COVID-19 Detection in CT/X-ray Imagery Using Vision Transformers
Abstract
1. Introduction
- (1)
- Unlike previous deep learning frameworks that only used one type of data, this work uses both CXR and CT images.
- (2)
- For the automatic detection and classification of COVID-19, we propose a new approach based on Vision Transformer (ViT).
- (3)
- The development of a Siamese encoder that employs a distillation technique to classify original and augmented images.
2. Related Work
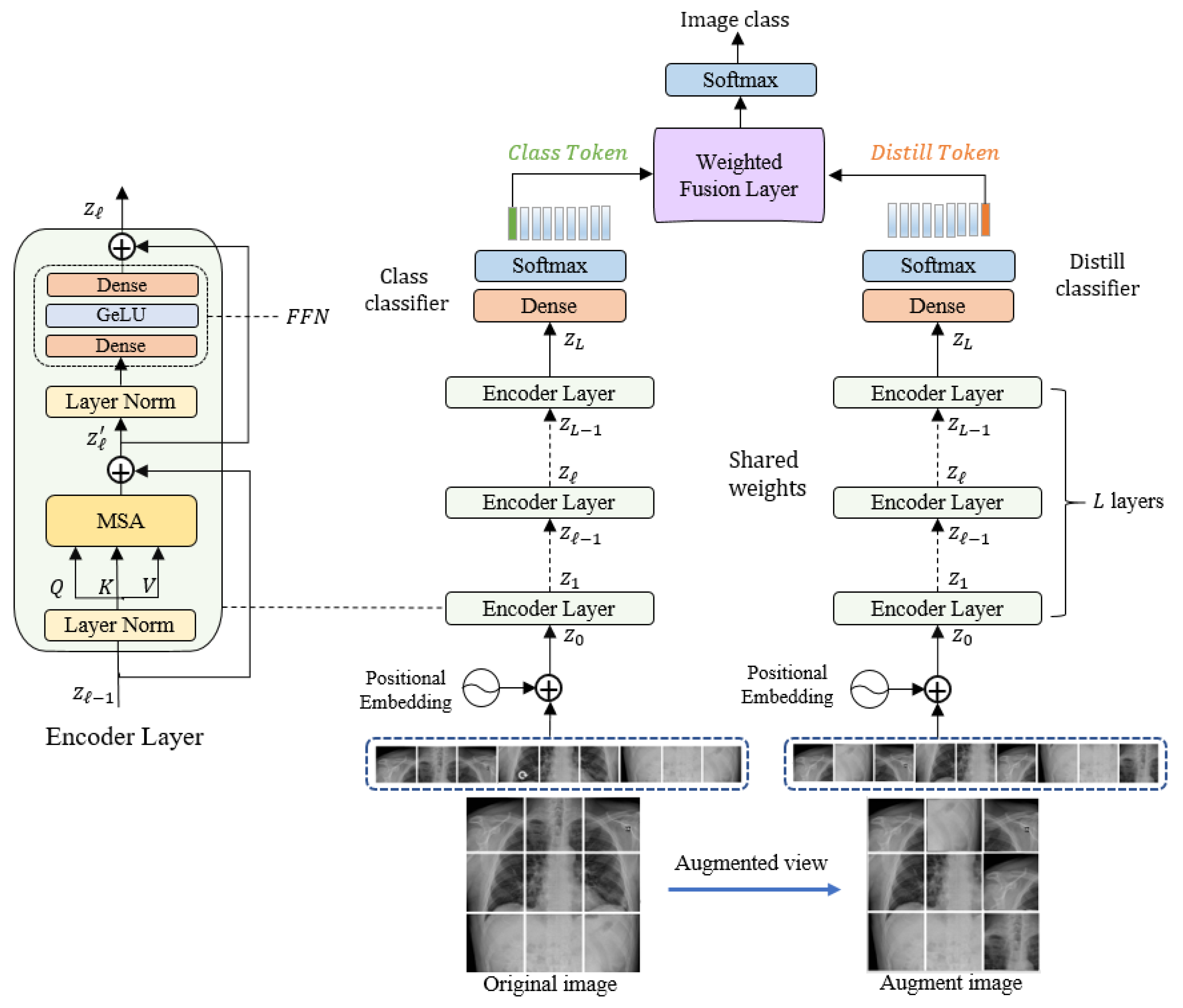
3. Methodology
3.1. Linear Embeddimg Layer
3.2. Siamese Encoder Module
3.3. Classification Layer
3.4. Network Optimization
| Algorithm 1: Main steps for training and testing the model. |
| Input: Training set of n chest images with corresponding ground-truth labels. Output: test images predicted class labels
|
4. Experiments
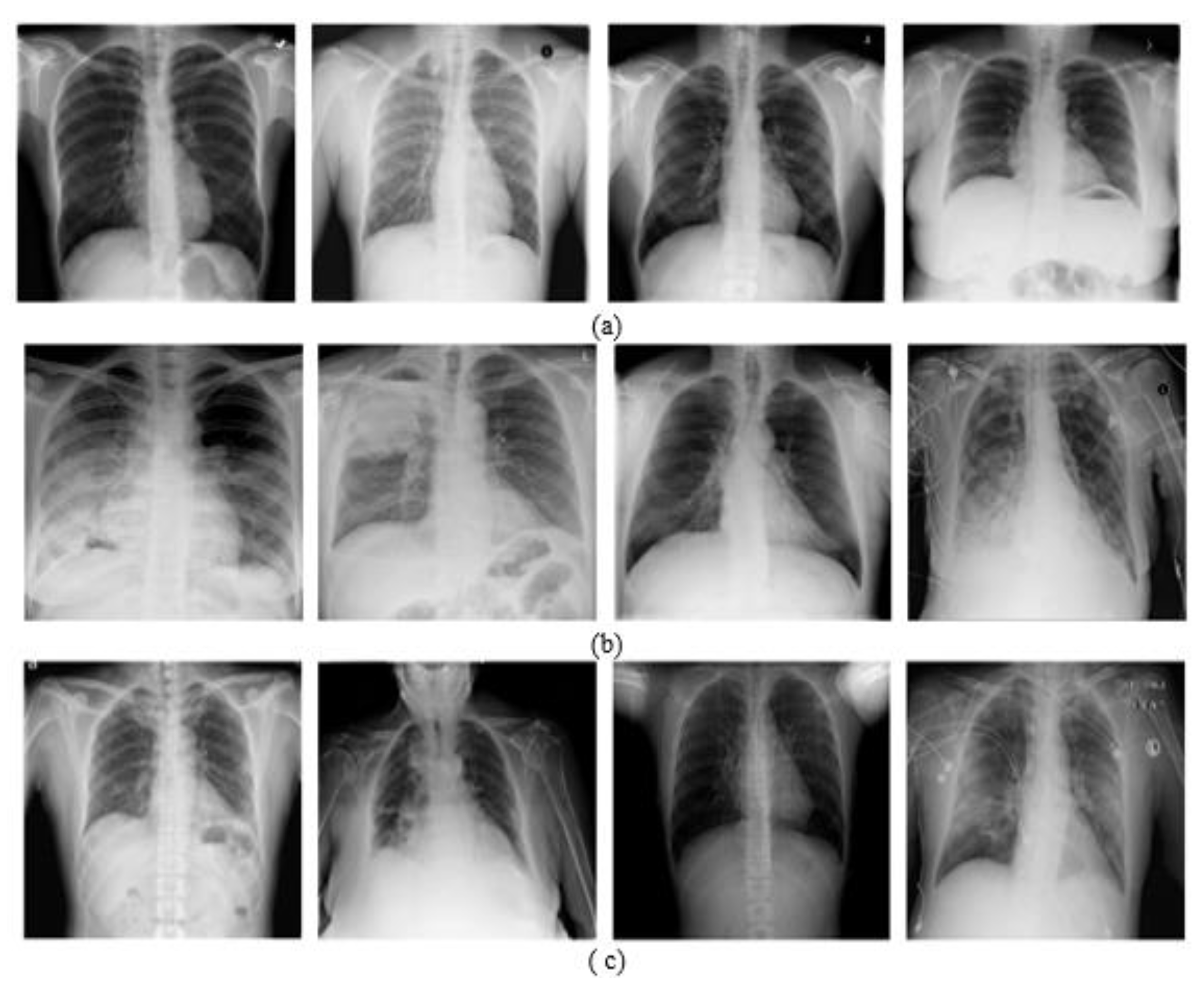
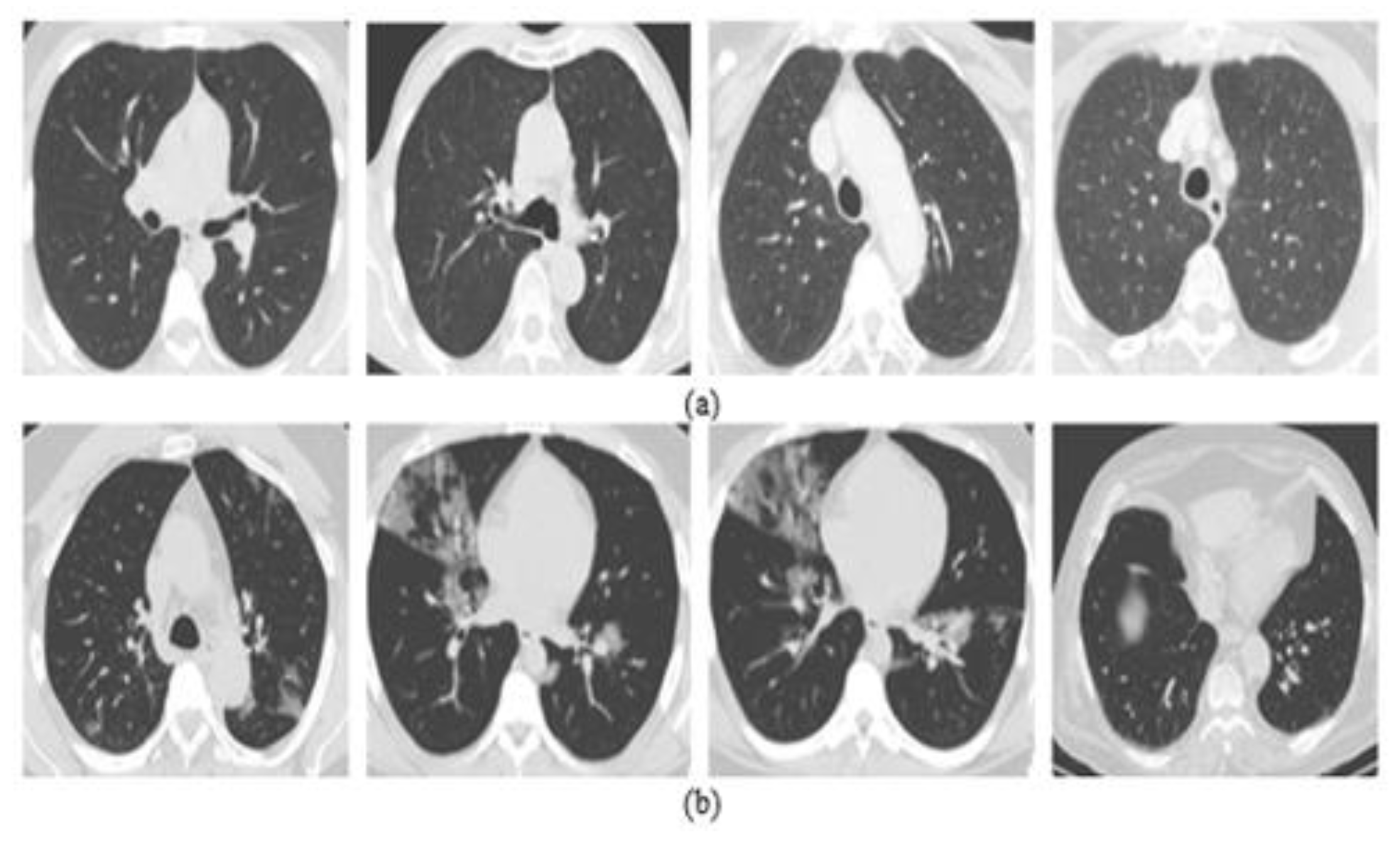
4.1. Datasets Description
4.1.1. Chest X-ray Dataset
4.1.2. Chest X-ray Dataset
4.2. Evaluation Measures
4.3. Experimental Setup
5. Results
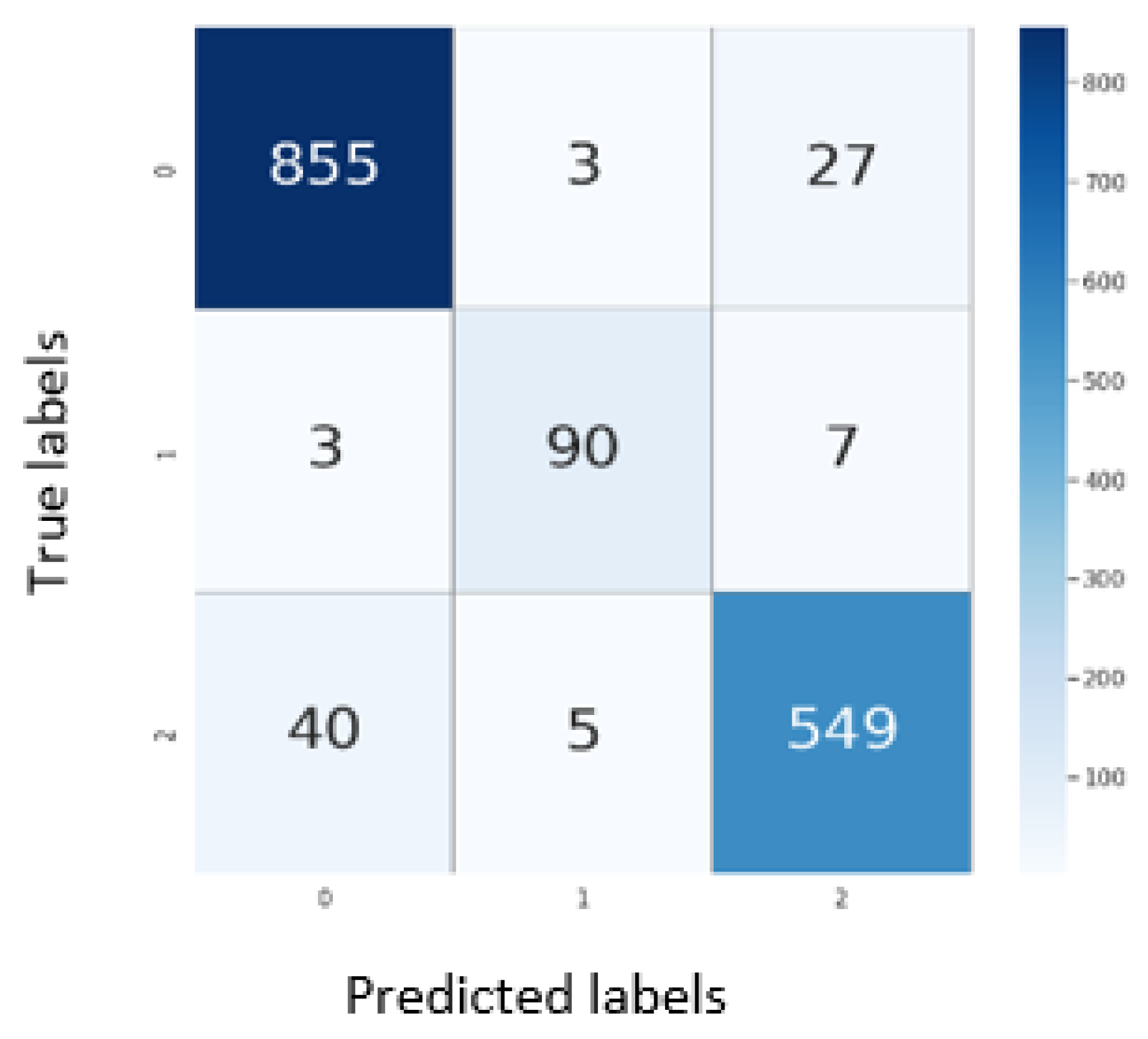
5.1. Results on CXR
5.2. Results on CT
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Jacofsky, D.; Jacofsky, E.M.; Jacofsky, M. Understanding Antibody Testing for COVID-19. J. Arthroplast. 2020, 35, S74–S81. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Ardebili, A. Real-Time RT-PCR in COVID-19 Detection: Issues Affecting the Results. Expert Rev. Mol. Diagn. 