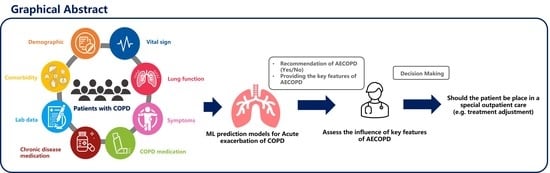
Explainable Machine Learning Model for Predicting First-Time Acute Exacerbation in Patients with Chronic Obstructive Pulmonary Disease
Abstract
:1. Introduction
2. Materials and Methods
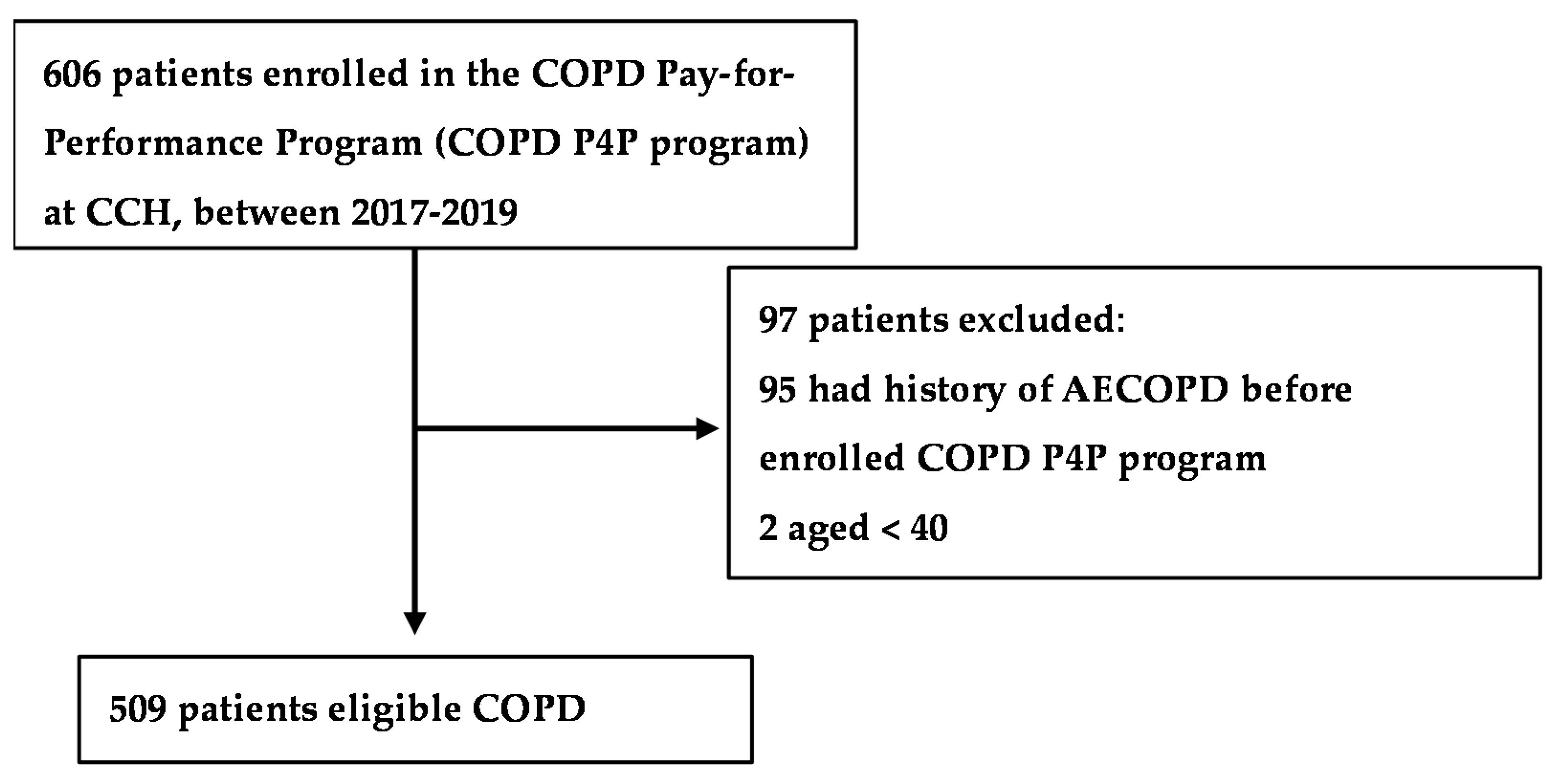
2.1. Study Participants
2.2. Outcome
