 2019, 338, 321–348. [Google Scholar] [CrossRef]
- Zhu, H.; Meng, F.; Cai, J.; Lu, S. Beyond pixels: A comprehensive survey from bottom-up to semantic image segmentation and cosegmentation. J. Vis. Commun. Image Represent. 2016, 34, 12–27. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.A.; Rajinikanth, V.; Satapathy, S.C.; Taniar, D.; Mohanty, J.R.; Tariq, U.; Damaševičius, R. VGG19 Network Assisted Joint Segmentation and Classification of Lung Nodules in CT Images. Diagnostics 2021, 11, 2208. [Google Scholar] [CrossRef]
- Zebari, D.A.; Ibrahim, D.A.; Zeebaree, D.Q.; Haron, H.; Salih, M.S.; Damaševičius, R.; Mohammed, M.A. Systematic Review of Computing Approaches for Breast Cancer Detection Based Computer Aided Diagnosis Using Mammogram Images. Appl. Artif. Intell. 2021, 1–47. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. arXiv 2015, arXiv:1505.04597. Available online: https://arxiv.org/abs/1505.04597 (accessed on 15 September 2021).
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Buşoniu, L.; Babuška, R.; De Schutter, B. Multi-Agent Reinforcement Learning: An Overview; Springer International Publishing: Berlin/Heidelberg, Germany, 2010; pp. 183–221. [Google Scholar] [CrossRef]
- Zhang, K.; Yang, Z.; Başar, T. Multi-Agent Reinforcement Learning: A Selective Overview of Theories and Algorithms; Springer International Publishing: Berlin/Heidelberg, Germany, 2021; pp. 321–384. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 3rd International Conference on Learning Representations (ICLR2015), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2014, 39, 640–651. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet Classification with Deep Convolutional Neural Networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Garcia-Garcia, A.; Orts-Escolano, S.; Oprea, S.; Villena-Martinez, V.; Garcia-Rodriguez, J. A review on deep learning techniques applied to semantic segmentation. arXiv 2017, arXiv:1704.06857. Available online: https://arxiv.org/abs/1704.06857 (accessed on 20 September 2021).
- Zhang, W.; Zeng, S.; Wang, D.; Xue, X. Weakly supervised semantic segmentation for social images. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 2718–2726. [Google Scholar]
- Papandreou, G.; Chen, L.; Murphy, K.P.; Yuille, A.L. Weakly-and Semi-Supervised Learning of A Deep Convolutional Net-work for Semantic Image Segmentation. In Proceedings of the 2015 IEEE International Conference on Computer Vision, Santiago, Chile, 11–18 December 2015; pp. 1742–1750. [Google Scholar]
- Vezhnevets, A.; Buhmann, J.M. Towards weakly supervised semantic segmentation by means of multiple instance and multi-task learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), San Francisco, CA, USA, 13–18 June 2010; pp. 3249–3256. [Google Scholar]
- Xu, J.; Schwing, A.G.; Urtasun, R. Tell Me What You See and I Will Show You Where It Is. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Washington, DC, USA, 27 June–2 July 2004; pp. 3190–3197. [Google Scholar]
- Rajchl, M.; Lee, M.C.H.; Oktay, O.; Kamnitsas, K.; Passerat-Palmbach, J.; Bai, W.; Damodaram, M.; Rutherford, M.A.; Hajnal, J.V. Kainz, B.; et al. DeepCut: Object Segmentation From Bounding Box Annotations Using Convolutional Neural Networks. IEEE. Trans. Med. Imaging. 2017, 36, 674–683. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Jayasumana, S.; Romera-Paredes, B.; Vineet, V.; Su, Z.; Du, D.; Huang, C.; Torr, P.H. Conditional random fields as recurrent neural networks. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1529–1537. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Yan, Z.; Zhang, H.; Jia, Y.; Breuel, T.; Yu, Y. Combining the best of convolutional layers and recurrent layers: A hybrid network for semantic segmentation. arXiv 2016, arXiv:1603.04871. Available online: https://arxiv.org/abs/1603.04871 (accessed on 15 September 2021).