2020, 20, 453–454. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Xu, D.; Ye, G.; Xia, C.; Wang, S.; Li, Y.; Xu, H. Positive RT-PCR Test Results in Patients Recovered From COVID-19. JAMA 2020, 323, 1502. [Google Scholar] [CrossRef]
- False-Negative Results of Initial RT-PCR Assays for COVID-19: A Systematic Review. Available online: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0242958 (accessed on 14 February 2021).
- Loeffelholz, M.J.; Tang, Y.-W. Laboratory Diagnosis of Emerging Human Coronavirus Infections—The State of the Art. Emerg. Microbes Infect. 2020, 9, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Eskandarian, A. A Survey and Tutorial of EEG-Based Brain Monitoring for Driver State Analysis. IEEE/CAA J. Autom. Sin. 2021, 8, 1222–1242. [Google Scholar] [CrossRef]
- Rahhal, M.M.A.; Bazi, Y.; AlHichri, H.; Alajlan, N.; Melgani, F.; Yager, R.R. Deep Learning Approach for Active Classification of Electrocardiogram Signals. Inf. Sci. 2016, 345, 340–354. [Google Scholar] [CrossRef]
- Kumar, A.; Kim, J.; Lyndon, D.; Fulham, M.; Feng, D. An Ensemble of Fine-Tuned Convolutional Neural Networks for Medical Image Classification. IEEE J. Biomed. Health Inform. 2017, 21, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, K.; Sivaswamy, J. Segmentation of Retinal Cysts from Optical Coherence Tomography Volumes via Selective Enhancement. IEEE J. Biomed. Health Inform. 2019, 23, 273–282. [Google Scholar] [CrossRef]
