DNA Methylation and Asthma Acquisition during Adolescence and Post-Adolescence, an Epigenome-Wide Longitudinal Study
Abstract
:1. Introduction
2. Methods
2.1. Study Population
2.2. Asthma Acquisition
2.3. DNA Methylation
2.4. Covariates
2.5. Forced Exhaled Nitric Oxide (FeNO) and Lung Function Measurement
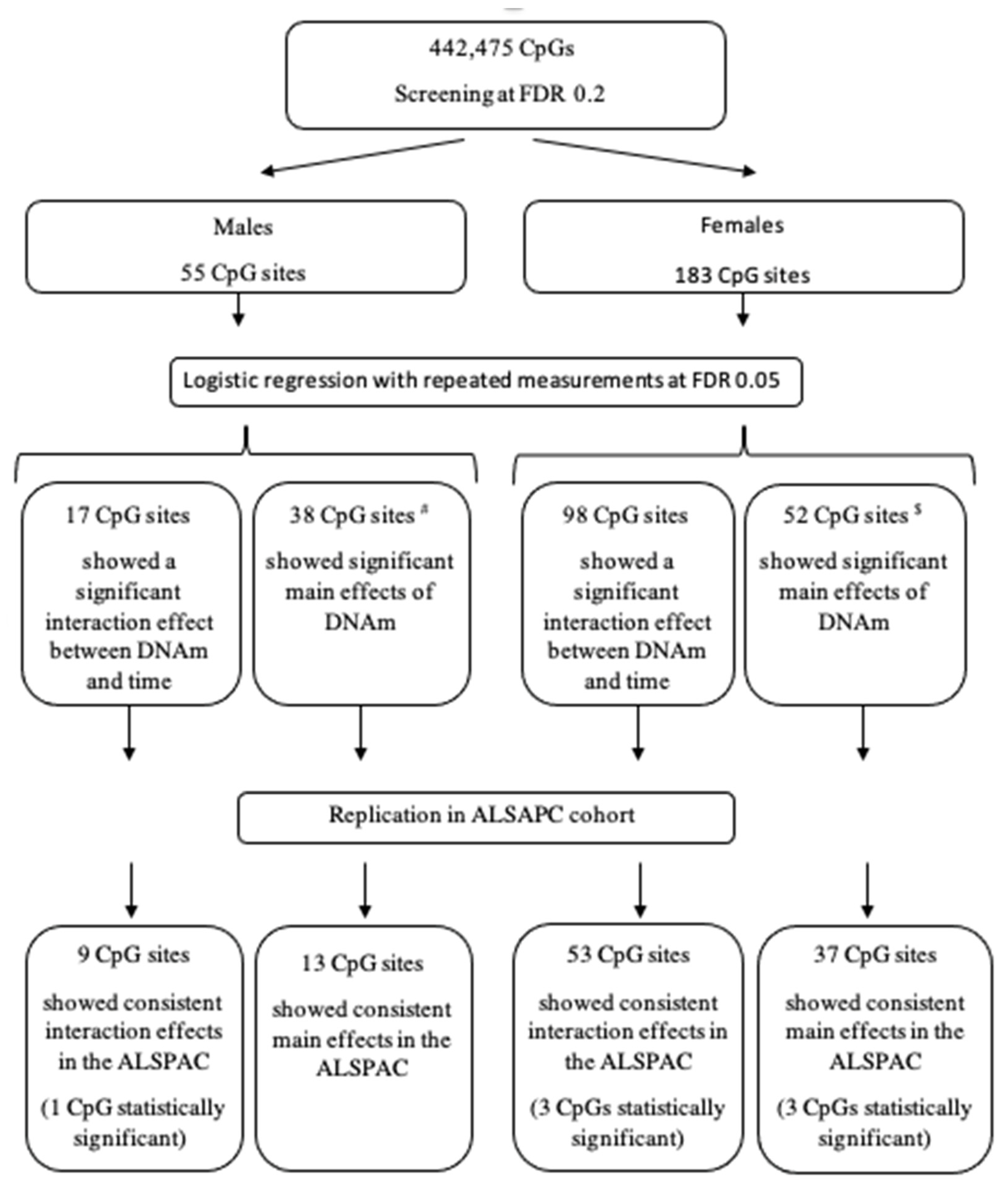
2.6. Statistical Analyses
2.7. Replication Cohort: ALSPAC
2.8. Association Analysis for FeNO and the FEV1/FVC Ratio
2.9. Detection of Differentially Methylated Regions (DMR)
2.10. Pathway Enrichment Analyses
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Declarations
Abbreviations
| CpG | 5′-C-Phosphate-G-3′ |
| DNAm | Deoxyribonucleic acid methylation |
| DMRs | Differentially Methylated Regions |
| FDR | False Discovery Rate |
References
- Fuchs, O.; Bahmer, T.; Rabe, K.F.; von Mutius, E. Asthma transition from childhood into adulthood. Lancet Respir. Med. 2017, 5, 224–234. [Google Scholar] [CrossRef]
- Asthma, G.I.F. Global Strategy for Asthma Management and Prevention; GINA: Fontana, WI, USA, 2020. [Google Scholar]
- Castro-Rodriguez, J.A.; Forno, E.; Rodriguez-Martinez, C.E.; Celedón, J.C. Risk and Protective Factors for Childhood Asthma: What Is the Evidence? J. Allergy Clin. Immunol. 2016, 4, 1111–1122. [Google Scholar] [CrossRef] [Green Version]
- Loftus, P.A.; Wise, S.K. Epidemiology of asthma. Curr. Opin. Otolaryngol. Head Neck Surg. 2016, 24, 245–249. [Google Scholar] [CrossRef]
- Murrison, L.B.; Brandt, E.B.; Myers, J.B.; Hershey, G.K.K. Environmental exposures and mechanisms in allergy and asthma development. J. Clin. Investig. 2019, 129, 1504–1515. [Google Scholar] [CrossRef] [Green Version]
- Joubert, B.R.; Felix, J.F.; Yousefi, P.; Bakulski, K.M.; Just, A.C.; Breton, C.; Reese, S.E.; Markunas, C.A.; Richmond, R.C.; Xu, C.J.; et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. Am. J. Hum. Genet. 2016, 98, 680–696. [Google Scholar] [CrossRef] [Green Version]
- DeVries, A.; Vercelli, D. Early predictors of asthma and allergy in children: The role of epigenetics. Curr. Opin. Allergy Clin. Immunol. 2015, 15, 435–439. [Google Scholar] [CrossRef] [Green Version]
- Patel, R.; Solatikia, F.; Zhang, H.; Wolde, A.; Kadalayil, L.; Karmaus, W.; Ewart, S.; Arathimos, R.; Relton, C.; Ring, S.; et al. Sex-specific associations of asthma acquisition with changes in DNA methylation during adolescence. Clin. Exp. Allergy 2021, 51, 318–328. [Google Scholar] [CrossRef]
- Rathod, A.; Duan, J.; Zhang, H.; Holloway, J.W.; Ewart, S.; Arshad, S.H.; Karmaus, W. Interweaving Between Genetic and Epigenetic Studies on Childhood Asthma. Epigenet. Insights 2020, 13, 2516865720923395. [Google Scholar] [CrossRef]