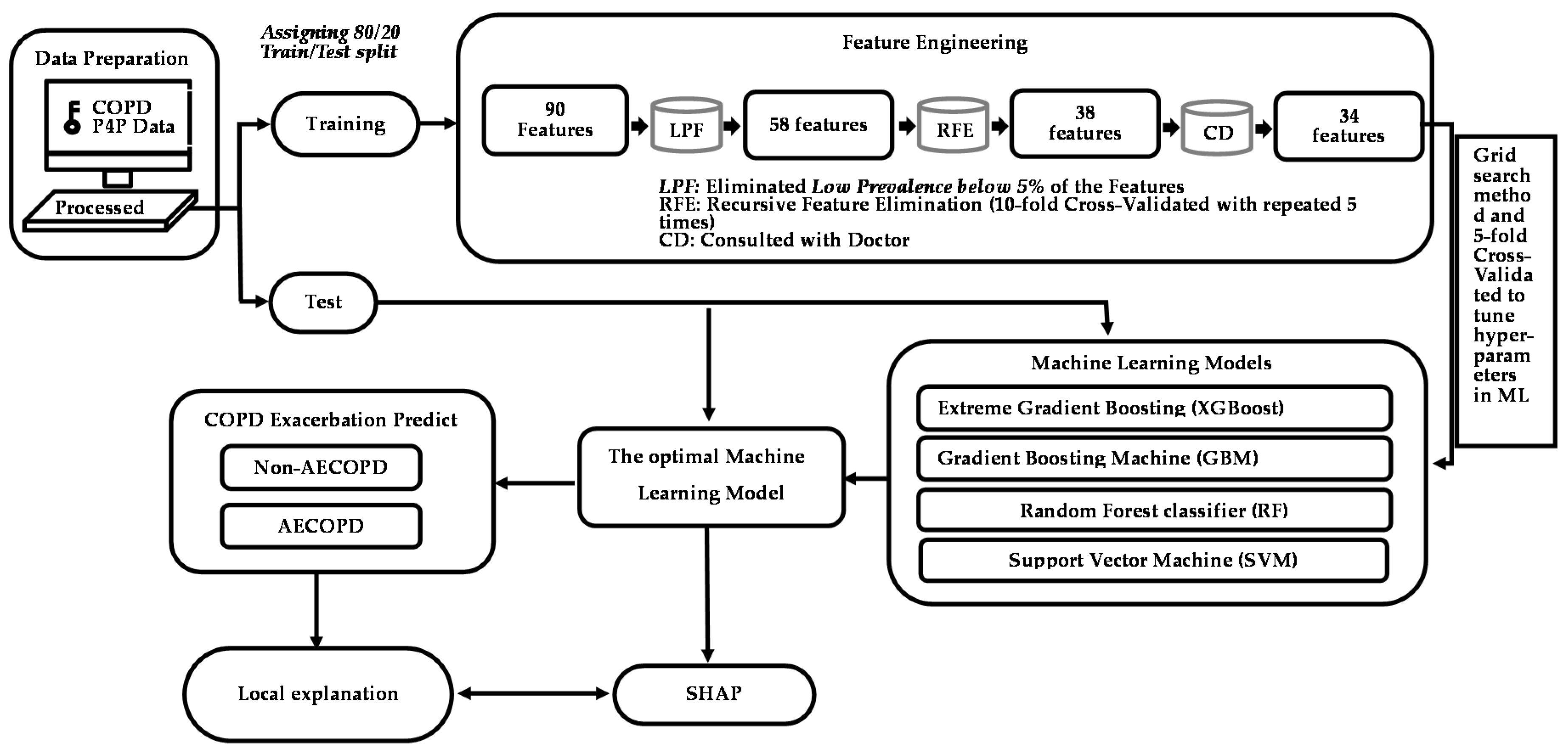
2.3. Feature Engineering
2.4. Statistical Analysis and ML Algorithms
3. Results
3.1. Study Population Characteristics
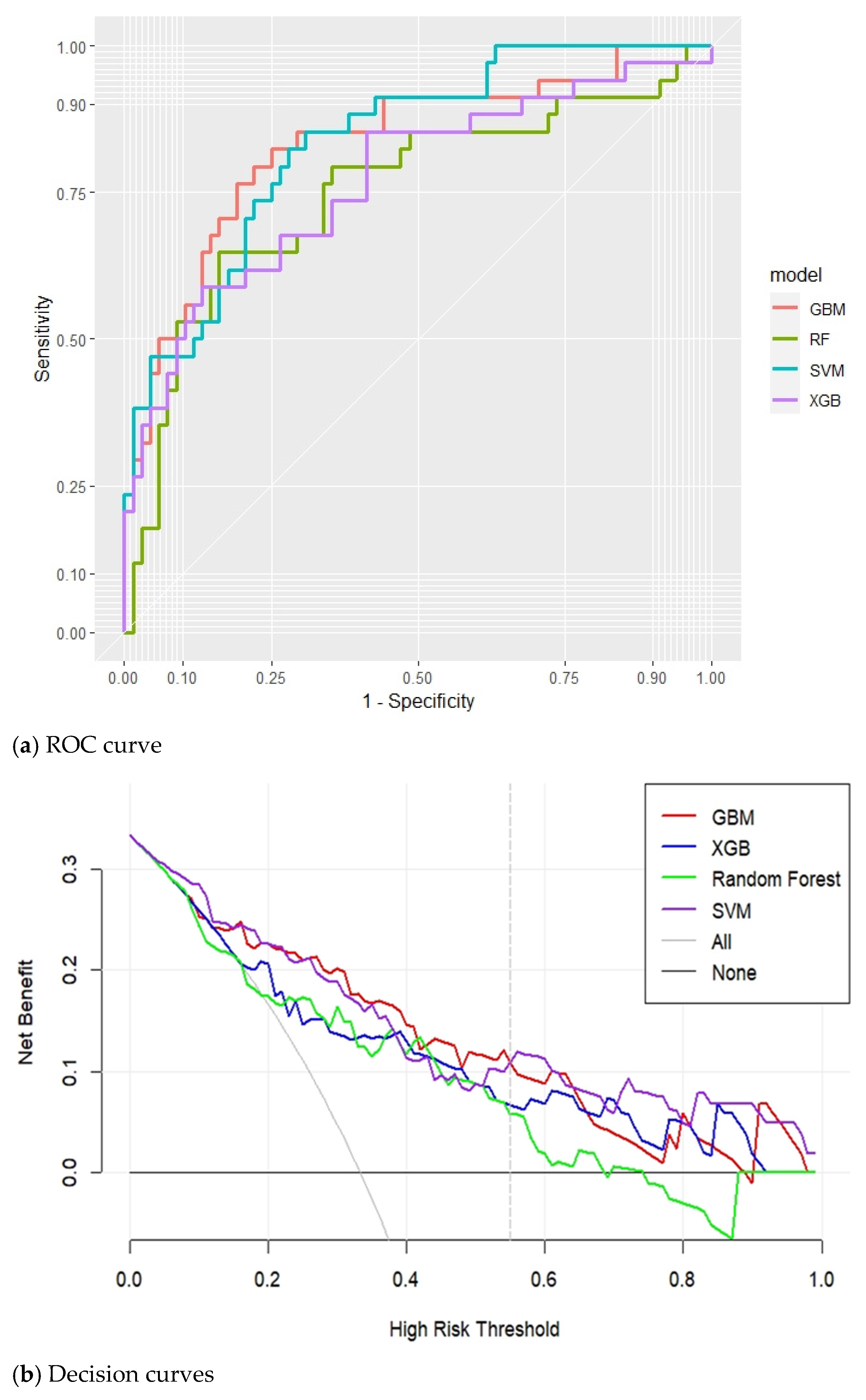
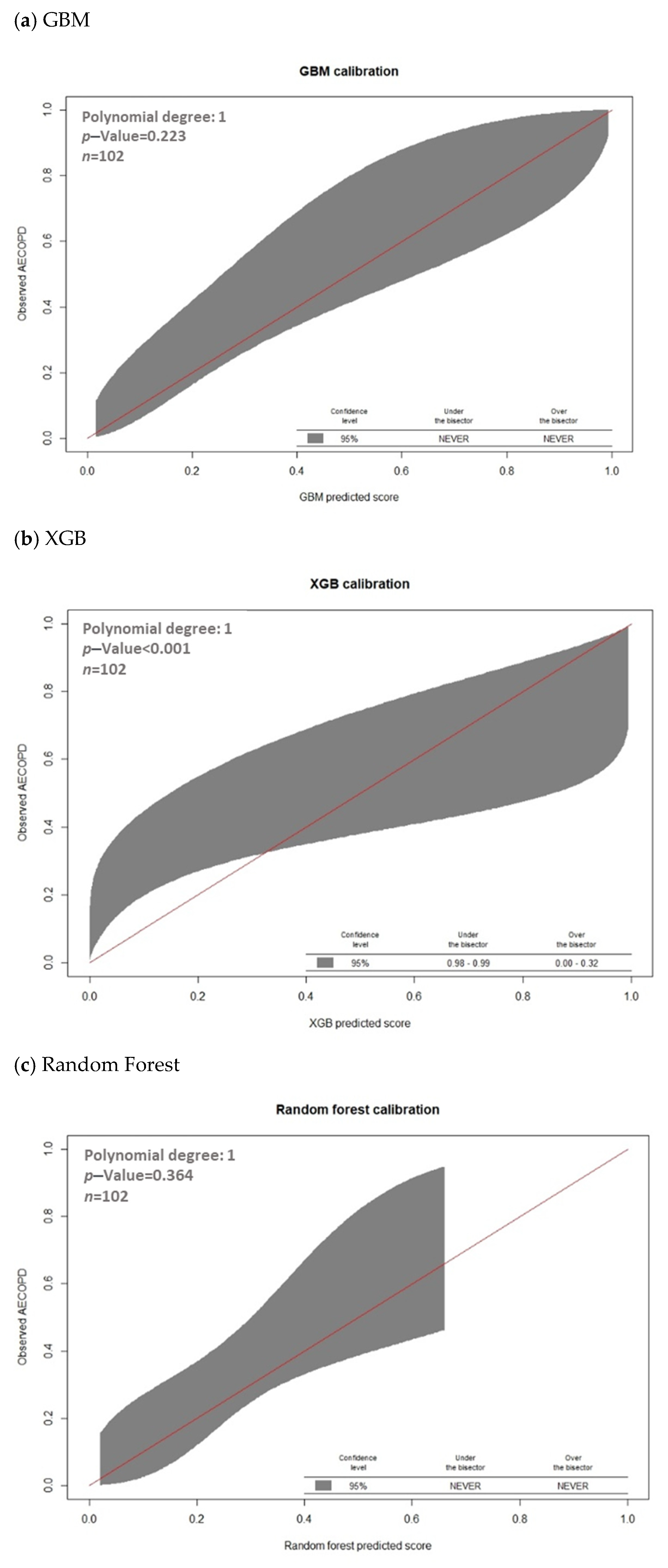
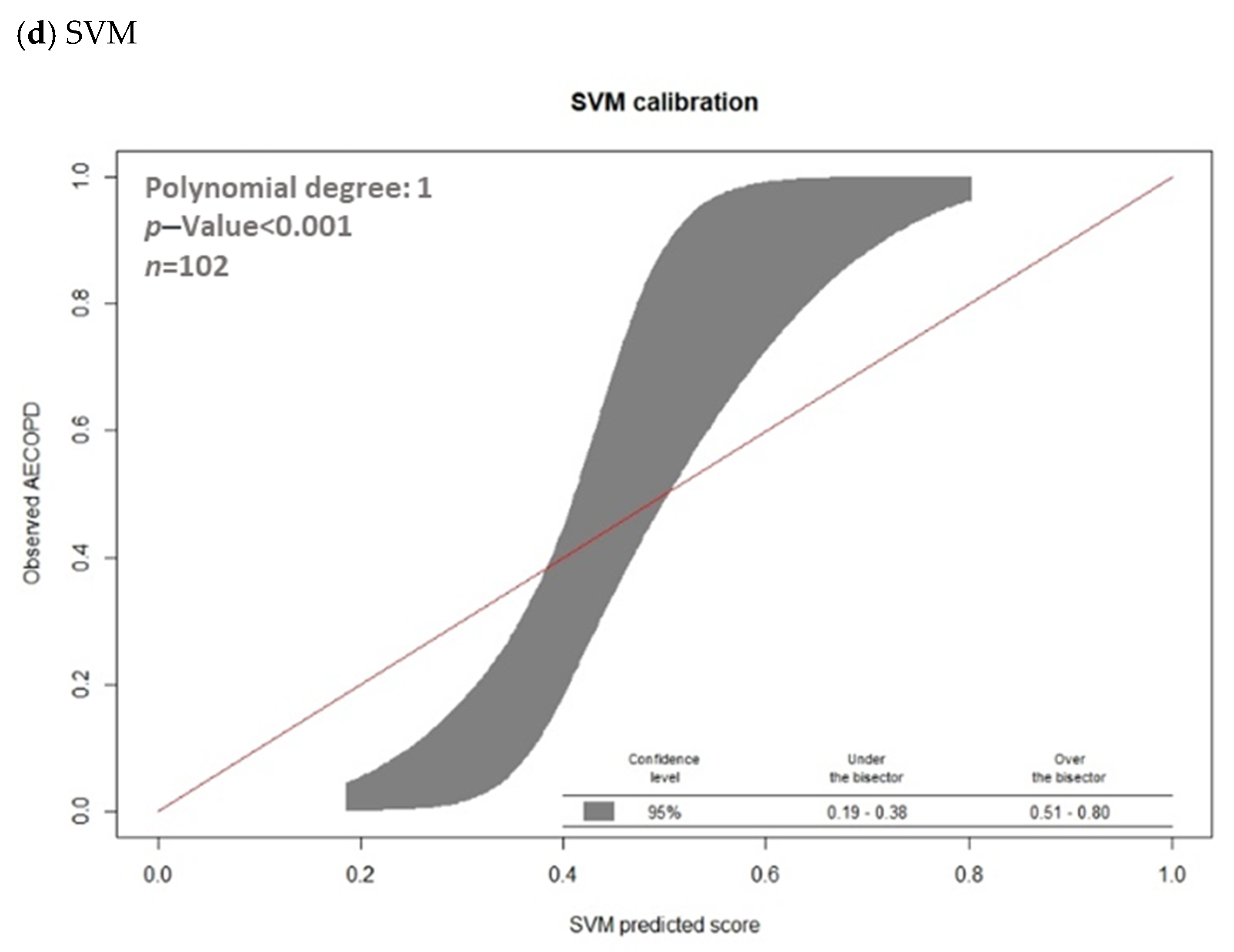
3.2. Model Prediction of AECOPD