- Vásquez-Correa, J.C.; Arias-Vergara, T.; Orozco-Arroyave, J.R.; Eskofier, B.; Klucken, J.; Nöth, E. Multimodal Assessment of Parkinson’s Disease: A Deep Learning Approach. IEEE J. Biomed. Health Inform. 2019, 23, 1618–1630. [Google Scholar] [CrossRef]
- Borghesi, A.; Zigliani, A.; Golemi, S.; Carapella, N.; Maculotti, P.; Farina, D.; Maroldi, R. Chest X-ray Severity Index as a Predictor of in-Hospital Mortality in Coronavirus Disease 2019: A Study of 302 Patients from Italy. Int. J. Infect. Dis. 2020, 96, 291–293. [Google Scholar] [CrossRef]
- Cozzi, D.; Albanesi, M.; Cavigli, E.; Moroni, C.; Bindi, A.; Luvarà, S.; Lucarini, S.; Busoni, S.; Mazzoni, L.N.; Miele, V. Chest X-ray in New Coronavirus Disease 2019 (COVID-19) Infection: Findings and Correlation with Clinical Outcome. Radiol. Med. 2020, 125, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Marggrander, D.T.; Borgans, F.; Jacobi, V.; Neb, H.; Wolf, T. Lung Ultrasound Findings in Patients with COVID-19. SN Compr. Clin. Med. 2020, 2, 2151–2157. [Google Scholar] [CrossRef] [PubMed]
- Ultrasound on the Frontlines of COVID-19: Report from an International Webinar—Liu—2020—Academic Emergency Medicine—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/full/10.1111/acem.14004 (accessed on 21 March 2021).