- Visin, F.; Romero, A.; Cho, K.; Matteucci, M.; Ciccone, M.; Kastner, K.; Bengio, Y.; Courville, A. ReSeg: A Recurrent Neural Network-Based Model for Semantic Segmentation. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Las Vegas, NV, USA, 28 June–1 July 2016; pp. 426–433. [Google Scholar]
- Pinheiro, P.H.; Collobert, R. Recurrent convolutional neural networks for scene labeling. In Proceedings of the 31st International Conference on Machine Learning (ICML), Beijing, China, 21–26 June 2014. [Google Scholar]
- Hu, Q.; Souza, L.F.D.F.; Holanda, G.B.; Alves, S.S.; Silva, F.H.D.S.; Han, T.; Filho, P.P.R. An effective approach for CT lung segmentation using mask region-based convolutional neural networks. Artif. Intell. Med. 2020, 103, 101792. [Google Scholar] [CrossRef]
- Savelli, B.; Bria, A.; Molinara, M.; Marrocco, C.; Tortorella, F. A multi-context CNN ensemble for small lesion detection. Artif. Intell. Med. 2019, 103, 101749. [Google Scholar] [CrossRef]
- Piantadosi, G.; Sansone, M.; Fusco, R.; Sansone, C. Multi-planar 3D breast segmentation in MRI via deep convolutional neural networks. Artif. Intell. Med. 2019, 103, 101781. [Google Scholar] [CrossRef]
- Kadry, S.; Rajinikanth, V.; Taniar, D.; Damaševičius, R.; Valencia, X.P.B. Automated segmentation of leukocyte from hematological images—A study using various CNN schemes. J. Supercomput. 2021, 1–21. [Google Scholar] [CrossRef]
- Zebari, D.A.; Ibrahim, D.A.; Zeebaree, D.Q.; Mohammed, M.A.; Haron, H.; Zebari, N.A.; Damaševičius, R.; Maskeliūnas, R. Breast Cancer Detection Using Mammogram Images with Improved Multi-Fractal Dimension Approach and Feature Fusion. Appl. Sci. 2021, 11, 12122. [Google Scholar] [CrossRef]
- Jabeen, K.; Khan, M.A.; Alhaisoni, M.; Tariq, U.; Zhang, Y.-D.; Hamza, A.; Mickus, A.; Damaševičius, R. Breast Cancer Classification from Ultrasound Images Using Probability-Based Optimal Deep Learning Feature Fusion. Sensors 2022, 22, 807. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Wang, J.; Shi, J.; Wu, Z.; Wang, Q.; Tang, Z.; He, K.; Shi, Y.; Shen, D. Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation, and Diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020, 14, 4–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Deng, X.; Fu, Q.; Zhou, Q.; Feng, J.; Ma, H.; Liu, W.; Zheng, C. A Weakly-Supervised Framework for COVID-19 Classification and Lesion Localization from Chest CT. IEEE Trans. Med. Imaging 2020, 39, 2615–2625. [Google Scholar] [CrossRef] [PubMed]
- Gozes, O.; Frid-Adar, M.; Greenspan, H.; Browning, P.D.; Zhang, H.; Ji, W.; Bernheim, A.; Siegel, E. Rapid AI Development Cycle for the Coronavirus (COVID-19) Pandemic: Initial Results for Automated Detection & Patient Monitoring Using Deep Learning CT Image Analysis. arXiv 2020, arXiv:2003.05037. Available online: https://arxiv.org/abs/2003.05037 (accessed on 1 July 2021).
- Wang, B.; Jin, S.; Yan, Q.; Xu, H.; Luo, C.; Wei, L.; Zhao, W.; Hou, X.; Ma, W.; Xu, Z.; et al. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system. Appl. Soft Comput. 2020, 98, 106897. [Google Scholar] [CrossRef]
- Li, L.; Qin, L.; Xu, Z.; Yin, Y.; Wang, X.; Kong, B.; Bai, J.; Lu, Y.; Fang, Z.; Song, Q.; et al. Artificial Intelligence Distinguishes COVID-19 from Community Acquired Pneumonia on Chest CT. Radiology 2020, 200905. [Google Scholar] [CrossRef]
- Chen, J.; Wu, L.; Zhang, J.; Zhang, L.; Gong, D.; Zhao, Y.; Chen, Q.; Huang, S.; Yang, M.; Yang, X.; et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020, 10, 19196. [Google Scholar] [CrossRef]
- Akram, T.; Attique, M.; Gul, S.; Shahzad, A.; Altaf, M.; Naqvi, S.S.R.; Damaševičius, R.; Maskeliūnas, R. A novel framework for rapid diagnosis of COVID-19 on computed tomography scans. Pattern Anal. Appl. 2021, 24, 951–964. [Google Scholar] [CrossRef]
- Khan, M.A.; Alhaisoni, M.; Tariq, U.; Hussain, N.; Majid, A.; Damaševičius, R.; Maskeliūnas, R. COVID-19 Case Recognition from Chest CT Images by Deep Learning, Entropy-Controlled Firefly Optimization, and Parallel Feature Fusion. Sensors 2021, 21, 7286. [Google Scholar] [CrossRef]