- Vercelli, D. Does epigenetics play a role in human asthma? Allergol. Int. Off. J. Jpn. Soc. Allergol. 2016, 65, 123–126. [Google Scholar] [CrossRef]
- Reese, S.E.; Xu, C.J.; den Dekker, H.T.; Lee, M.K.; Sikdar, S.; Ruiz-Arenas, C.; Merid, S.K.; Rezwan, F.I.; Page, C.M.; Ullemar, V.; et al. Epigenome-wide meta-analysis of DNA methylation and childhood asthma. J. Allergy Clin. Immunol. 2019, 143, 2062–2074. [Google Scholar] [CrossRef] [Green Version]
- Arathimos, R.; Suderman, M.; Sharp, G.C.; Burrows, K.; Granell, R.; Tilling, K.; Gaunt, T.R.; Henderson, J.; Ring, S.; Richmond, R.C.; et al. Epigenome-wide association study of asthma and wheeze in childhood and adolescence. Clin. Epigenet. 2017, 9, 112. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.J.; Soderhall, C.; Bustamante, M.; Baiz, N.; Gruzieva, O.; Gehring, U.; Mason, D.; Chatzi, L.; Basterrechea, M.; Llop, S.; et al. DNA methylation in childhood asthma: An epigenome-wide meta-analysis. Lancet Respir. Med. 2018, 6, 379–388. [Google Scholar] [CrossRef] [Green Version]
- Rathod, R.; Zhang, H.; Karmaus, W.; Ewart, S.; Kadalayil, L.; Relton, C.; Ring, S.; Arshad, S.H.; Holloway, J.W. BMI trajectory in childhood is associated with asthma incidence at young adulthood mediated by DNA methylation. Allergy Asthma Clin. Immunol. Off. J. Can. Soc. Allergy Clin. Immunol. 2021, 17, 77. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.D.; Wright, A.L.; Taussig, L.M.; Holberg, C.J.; Halonen, M.; Morgan, W.J. Asthma and wheezing in the first six years of life. The Group Health Medical Associates. N. Engl. J. Med. 1995, 332, 133–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koper, I.; Hufnagl, K.; Ehmann, R. Gender aspects and influence of hormones on bronchial asthma—Secondary publication and update. World Allergy Organ. J. 2017, 10, 46. [Google Scholar] [CrossRef]
- Pignataro, F.S.; Bonini, M.; Forgione, A.; Melandri, S.; Usmani, O.S. Asthma and gender: The female lung. Pharmacol. Res. 2017, 119, 384–390. [Google Scholar] [CrossRef]
- Osman, M.; Hansell, A.L.; Simpson, C.R.; Hollowell, J.; Helms, P.J. Gender-specific presentations for asthma, allergic rhinitis and eczema in primary care. Prim. Care Respir. J. Gen. Pract. Airw. Group 2007, 16, 28–35. [Google Scholar] [CrossRef]
- Arshad, S.H.; Holloway, J.W.; Karmaus, W.; Zhang, H.; Ewart, S.; Mansfield, L.; Matthews, S.; Hodgekiss, C.; Roberts, G.; Kurukulaaratchy, R. Cohort Profile: The Isle Of Wight Whole Population Birth Cohort (IOWBC). Int. J. Epidemiol. 2018, 47, 1043–1044. [Google Scholar] [CrossRef]