- Radpour, A.; Bahrami-Motlagh, H.; Taaghi, M.T.; Sedaghat, A.; Karimi, M.A.; Hekmatnia, A.; Haghighatkhah, H.-R.; Sanei-Taheri, M.; Arab-Ahmadi, M.; Azhideh, A. COVID-19 Evaluation by Low-Dose High Resolution CT Scans Protocol. Acad. Radiol. 2020, 27, 901. [Google Scholar] [CrossRef] [PubMed]
- Contribution of CT Features in the Diagnosis of COVID-19. Available online: https://www.hindawi.com/journals/crj/2020/1237418/ (accessed on 21 March 2021).
- Sverzellati, N.; Ryerson, C.J.; Milanese, G.; Renzoni, E.A.; Volpi, A.; Spagnolo, P.; Bonella, F.; Comelli, I.; Affanni, P.; Veronesi, L.; et al. Chest X-ray or CT for COVID-19 Pneumonia? Comparative Study in a Simulated Triage Setting. Eur. Respir. 2021, 58, 2004188. [Google Scholar] [CrossRef] [PubMed]
- Borakati, A.; Perera, A.; Johnson, J.; Sood, T. Diagnostic Accuracy of X-ray versus CT in COVID-19: A Propensity-Matched Database Study. BMJ Open 2020, 10, e042946. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, D.; Ahuja, K.; Grover, H.; Sharma, P.; Solanki, S.; Gupta, N.; Patel, L. Review of X-ray and Computed Tomography Scan Findings with a Promising Role of Point of Care Ultrasound in COVID-19 Pandemic. World J. Radiol. 2020, 12, 195–203. [Google Scholar] [CrossRef]
- Ross, N.E.; Pritchard, C.J.; Rubin, D.M.; Dusé, A.G. Automated Image Processing Method for the Diagnosis and Classification of Malaria on Thin Blood Smears. Med. Bio. Eng. Comput. 2006, 44, 427–436. [Google Scholar] [CrossRef]
- Walter, T.; Klein, J.; Massin, P.; Erginay, A. A Contribution of Image Processing to the Diagnosis of Diabetic Retinopathy-Detection of Exudates in Color Fundus Images of the Human Retina. IEEE Trans. Med. Imaging 2002, 21, 1236–1243. [Google Scholar] [CrossRef]
- Singh, A.; Dutta, M.K.; ParthaSarathi, M.; Uher, V.; Burget, R. Image Processing Based Automatic Diagnosis of Glaucoma Using Wavelet Features of Segmented Optic Disc from Fundus Image. Comput. Methods Programs Biomed. 2016, 124, 108–120. [Google Scholar] [CrossRef]
- Veropoulos, K.; Campbell, C.; Learmonth, G. Image Processing and Neural Computing Used in the Diagnosis of Tuberculosis. In Proceedings of the IEE Colloquium on Intelligent Methods in Healthcare and Medical Applications (Digest No. 1998/514), IET, York, UK, 20 October 1998. [Google Scholar] [CrossRef]
- Sadoughi, F.; Kazemy, Z.; Hamedan, F.; Owji, L.; Rahmanikatigari, M.; Azadboni, T.T. Artificial Intelligence Methods for the Diagnosis of Breast Cancer by Image Processing: A Review. Breast Cancer Targets Ther. 2018, 10, 219–230. [Google Scholar] [CrossRef]
- Gola Isasi, A.; García Zapirain, B.; Méndez Zorrilla, A. Melanomas Non-Invasive Diagnosis Application Based on the ABCD Rule and Pattern Recognition Image Processing Algorithms. Comput. Biol. Med. 2011, 41, 742–755. [Google Scholar] [CrossRef] [PubMed]
- Image Processing for Computer-aided Diagnosis of Lung Cancer by CT (LSCT)—Yamamoto—1994—Systems and Computers in Japan—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/scj.4690250207 (accessed on 21 March 2021).
- Stoitsis, J.; Valavanis, I.; Mougiakakou, S.G.; Golemati, S.; Nikita, A.; Nikita, K.S. Computer Aided Diagnosis Based on Medical Image Processing and Artificial Intelligence Methods. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2006, 569, 591–595. [Google Scholar] [CrossRef]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Akagi, M.; Nakamura, Y.; Higaki, T.; Narita, K.; Honda, Y.; Zhou, J.; Yu, Z.; Akino, N.; Awai, K. Deep Learning Reconstruction Improves Image Quality of Abdominal Ultra-High-Resolution CT. Eur. Radiol. 2019, 29, 6163–6171. [Google Scholar] [CrossRef]
- Nardelli, P.; Jimenez-Carretero, D.; Bermejo-Pelaez, D.; Washko, G.R.; Rahaghi, F.N.; Ledesma-Carbayo, M.J.; Estépar, R.S.J. Pulmonary Artery–Vein Classification in CT Images Using Deep Learning. IEEE Trans. Med Imaging 2018, 37, 2428–2440. [Google Scholar] [CrossRef]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-End Lung Cancer Screening with Three-Dimensional Deep Learning on Low-Dose Chest Computed Tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef]
- AnatomyNet: Deep Learning for Fast and Fully Automated Whole-volume Segmentation of Head and Neck Anatomy—Zhu—2019—Medical Physics—Wiley Online Library. Available online: https://aapm.onlinelibrary.wiley.com/doi/full/10.1002/mp.13300 (accessed on 23 February 2021).
- Deep Learning at Chest Radiography: Automated Classification of Pulmonary Tuberculosis by Using Convolutional Neural Networks|Radiology. Available online: https://pubs.rsna.org/doi/10.1148/radiol.2017162326 (accessed on 23 February 2021).