- Rehman, N.-U.; Zia, M.S.; Meraj, T.; Rauf, H.T.; Damaševičius, R.; El-Sherbeeny, A.M.; El-Meligy, M.A. A Self-Activated CNN Approach for Multi-Class Chest-Related COVID-19 Detection. Appl. Sci. 2021, 11, 9023. [Google Scholar] [CrossRef]
- Mnih, V.; Kavukcuoglu, K.; Silver, D.; Graves, A.; Antonoglou, I.; Wierstra, D.; Riedmiller, M.A. Playing Atari with Deep Reinforcement Learning. arXiv 2013, arXiv:1312.5602. [Google Scholar]
- Zhou, K.; Qiao, Y.; Xiang, T. Deep reinforcement learning for unsupervised video summarization with diversity-representativeness reward. In Proceedings of the AAAI Conference on Artificial Intelligence, Palo Alto, CA, USA, 2–9 February 2018. [Google Scholar]
- Yun, S.; Choi, J.; Yoo, Y.; Yun, K.; Choi, J.Y. Action-Decision Networks for Visual Tracking with Deep Reinforcement Learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Caicedo, J.C.; Lazebnik, S. Active Object Localization with Deep Reinforcement Learning. In Proceedings of the International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 2488–2496. [Google Scholar]
- Han, J.; Yang, L.; Zhang, D.; Chang, X.; Liang, X. Reinforcement cutting-agent learning for video object segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 9080–9089. [Google Scholar]
- Lee, K.M.; Myeong, H.; Song, G. SeedNet: Automatic Seed Generation with Deep Reinforcement Learning for Robust Interactive Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 1760–1768. [Google Scholar] [CrossRef]
- Acuna, D.; Ling, H.; Kar, A.; Fidler, S. Efficient interactive annotation of segmentation datasets with polygon-rnn++. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 859–868. [Google Scholar]
- Rudovic, O.; Zhang, M.; Schuller, B.; Picard, R. Multi-modal Active Learning From Human Data: A Deep Reinforcement Learning Approach. In Proceedings of the 2019 International Conference on Multimodal Interaction, Suzhou, China, 14–18 October 2019; pp. 6–15. [Google Scholar] [CrossRef] [Green Version]
- Yazdani, R.; Ruwase, O.; Zhang, M.; He, Y.; Arnau, J.; González, A. LSTM-Sharp: An Adaptable, Energy-Efficient Hardware Accelerator for Long Short-Term Memory. arXiv 2019, arXiv:1911.01258. Available online: https://arxiv.org/abs/1911.01258 (accessed on 15 September 2021).
- Fang, M.; Li, Y.; Cohn, T. Learning how to active learn: A deep reinforcement learning approach. In Proceedings of the Conference on Empirical Methods in Natural Language Processing, Copenhagen, Denmark, 7–11 September 2017; Association for Computational Linguistics (ACL): Stroudsburg, PA, USA, 2017; pp. 595–605. [Google Scholar]
- Konyushkova, K.; Sznitman, R.; Fua, P. Discovering General-Purpose Active Learning Strategies. arXiv 2018, arXiv:1810.04114. Available online: https://arxiv.org/abs/1810.04114 (accessed on 1 July 2021).
- Boutilier, C. Sequential optimality and coordination in multiagent systems. In Proceedings of the International Joint Conference on Artificial Intelligence (IJCAI), Stockholm, Sweden, 31 July–6 August 1999; pp. 478–485. [Google Scholar]
- Sutton, R.S.; Barto, A.G. Reinforcement Learning: An Introduction; MIT Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Van Hasselt, H.; Guez, A.; Silver, D. Deep reinforcement learning with double Q-learning. In Proceedings of the Thirtieth AAAI Conference on Artificial Intelligence, Phoenix, AZ, USA, 2 March 2016. [Google Scholar]
- Yang, J.; Veeraraghavan, H.; Armato, S.G.; Farahani, K.; Kirby, J.S.; Kalpathy-Kramer, J.; Van Elmpt, W.; Dekker, A.; Han, X.; Feng, X.; et al. Autosegmentation for thoracic radiation treatment planning: A grand challenge at AAPM 2017. Med. Phys. 2018, 45, 4568–4581. [Google Scholar] [CrossRef]
- Ma, J.G. COVID-19 CT Lung and Infection Segmentation Dataset (Version 1.0). 2020. Available online: https://zenodo.org/record/3757476#.Yg5dC4TMJPY (accessed on 12 February 2022).