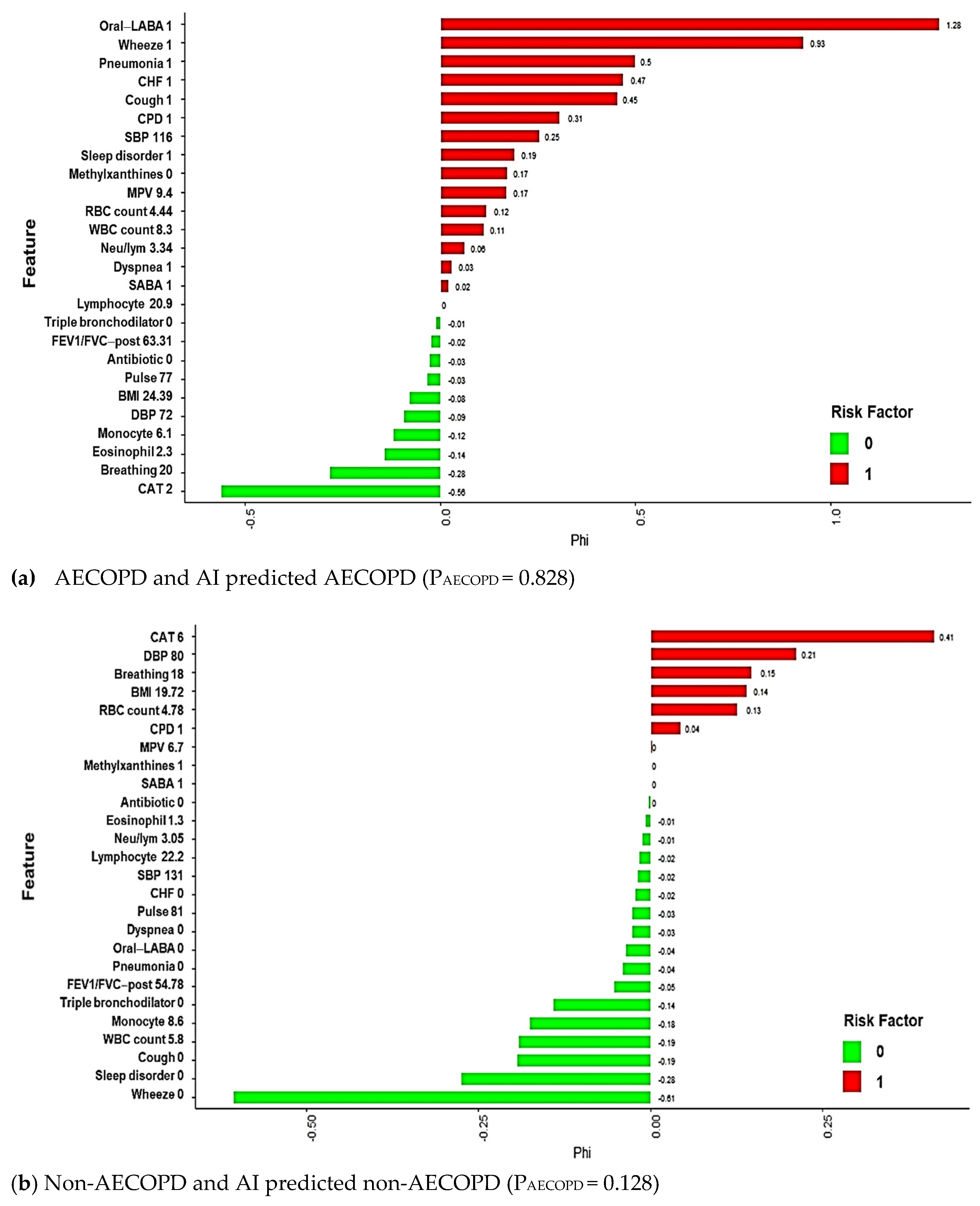
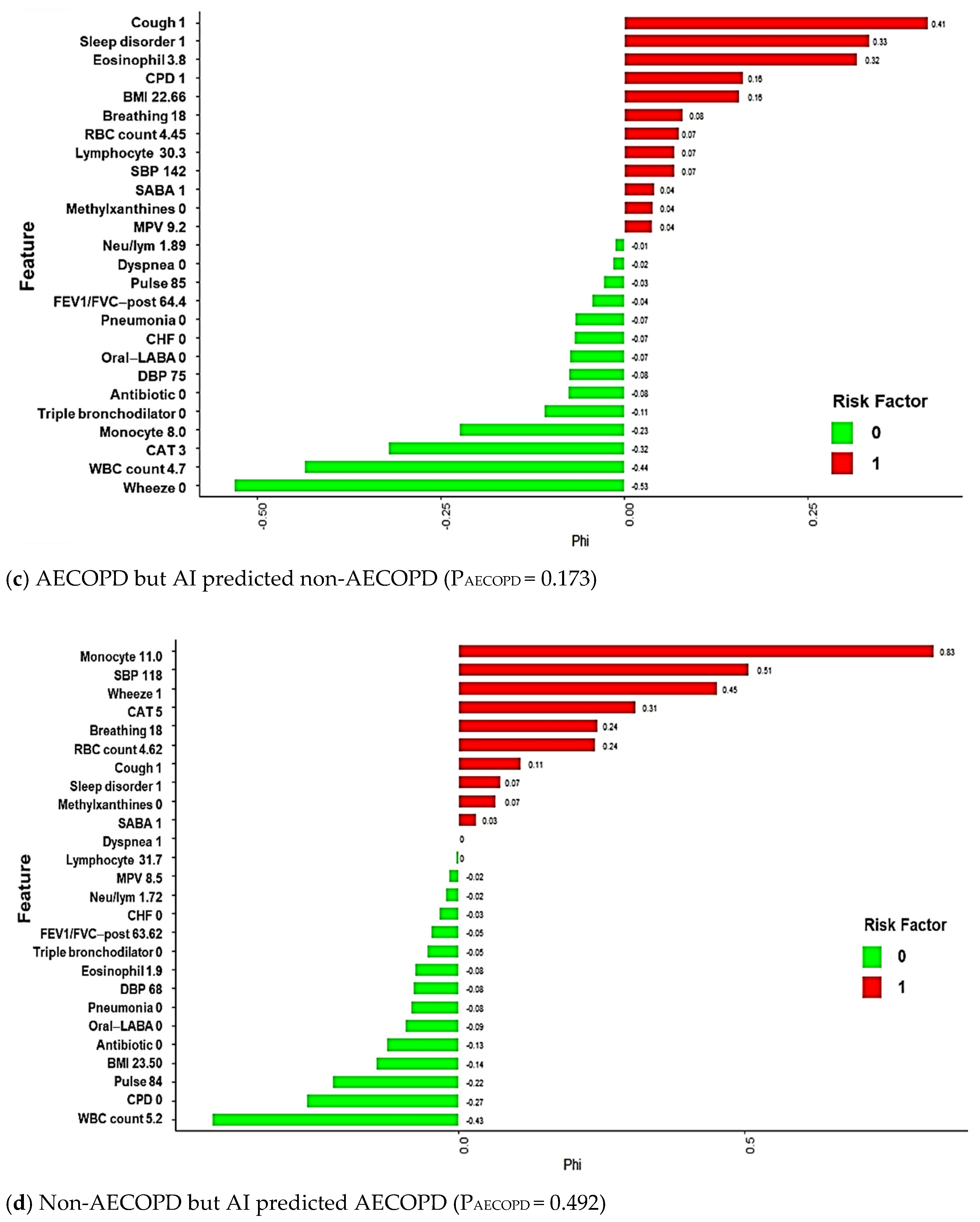
3.3. Model Explanations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Code Availability
References
- López-Campos, J.L.; Tan, W.; Soriano, J.B. Global burden of COPD. Respirology 2016, 21, 14–23. [Google Scholar] [CrossRef]
- Blasi, F.; Cesana, G.; Conti, S.; Chiodini, V.; Aliberti, S.; Fornari, C.; Mantovani, L.G. The Clinical and Economic Impact of Exacerbations of Chronic Obstructive Pulmonary Disease: A Cohort of Hospitalized Patients. PLoS ONE 2014, 9, e101228. [Google Scholar] [CrossRef]
- Global Initiative for Chronic Obstructive Lung Disease. Global Strategy for The Diagnosis, Management, and Prevention of Chronic Obstructive Pulmonary Disease (2021 Report). 2020. Available online: https://goldcopd.org/wp-content/uploads/2020/11/GOLD-REPORT-2021-v1.1-25Nov20_WMV.pdf (accessed on 2 February 2021).