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Kanne, J.P.; Little, B.P.; Chung, J.H.; Elicker, B.M.; Ketai, L.H. Essentials for Radiologists on COVID-19: An Update—Radiology Scientific Expert Panel. Radiology 2020, 296, E113–E114. [Google Scholar] [CrossRef]
- Kanne, J.P.; Bai, H.; Bernheim, A.; Chung, M.; Haramati, L.B.; Kallmes, D.F.; Little, B.P.; Rubin, G.D.; Sverzellati, N. COVID-19 Imaging: What We Know Now and What Remains Unknown. Radiology 2021, 299, E262–E279. [Google Scholar] [CrossRef]
- Schmitt, W.; Marchiori, E. Covid-19: Round and Oval Areas of Ground-Glass Opacity. Pulmonology 2020, 26, 246–247. [Google Scholar] [CrossRef]
- Roberts, M.; Driggs, D.; Thorpe, M.; Gilbey, J.; Yeung, M.; Ursprung, S.; Aviles-Rivero, A.I.; Etmann, C.; McCague, C.; Beer, L.; et al. Common Pitfalls and Recommendations for Using Machine Learning to Detect and Prognosticate for COVID-19 Using Chest Radiographs and CT Scans. Nat. Mach. Intell. 2021, 3, 199–217. [Google Scholar] [CrossRef]
- Automatic X-ray COVID-19 Lung Image Classification System Based on Multi-Level Thresholding and Support Vector Machine|MedRxiv. Available online: https://www.medrxiv.org/content/10.1101/2020.03.30.20047787v1 (accessed on 21 January 2021).
- Barstugan, M.; Ozkaya, U.; Ozturk, S. Coronavirus (COVID-19) Classification Using CT Images by Machine Learning Methods. arXiv 2020, arXiv:2003.09424. [Google Scholar]
- Marques, G.; Agarwal, D.; de la Torre Díez, I. Automated Medical Diagnosis of COVID-19 through EfficientNet Convolutional Neural Network. Appl. Soft Comput. 2020, 96, 106691. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.Z.; Islam, M.M.; Asraf, A. A Combined Deep CNN-LSTM Network for the Detection of Novel Coronavirus (COVID-19) Using X-ray Images. Inform. Med. Unlocked 2020, 20, 100412. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Wang, S.; Wang, J.; Liu, J.; Jin, Q.; Sun, L. Multiscale Attention Guided Network for COVID-19 Diagnosis Using Chest X-ray Images. IEEE J. Biomed. Health Inform. 2021, 25, 1336–1346. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.K.; Singh, A. Diagnosis of COVID-19 from Chest X-ray Images Using Wavelets-Based Depthwise Convolution Network. Big Data Min. Anal. 2021, 4, 84–93. [Google Scholar] [CrossRef]
- Calderon-Ramirez, S.; Yang, S.; Moemeni, A.; Colreavy-Donnelly, S.; Elizondo, D.A.; Oala, L.; Rodríguez-Capitán, J.; Jiménez-Navarro, M.; López-Rubio, E.; Molina-Cabello, M.A. Improving Uncertainty Estimation With Semi-Supervised Deep Learning for COVID-19 Detection Using Chest X-ray Images. IEEE Access 2021, 9, 85442–85454. [Google Scholar] [CrossRef]
- Tang, S.; Wang, C.; Nie, J.; Kumar, N.; Zhang, Y.; Xiong, Z.; Barnawi, A. EDL-COVID: Ensemble Deep Learning for COVID-19 Case Detection From Chest X-ray Images. IEEE Trans. Ind. Inform. 2021, 17, 6539–6549. [Google Scholar] [CrossRef]
- Improving the Performance of CNN to Predict the Likelihood of COVID-19 Using Chest X-ray Images with Preprocessing Algorithms—Google Search. Available online: https://www.google.com/search?q=Improving+the+performance+of+CNN+to+predict+the+likelihood+of+COVID-19+using+chest+X-ray+images+with+preprocessing+algorithms&rlz=1C1CHBD_enSA940SA940&oq=Improving+the+performance+of+CNN+to+predict+the+likelihood+of+COVID-19+using+chest+X-ray+images+with+preprocessing+algorithms&aqs=chrome..69i57.182j0j4&sourceid=chrome&ie=UTF-8 (accessed on 21 March 2021).
- Abbas, A.; Abdelsamea, M.M.; Gaber, M.M. Classification of COVID-19 in Chest X-ray Images Using DeTraC Deep Convolutional Neural Network. Appl. Intell. 2021, 51, 854–864. [Google Scholar] [CrossRef]
- Stephanie, S.; Shum, T.; Cleveland, H.; Challa, S.R.; Herring, A.; Jacobson, F.L.; Hatabu, H.; Byrne, S.C.; Shashi, K.; Araki, T.; et al. Determinants of Chest X-ray Sensitivity for COVID-19: A Multi-Institutional Study in the United States. Radiol. Cardiothorac. Imaging 2020, 2, e200337. [Google Scholar] [CrossRef]
- Benmalek, E.; Elmhamdi, J.; Jilbab, A. Comparing CT Scan and Chest X-ray Imaging for COVID-19 Diagnosis. Biomed. Eng. Adv. 2021, 1, 100003. [Google Scholar] [CrossRef] [PubMed]
- Amyar, A.; Modzelewski, R.; Li, H.; Ruan, S. Multi-Task Deep Learning Based CT Imaging Analysis for COVID-19 Pneumonia: Classification and Segmentation. Comput. Biol. Med. 2020, 126, 104037. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Jiang, X.; Ma, C.; Du, P.; Li, X.; Lv, S.; Yu, L.; Ni, Q.; Chen, Y.; Su, J.; et al. A Deep Learning System to Screen Novel Coronavirus Disease 2019 Pneumonia. Engineering 2020, 6, 1122–1129. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Mo, Z.; Yan, F.; Xia, L.; Shan, F.; Ding, Z.; Song, B.; Gao, W.; Shao, W.; Shi, F.; et al. Adaptive Feature Selection Guided Deep Forest for COVID-19 Classification With Chest CT. IEEE J. Biomed. Health Inform. 2020, 24, 2798–2805. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.; Chung, H.; Kang, W.S.; Kim, K.W.; Shin, Y.; Kang, S.J.; Lee, J.H.; Kim, Y.J.; Kim, N.Y.; Jung, H.; et al. COVID-19 Pneumonia Diagnosis Using a Simple 2D Deep Learning Framework With a Single Chest CT Image: Model Development and Validation. J. Med. Internet Res. 2020, 22, e19569. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-H.; Gao, S.-H.; Mei, J.; Xu, J.; Fan, D.-P.; Zhao, C.-W.; Cheng, M.-M. JCS: An Explainable COVID-19 Diagnosis System by Joint Classification and Segmentation. arXiv 2020, arXiv:2004.07054. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, H.; Loew, M.; Ko, H. COVID-19 CT Image Synthesis With a Conditional Generative Adversarial Network. IEEE J. Biomed. Health Inform. 2021, 25, 441–452. [Google Scholar] [CrossRef]
- COVID-19 Detection Through Transfer Learning Using Multimodal Imaging Data—IEEE Journals & Magazine. Available online: https://ieeexplore.ieee.org/document/9167243 (accessed on 21 January 2021).