- MedSeg. COVID-19 CT Segmentation Dataset. Available online: https://htmlsegmentation.s3.eu-north-1.amazonaws.com/index.html (accessed on 12 February 2022).
- Coronacases. Available online: https://coronacases.org (accessed on 12 February 2022).
- Gal, Y.; Islam, R.; Ghahramani, Z. Deep Bayesian Active Learning with Image Data. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 1183–1192. [Google Scholar]


| DataSet | %Slice with Infection |
|---|---|
| COVID-19-A [61] | _ |
| COVID-19-B [62] | 100% |
| COVID-19-C [63] | 44.9% |
| COVID-19-D [64] | 52.3% |
| Metrics | Formulas | Description |
|---|---|---|
| Accuracy (ACC) | The ratio of correctly predicted pixels to the total number of pixels in the processed image. | |
| Precision (Pc) | The ratio of correctly predicted lesion pixels to the total of expected lesion pixels. | |
| Sensitivity (Sen) | The ratio of the correctly predicted lesion pixels to the total number of real lesion pixels. | |
| F1 score (F1) | The ratio obtained from a combination of both precision and sensitivity results. | |
| Specificity (Sp) | The ratio of correctly predicted normal pixels to the total number of actual normal pixels. | |
| Dice coefficient (DC) | The similarity between the method output (Y) and the ground truth (X). | |
| Structural metric (Sm) | Sm = (1 − β).Sos(Sop,Sgt) + β.Sor(Sop,Sgt) | The structural similarity between the prediction map and ground truth mask. |
| Mean Absolute Error (MAE) | ∑wi∑hj|Sop(i,j) − Sgt(i,j)| | Measures the pixel-wise difference. |
| ACC | DC | Sen | Sp | Pc | F1 | Sm | MAE | |
|---|---|---|---|---|---|---|---|---|
| Our approach | 0.9712 | 0.8081 | 0.7997 | 0.9948 | 0.8621 | 0.8301 | 0.8438 | 0.0086 |
| U-Net++ [43] | 0.9687 | 0.7972 | 0.7845 | 0.9952 | 0.8437 | 0.8206 | 0.8623 | 0.0085 |
| COVNet [42] | 0.9698 | 0.7754 | 0.7400 | 0.9959 | 0.8470 | 0.7930 | 0.8334 | 0.0094 |
| DeCoVNet [39] | 0.9697 | 0.8020 | 0.8106 | 0.9962 | 0.8347 | 0.8116 | 0.8511 | 0.0107 |
| AlexNet [20] | 0.8900 | 0.6910 | 0.8110 | 0.9930 | 0.9500 | 0.8062 | 0.8475 | 0.0125 |
| ResNet [18] | 0.8984 | 0.7408 | 0.7608 | 0.9937 | 0.7549 | 0.7558 | 0.8080 | 0.0157 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Allioui, H.; Mohammed, M.A.; Benameur, N.; Al-Khateeb, B.; Abdulkareem, K.H.; Garcia-Zapirain, B.; Damaševičius, R.; Maskeliūnas, R. A Multi-Agent Deep Reinforcement Learning Approach for Enhancement of COVID-19 CT Image Segmentation. J. Pers. Med. 2022, 12, 309. https://doi.org/10.3390/jpm12020309
Allioui H, Mohammed MA, Benameur N, Al-Khateeb B, Abdulkareem KH, Garcia-Zapirain B, Damaševičius R, Maskeliūnas R. A Multi-Agent Deep Reinforcement Learning Approach for Enhancement of COVID-19 CT Image Segmentation. Journal of Personalized Medicine. 2022; 12(2):309. https://doi.org/10.3390/jpm12020309
Chicago/Turabian StyleAllioui, Hanane, Mazin Abed Mohammed, Narjes Benameur, Belal Al-Khateeb, Karrar Hameed Abdulkareem, Begonya Garcia-Zapirain, Robertas Damaševičius, and Rytis Maskeliūnas. 2022. "A Multi-Agent Deep Reinforcement Learning Approach for Enhancement of COVID-19 CT Image Segmentation" Journal of Personalized Medicine 12, no. 2: 309. https://doi.org/10.3390/jpm12020309
APA StyleAllioui, H., Mohammed, M. A., Benameur, N., Al-Khateeb, B., Abdulkareem, K. H., Garcia-Zapirain, B., Damaševičius, R., & Maskeliūnas, R. (2022). A Multi-Agent Deep Reinforcement Learning Approach for Enhancement of COVID-19 CT Image Segmentation. Journal of Personalized Medicine, 12(2), 309. https://doi.org/10.3390/jpm12020309