- Agustí, A.; Calverley, P.M.; Decramer, M.; Stockley, R.A.; Wedzicha, J.A. Prevention of Exacerbations in Chronic Obstructive Pulmonary Disease: Knowns and Unknowns. Chronic Obstr. Pulm. Dis. 2014, 1, 166–184. [Google Scholar] [CrossRef]
- Jiang, L.; Gershon, A.S. Using Health Administrative Data to Predict Chronic Obstructive Pulmonary Disease Exacerbations. Ann. Am. Thorac. Soc. 2020, 17, 1056–1057. [Google Scholar] [CrossRef]
- Halpin, D.M.; Miravitlles, M.; Metzdorf, N.; Celli, B. Impact and prevention of severe exacerbations of COPD: A review of the evidence. Int. J. Chronic Obstr. Pulm. Dis. 2017, 12, 2891–2908. [Google Scholar] [CrossRef] [Green Version]
- Balcells, E.; Antó, J.M.; Gea, J.; Gómez, F.P.; Rodríguez, E.; Marin, A.; Ferrer, A.; de Batlle, J.; Farrero, E.; Benet, M.; et al. Characteristics of patients admitted for the first time for COPD exacerbation. Respir. Med. 2009, 103, 1293–1302. [Google Scholar] [CrossRef] [Green Version]
- Halpin, D.M.G.; Decramer, M.; Celli, B.; Kesten, S.; Leimer, I.; Tashkin, D.P. Risk of Nonlower Respiratory Serious Adverse Events Following COPD Exacerbations in the 4-year UPLIFT® Trial. Lung 2011, 189, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, S.P. COPD exacerbations: Finally, a more than acceptable risk score. Lancet Respir. Med. 2020, 8, 939–941. [Google Scholar] [CrossRef]
- Peng, J.; Chen, C.; Zhou, M.; Xie, X.; Zhou, Y.; Luo, C.H. A Machine-learning Approach to Forecast Aggravation Risk in Patients with Acute Exacerbation of Chronic Obstructive Pulmonary Disease with Clinical Indicators. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Tavakoli, H.; Chen, W.; Sin, D.D.; Fitzgerald, J.M.; Sadatsafavi, M. Predicting Severe Chronic Obstructive Pulmonary Disease Exacerbations. Developing a Population Surveillance Approach with Administrative Data. Ann. Am. Thorac. Soc. 2020, 17, 1069–1076. [Google Scholar] [CrossRef]
- The Lancet Respiratory Medicine. Opening the black box of machine learning. Lancet Respir. Med. 2018, 6, 801. [Google Scholar] [CrossRef]
- Kundu, S. AI in medicine must be explainable. Nat. Med. 2021, 27, 1328. [Google Scholar] [CrossRef]
- Cabitza, F.; Rasoini, R.; Gensini, G.F. Unintended Consequences of Machine Learning in Medicine. JAMA 2017, 318, 517–518. [Google Scholar] [CrossRef]
- Cheng, S.-L.; Li, Y.-R.; Huang, N.; Yu, C.-J.; Wang, H.-C.; Lin, M.-C.; Chiu, K.-C.; Hsu, W.-H.; Chen, C.-Z.; Sheu, C.-C.; et al. Effectiveness of Nationwide COPD Pay-for-Performance Program on COPD Exacerbations in Taiwan. Int. J. Chronic Obstr. Pulm. Dis. 2021, 16, 2869–2881. [Google Scholar] [CrossRef]
- Lee, J.; Jung, H.M.; Kim, S.K.; Yoo, K.H.; Jung, K.-S.; Lee, S.H.; Rhee, C.K. Factors associated with chronic obstructive pulmonary disease exacerbation, based on big data analysis. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Ogundimu, E.O.; Altman, D.G.; Collins, G.S. Adequate sample size for developing prediction models is not simply related to events per variable. J. Clin. Epidemiol. 2016, 76, 175–182. [Google Scholar] [CrossRef] [Green Version]
- Sadatsafavi, M.; McCormack, J.; Petkau, J.; Lynd, L.D.; Lee, T.Y.; Sin, D.D. Should the number of acute exacerbations in the previous year be used to guide treatments in COPD? Eur. Respir. J. 2021, 57, 2002122. [Google Scholar] [CrossRef]
- Hussain, A.; Choi, H.-E.; Kim, H.-J.; Aich, S.; Saqlain, M.; Kim, H.-C. Forecast the Exacerbation in Patients of Chronic Obstructive Pulmonary Disease with Clinical Indicators Using Machine Learning Techniques. Diagnostics 2021, 11, 829. [Google Scholar] [CrossRef]
- Bzdok, D.; Altman, N.; Krzywinski, M. Statistics versus machine learning. Nat. Methods 2018, 15, 233–234. [Google Scholar] [CrossRef]
- Wang, C.; Chen, X.; Du, L.; Zhan, Q.; Yang, T.; Fang, Z. Comparison of machine learning algorithms for the identification of acute exacerbations in chronic obstructive pulmonary disease. Comput. Methods Programs Biomed. 2019, 188, 105267. [Google Scholar] [CrossRef]