- Das, K.M.; Alkoteesh, J.A.; Al Kaabi, J.; Al Mansoori, T.; Winant, A.J.; Singh, R.; Paraswani, R.; Syed, R.; Sharif, E.M.; Balhaj, G.B.; et al. Comparison of Chest Radiography and Chest CT for Evaluation of Pediatric COVID-19 Pneumonia: Does CT Add Diagnostic Value? Pediatric Pulmonol. 2021, 56, 1409–1418. [Google Scholar] [CrossRef]
- Wang, Y.X.J.; Liu, W.-H.; Yang, M.; Chen, W. The Role of CT for Covid-19 Patient’s Management Remains Poorly Defined. Ann. Transl. Med. 2020, 8, 145. [Google Scholar] [CrossRef]
- López-Cabrera, J.D.; Orozco-Morales, R.; Portal-Diaz, J.A.; Lovelle-Enríquez, O.; Pérez-Díaz, M. Current Limitations to Identify COVID-19 Using Artificial Intelligence with Chest X-ray Imaging. Health Technol. 2021, 11, 411–424. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. Adv. Neural Inf. Process. Syst. 2017, 30, 5998–6008. [Google Scholar]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. BERT: Pre-Training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), Association for Computational Linguistics, Minneapolis, MN, USA, 2–7 June 2019; pp. 4171–4186. [Google Scholar]
- Hendrycks, D.; Gimpel, K. Gaussian Error Linear Units (GELUs). arXiv 2020, arXiv:1606.08415. [Google Scholar]
- Khalifa, N.E.M.; Taha, M.H.N.; Hassanien, A.E.; Elghamrawy, S. Detection of Coronavirus (COVID-19) Associated Pneumonia Based on Generative Adversarial Networks and a Fine-Tuned Deep Transfer Learning Model Using Chest X-ray Dataset. arXiv 2020, arXiv:2004.01184. [Google Scholar]
- Loey, M.; Smarandache, F.; Khalifa, N.E.M. Within the Lack of Chest COVID-19 X-ray Dataset: A Novel Detection Model Based on GAN and Deep Transfer Learning. Symmetry 2020, 12, 651. [Google Scholar] [CrossRef]
- Mobiny, A.; Cicalese, P.A.; Zare, S.; Yuan, P.; Abavisani, M.; Wu, C.C.; Ahuja, J.; de Groot, P.M.; Van Nguyen, H. Radiologist-Level COVID-19 Detection Using CT Scans with Detail-Oriented Capsule Networks. arXiv 2020, arXiv:2004.07407. [Google Scholar]
- Al Rahhal, M.; Bazi, Y.; Jomaa, R.M.; Zuair, M.; Alajlan, N. Deep Learning Approach for COVID-19 Detection in Computed Tomography Images. Cmc Comput. Mater. Contin. 2021, 67, 2093–2110. [Google Scholar] [CrossRef]
- DeVries, T.; Taylor, G.W. Improved Regularization of Convolutional Neural Networks with Cutout. arXiv 2017, arXiv:1708.04552. [Google Scholar]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. Mixup: Beyond Empirical Risk Minimization. arXiv 2018, arXiv:1710.09412. [Google Scholar]
- Yun, S.; Han, D.; Chun, S.; Oh, S.J.; Yoo, Y.; Choe, J. CutMix: Regularization Strategy to Train Strong Classifiers with Localizable Features. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Korea, 27 October–2 November 2019; pp. 6022–6031. [Google Scholar]
- Wang, L.; Lin, Z.Q.; Wong, A. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-ray Images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef]
- Cohen, J.P.; Morrison, P.; Dao, L. COVID-19 Image Data Collection. arXiv Prepr. 2020, arXiv:2003.11597. [Google Scholar]
- Chung, A. Figure 1 COVID-19 Chest X-ray Data Initiative. 2020. Available online: https://github.com/agchung/Figure1-COVID-chestxray-dataset (accessed on 1 November 2021).
- Chung, A. Actualmed COVID-19 Chest X-ray Data Initiative. 2020. Available online: https://github.com/agchung/Actualmed-COVID-chestxray-dataset (accessed on 1 November 2021).
- Radiological Society of North America. RSNA Pneumonia Detection Challenge. 2018. Available online: https://kaggle.com/c/rsna-pneumonia-detection-challenge (accessed on 1 November 2021).
- COVID-19 Radiography Database. 2020. Available online: https://kaggle.com/tawsifurrahman/covid19-radiography-database (accessed on 1 November 2021).
- Soares, E.; Angelov, P.; Biaso, S.; Froes, M.H.; Abe, D.K. SARS-CoV-2 CT-Scan Dataset: A Large Dataset of Real Patients CT Scans for SARS-CoV-2 Identification. medRxiv 2020. [Google Scholar] [CrossRef]
- Šimundić, A.-M. Measures of Diagnostic Accuracy: Basic Definitions. EJIFCC 2009, 19, 203–211. [Google Scholar] [PubMed]
- Zhuang, J.; Tang, T.; Ding, Y.; Tatikonda, S.; Dvornek, N.; Papademetris, X.; Duncan, J.S. AdaBelief Optimizer: Adapting Stepsizes by the Belief in Observed Gradients. arXiv 2020, arXiv:2010.07468. [Google Scholar]
- Bernheim, A.; Mei, X.; Huang, M.; Yang, Y.; Fayad, Z.A.; Zhang, N.; Diao, K.; Lin, B.; Zhu, X.; Li, K.; et al. Chest CT Findings in Coronavirus Disease-19 (COVID-19): Relationship to Duration of Infection. Radiology 2020, 295, 200463. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.; Luz, E.; Silva, G.; Moreira, G.; Silva, R.; Lucio, D.; Menotti, D. COVID-19 Detection in CT Images with Deep Learning: A Voting-Based Scheme and Cross-Datasets Analysis. Inform. Med. Unlocked 2020, 20, 100427. [Google Scholar] [CrossRef]
- Pathak, Y.; Shukla, P.K.; Arya, K.V. Deep Bidirectional Classification Model for COVID-19 Disease Infected Patients. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 18, 1234–1241. [Google Scholar] [CrossRef]
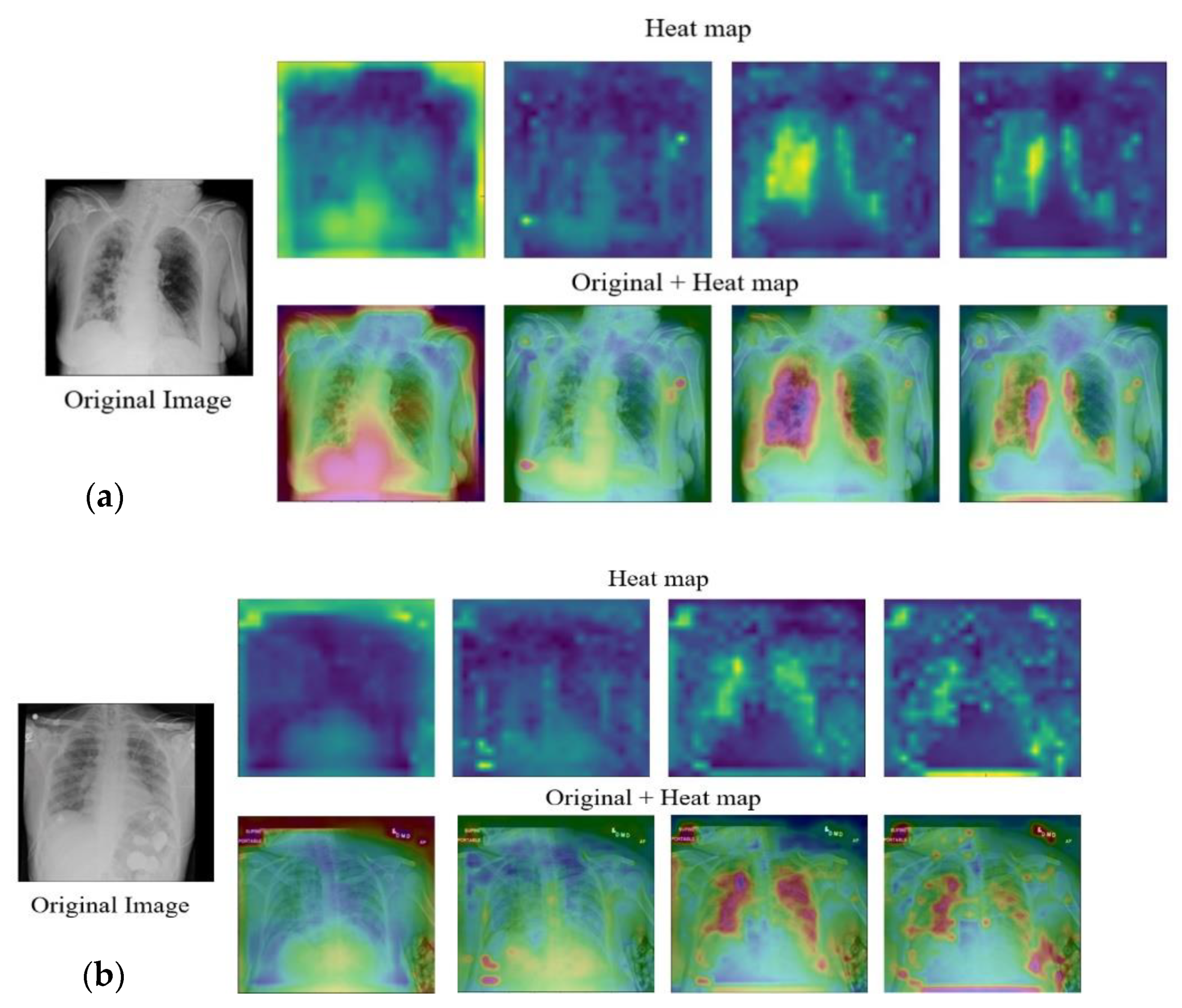
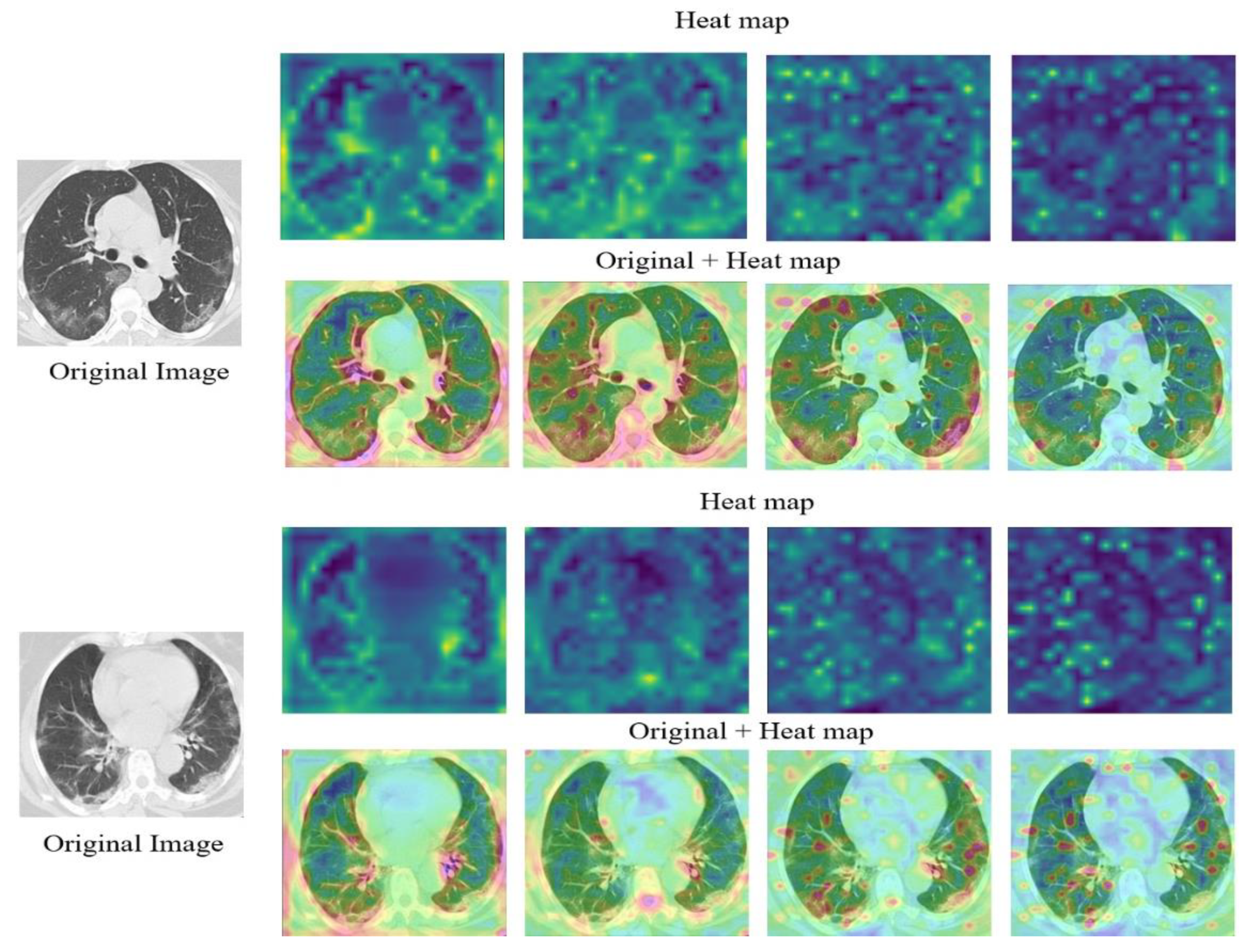
| Normal | Pneumonia | COVID-19 | |
|---|---|---|---|
| Train | 66 | 5438 | 258 |
| Test | 100 | 100 | 100 |
| Total | 8066 | 5538 | 358 |
| Overall | Per Class | |||
|---|---|---|---|---|
| Normal | COVID-19 | Pneumonia | ||
| Accuracy | 94.62 | 96.61 | 90 | 92.42 |
| Precision | 96.77 | 95.21 | 92.84 | 94.17 |
| Recall | 96.77 | 97.61 | 90 | 92.42 |
| Specificity | 99.65 | 93.7 | 99.43 | 96.53 |
| F1 | 96.77 | 95.91 | 90.91 | 93.29 |
| Trial 1 | Trial 2 | Trial 3 | Avg ± sd. | |
|---|---|---|---|---|
| Accuracy | 99.09 | 99.19 | 99.59 | 99.29 ± 0.26 |
| Precision | 98.57 | 99.39 | 99.41 | 99.12 ± 0.48 |
| Recall | 99.58 | 98.98 | 99.8 | 99.45 ± 0.42 |
| Specificity | 98.61 | 99.39 | 99.38 | 99.13 ± 0.45 |
| F1 | 99.08 | 99.18 | 99.68 | 99.31 ± 0.32 |
| Trial 1 | Trial 2 | Trial 3 | Avg ± sd. | |
|---|---|---|---|---|
| Accuracy | 99.40 | 98.99 | 98.99 | 99.13 ± 0.23 |