- Xu, M.; Tantisira, K.G.; Wu, A.; A Litonjua, A.; Chu, J.-H.; E Himes, B.; Damask, A.; Weiss, S.T. Genome Wide Association Study to predict severe asthma exacerbations in children using random forests classifiers. BMC Med. Genet. 2011, 12, 90. [Google Scholar] [CrossRef] [Green Version]
- Agusti, A. The path to personalised medicine in COPD. Thorax 2014, 69, 857–864. [Google Scholar] [CrossRef] [Green Version]
- Agusti, A.; MacNee, W. The COPD control panel: Towards personalised medicine in COPD. Thorax 2012, 68, 687–690. [Google Scholar] [CrossRef] [Green Version]
- Agustí, A.; Antó, J.M.; Auffray, C.; Barbé, F.; Barreiro, E.; Dorca, J.; Escarrabill, J.; Faner, R.; Furlong, L.I.; Garcia-Aymerich, J.; et al. Personalized Respiratory Medicine: Exploring the Horizon, Addressing the Issues. Summary of a BRN-AJRCCM Workshop Held in Barcelona on June 12, 2014. Am. J. Respir. Crit. Care Med. 2015, 191, 391–401. [Google Scholar] [CrossRef]
- Agusti, A.; Bel, E.; Thomas, M.; Vogelmeier, C.; Brusselle, G.; Holgate, S.; Humbert, M.; Jones, P.; Gibson, P.G.; Vestbo, J.; et al. Treatable traits: Toward precision medicine of chronic airway diseases. Eur. Respir. J. 2016, 47, 410–419. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, M.; Wojtusciszyn, A.; Favre, L.; Boughorbel, S.; Shan, J.; Letaief, K.B.; Pitteloud, N.; Chouchane, L. Precision medicine in the era of artificial intelligence: Implications in chronic disease management. J. Transl. Med. 2020, 18, 1–12. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4768–4777. [Google Scholar]
- Oh, T.R.; Song, S.H.; Choi, H.S.; Suh, S.H.; Kim, C.S.; Jung, J.Y.; Choi, K.H.; Oh, K.-H.; Ma, S.K.; Bae, E.H.; et al. Predictive Model for High Coronary Artery Calcium Score in Young Patients with Non-Dialysis Chronic Kidney Disease. J. Pers. Med. 2021, 11, 1372. [Google Scholar] [CrossRef]
- Lu, C.; Song, J.; Li, H.; Yu, W.; Hao, Y.; Xu, K.; Xu, P. Predicting Venous Thrombosis in Osteoarthritis Using a Machine Learning Algorithm: A Population-Based Cohort Study. J. Pers. Med. 2022, 12, 114. [Google Scholar] [CrossRef]
- Yang, J.; Qiao, M.; Li, Y.; Hu, G.; Song, C.; Xue, L.; Bai, H.; Yang, J.; Yang, X. Expansion of a Population of Large Monocytes (Atypical Monocytes) in Peripheral Blood of Patients with Acute Exacerbations of Chronic Obstructive Pulmonary Diseases. Mediat. Inflamm. 2018, 2018, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Hsu, C.-L.; Dong, Y.-H.; Li, Y.-Y.; Chang, C.-H.; Lai, M.-S. Bronchodilators use in patients with COPD. Int. J. Chronic Obstr. Pulm. Dis. 2015, 10, 1769–1779. [Google Scholar] [CrossRef] [Green Version]
- Roche, N. Systemic Medications in Chronic Obstructive Pulmonary Disease: Use and Outcomes. Clin. Chest Med. 2020, 41, 485–494. [Google Scholar] [CrossRef]
- Tseng, C.-M.; Chen, Y.-T.; Ou, S.-M.; Hsiao, Y.-H.; Li, S.-Y.; Wang, S.-J.; Yang, A.C.-C.; Chen, T.-J.; Perng, D.-W. The Effect of Cold Temperature on Increased Exacerbation of Chronic Obstructive Pulmonary Disease: A Nationwide Study. PLoS ONE 2013, 8, e57066. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.-M.; Liu, W.-P.; Kuo, H.-W. Diurnal temperature range and emergency room admissions for chronic obstructive pulmonary disease in Taiwan. Int. J. Biometeorol. 2008, 53, 17–23. [Google Scholar] [CrossRef]


| COPD Patient Data | SPLIT DATA | |||||
|---|---|---|---|---|---|---|
| Non-AECOPD (n = 354) | AECOPD (n = 155) | p-Value | Train Data (n = 407) | Test Data (n = 102) | p-Value | |
| Demographic | ||||||
| Age | 72 ± 10 | 73 ± 10 | 0.557 | 73 ± 10 | 72 ± 11 | 0.882 |
| BMI | 24 ± 4 | 24 ± 4 | 0.593 | 24 ± 4 | 24 ± 4 | 0.962 |
| Vital sign | ||||||
| Pulse | 84 ± 13 | 86 ± 15 | 0.081 | 84 ± 14 | 84 ± 13 | 0.817 |
| Breathing | 19 ± 1 | 19 ± 1 | 0.490 | 19 ± 1 | 19 ± 1 | 0.650 |
| SBP | 135 ± 18 | 135 ± 17 | 0.856 | 135 ± 18 | 137 ± 17 | 0.184 |
| DBP | 75 ± 10 | 76 ± 11 | 0.110 | 75 ± 11 | 77 ± 10 | 0.072 |
| Lung function | ||||||
| FEV1/FVC_post | 60 ± 10 | 57 ± 11 | 0.014 | 59 ± 11 | 60 ± 10 | 0.289 |