| Precision | 98.77 | 99.60 | 100.00 | 99.46 ± 0.63 |
| Recall | 100.00 | 98.41 | 99.5 | 98.82 ± 1.04 |
| Specificity | 99.82 | 99.59 | 100.00 | 99.47 ± 0.6 |
| F1 | 99.38 | 99.00 | 99.01 | 99.13 ± 0.22 |
| Trial 1 | Trial 2 | Trial 3 | Avg ± sd. | |
|---|---|---|---|---|
| Accuracy | 99.55 | 99.01 | 99.55 | 99.37 ± 0.31 |
| Precision | 99.6 | 98.18 | 99.6 | 99.13 ± 0.82 |
| Recall | 99.5 | 99.79 | 99.5 | 99.6 ± 0.17 |
| Specificity | 99.6 | 98.28 | 99.6 | 99.16 ± 0.76 |
| F1 | 99.55 | 98.98 | 99.55 | 99.36 ± 0.33 |
| Training-to-Testing Ratio (%) | Accuracy | Precision | Recall | F1 | |
|---|---|---|---|---|---|
| Alrahhal et al. [67] | 80:20 | 99.24 | 99.16 | 99.25 | 99.21 |
| Soares et al. [77] | 97.38 | 99.16 | 95.53 | 97.31 | |
| Silva et al. [81] | 98.99 | 99.20 | 98.80 | 98.99 | |
| Proposed | 99.13 | 99.46 | 98.82 | 99.13 | |
| Alrahhal et al. [67] | 60:40 | 98.65 | 97.81 | 99.41 | 98.60 |
| Pathak et al. [82] | 98.37 | 98.74 | 98.87 | 98.14 | |
| Proposed | 99.29 | 99.12 | 99.45 | 99.31 | |
| Alrahhal et al. [67] | 20:80 | 96.16 | 96.90 | 95.41 | 96.15 |
| Proposed | 99.37 | 99.13 | 99.60 | 99.36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Rahhal, M.M.; Bazi, Y.; Jomaa, R.M.; AlShibli, A.; Alajlan, N.; Mekhalfi, M.L.; Melgani, F. COVID-19 Detection in CT/X-ray Imagery Using Vision Transformers. J. Pers. Med. 2022, 12, 310. https://doi.org/10.3390/jpm12020310
Al Rahhal MM, Bazi Y, Jomaa RM, AlShibli A, Alajlan N, Mekhalfi ML, Melgani F. COVID-19 Detection in CT/X-ray Imagery Using Vision Transformers. Journal of Personalized Medicine. 2022; 12(2):310. https://doi.org/10.3390/jpm12020310
Chicago/Turabian StyleAl Rahhal, Mohamad Mahmoud, Yakoub Bazi, Rami M. Jomaa, Ahmad AlShibli, Naif Alajlan, Mohamed Lamine Mekhalfi, and Farid Melgani. 2022. "COVID-19 Detection in CT/X-ray Imagery Using Vision Transformers" Journal of Personalized Medicine 12, no. 2: 310. https://doi.org/10.3390/jpm12020310
APA StyleAl Rahhal, M. M., Bazi, Y., Jomaa, R. M., AlShibli, A., Alajlan, N., Mekhalfi, M. L., & Melgani, F. (2022). COVID-19 Detection in CT/X-ray Imagery Using Vision Transformers. Journal of Personalized Medicine, 12(2), 310. https://doi.org/10.3390/jpm12020310