| CAT | 4 ± 2 | 6 ± 3 | <0.001 | 4 ± 3 | 4 ± 3 | 0.678 |
| Symptoms | ||||||
| Cough | 177(50%) | 111(71.6%) | <0.001 | 230(56.5%) | 58(56.9%) | 0.949 |
| Dyspnea | 142(40.1%) | 87(56.1%) | 0.001 | 193(47.4%) | 36(35.3%) | 0.028 |
| Wheeze | 112(31.6%) | 93(60%) | <0.001 | 170(41.8%) | 35(34.3%) | 0.170 |
| Comorbidity within 1 year | ||||||
| Chronic pulmonary disease | 163(46.2%) | 101(65.2%) | <0.001 | 211(51.8%) | 53(52.5%) | 0.909 |
| Congestive heart failure | 17(4.8%) | 17(11%) | 0.011 | 31(7.6%) | 3(3%) | 0.094 |
| Sleep disorder | 112(31.7%) | 68(43.9%) | 0.008 | 140(34.4%) | 40(39.6%) | 0.328 |
| anxiety | 26(7.4%) | 15(9.7%) | 0.378 | 33(8.1%) | 8(7.9%) | 0.951 |
| Pneumonia | 35(9.9%) | 41(26.5%) | <0.001 | 61(15%) | 15(14.9%) | 0.973 |
| Hypertension | 159(44.9%) | 85(54.8%) | 0.039 | 196(48.2%) | 48(47.1%) | 0.843 |
| Cancer | 46(13%) | 31(20%) | 0.044 | 63(15.5%) | 14(13.9%) | 0.685 |
| COPD medication within 6 months | ||||||
| Short-acting bronchodilators | 216(61%) | 111(71.6%) | 0.022 | 259(63.6%) | 68(66.7%) | 0.568 |
| Dual bronchodilator | 58(16.4%) | 49(31.6%) | <0.001 | 84(20.6%) | 23(22.5%) | 0.672 |
| Triple bronchodilator | 204(57.6%) | 118(76.1%) | <0.001 | 262(64.4%) | 60(58.8%) | 0.298 |
| Chronic disease medication within 1 year | ||||||
| Antibiotic | 44(12.4%) | 47(30.3%) | <0.001 | 78(19.2%) | 13(12.7%) | 0.130 |
| Oral long-acting bronchodilator | 14(4%) | 25(16.1%) | <0.001 | 31(7.6%) | 8(7.8%) | 0.939 |
| Methylxanthines | 217(61.3%) | 122(78.7%) | <0.001 | 267(65.6%) | 72(70.6%) | 0.340 |
| Lab data within 6 months | ||||||
| WBC count | 7.7 ± 3.1 | 9.1 ± 3.9 | <0.001 | 8.3 ± 3.6 | 7.4 ± 2.4 | 0.003 |
| RBC count | 4.5 ± 0.7 | 4.5 ± 0.5 | 0.165 | 4.5 ± 0.6 | 4.5 ± 0.7 | 0.910 |
| Mean platelet volume | 8.1 ± 0.9 | 8.2 ± 1 | 0.480 | 8.1 ± 1 | 8.1 ± 0.8 | 0.606 |
| Monocyte | 2.6 ± 3.2 | 2.5 ± 2.7 | 0.753 | 9 ± 3.5 | 8.7 ± 3.8 | 0.424 |
| Eosinophil | 21.9 ± 11.3 | 20.5 ± 13.2 | 0.249 | 2.4 ± 2.5 | 3 ± 4.4 | 0.224 |
| Lymphocyte | 5.3 ± 7.1 | 7.2 ± 11.8 | 0.091 | 21.7 ± 12.2 | 20.5 ± 10.7 | 0.421 |
| Neutrophil/Lymphocyte | 8.9 ± 3.5 | 9 ± 3.7 | 0.716 | 5.9 ± 8.6 | 6 ± 10.1 | 0.887 |
| Outcome | ||||||
| AECOPD | 121(29.7%) | 34(33.3%) | 0.479 | |||
| Model | AUC | Threshold | Sensitivity | Specificity | PPV | NPV | F1 Score | Accurate |
|---|---|---|---|---|---|---|---|---|
| Gradient boosted machines (GBMs) | 0.8326 | 0.2614 | 79.41% | 77.94% | 64.29% | 88.33% | 71.05% | 78.43% |
| Extreme Gradient Boosting (XGBoost) | 0.7703 | 0.2961 | 58.82% | 86.76% | 68.97% | 80.82% | 63.49% | 77.45% |
| Random Forest (RF) | 0.7509 | 0.3237 | 64.71% | 83.82% | 66.67% | 82.61% | 65.68% | 77.45% |
| Support Vector Machine (SVM) | 0.8361 | 0.3913 | 82.35% | 69.12% | 57.14% | 88.68% | 67.47% | 73.53% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kor, C.-T.; Li, Y.-R.; Lin, P.-R.; Lin, S.-H.; Wang, B.-Y.; Lin, C.-H. Explainable Machine Learning Model for Predicting First-Time Acute Exacerbation in Patients with Chronic Obstructive Pulmonary Disease. J. Pers. Med. 2022, 12, 228. https://doi.org/10.3390/jpm12020228
Kor C-T, Li Y-R, Lin P-R, Lin S-H, Wang B-Y, Lin C-H. Explainable Machine Learning Model for Predicting First-Time Acute Exacerbation in Patients with Chronic Obstructive Pulmonary Disease. Journal of Personalized Medicine. 2022; 12(2):228. https://doi.org/10.3390/jpm12020228
Chicago/Turabian StyleKor, Chew-Teng, Yi-Rong Li, Pei-Ru Lin, Sheng-Hao Lin, Bing-Yen Wang, and Ching-Hsiung Lin. 2022. "Explainable Machine Learning Model for Predicting First-Time Acute Exacerbation in Patients with Chronic Obstructive Pulmonary Disease" Journal of Personalized Medicine 12, no. 2: 228. https://doi.org/10.3390/jpm12020228
APA StyleKor, C.-T., Li, Y.-R., Lin, P.-R., Lin, S.-H., Wang, B.-Y., & Lin, C.-H. (2022). Explainable Machine Learning Model for Predicting First-Time Acute Exacerbation in Patients with Chronic Obstructive Pulmonary Disease. Journal of Personalized Medicine, 12(2), 228. https://doi.org/10.3390/jpm12020228