- Zhang, H.; Kaushal, A.; Merid, S.K.; Melen, E.; Pershagen, G.; Rezwan, F.I.; Han, L.; Ewart, S.; Arshad, S.H.; Karmaus, W.; et al. DNA methylation and allergic sensitizations: A genome-scale longitudinal study during adolescence. Allergy 2019, 74, 1166–1175. [Google Scholar] [CrossRef] [Green Version]
- Soto-Ramirez, N.; Arshad, S.H.; Holloway, J.W.; Zhang, H.; Schauberger, E.; Ewart, S.; Patil, V.; Karmaus, W. The interaction of genetic variants and DNA methylation of the interleukin-4 receptor gene increase the risk of asthma at age 18 years. Clin. Epigenet. 2013, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Kaushal, A.; Soto-Ramirez, N.; Ziyab, A.H.; Ewart, S.; Holloway, J.W.; Karmaus, W.; Arshad, H. Acquisition, remission, and persistence of eczema, asthma, and rhinitis in children. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2018, 48, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Ziyab, A.H.; Karmaus, W.; Zhang, H.; Holloway, J.W.; Steck, S.E.; Ewart, S.; Arshad, S.H. Association of filaggrin variants with asthma and rhinitis: Is eczema or allergic sensitization status an effect modifier? Int. Arch. Allergy Immunol. 2014, 164, 308–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathod, A.; Rathod, R.; Zhang, H.; Rahimabad, P.K.; Karmaus, W.; Arshad, H. Association of Asthma and Rhinitis with Epigenetics of Coronavirus Related Genes. Epigenet. Insights 2021, 14, 25168657211039224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tong, X.; Holloway, J.W.; Rezwan, F.I.; Lockett, G.A.; Patil, V.; Ray, M.; Everson, T.M.; Soto-Ramírez, N.; Arshad, S.H.; et al. The interplay of DNA methylation over time with Th2 pathway genetic variants on asthma risk and temporal asthma transition. Clin. Epigenet. 2014, 6, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [Green Version]
- Rathod, R.; Rathod, A.; Rahimabad, P.K.; Duan, J.; Zhang, H.; Arshad, S.H.; Karmaus, W. Methylation of Host Genes Associated with Coronavirus Infection from Birth to 26 Years. Genes 2021, 12, 1198. [Google Scholar] [CrossRef] [PubMed]
- Lehne, B.; Drong, A.W.; Loh, M.; Zhang, W.; Scott, W.R.; Tan, S.T.; Afzal, U.; Scott, J.; Jarvelin, M.R.; Elliott, P.; et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015, 16, 37. [Google Scholar] [CrossRef] [Green Version]
- Reinius, L.E.; Acevedo, N.; Joerink, M.; Pershagen, G.; Dahlen, S.E.; Greco, D.; Soderhall, C.; Scheynius, A.; Kere, J. Differential DNA methylation in purified human blood cells: Implications for cell lineage and studies on disease susceptibility. PLoS ONE 2012, 7, e41361. [Google Scholar] [CrossRef]
- Koestler, D.C.; Christensen, B.; Karagas, M.R.; Marsit, C.J.; Langevin, S.M.; Kelsey, K.T.; Wiencke, J.K.; Houseman, E.A. Blood-based profiles of DNA methylation predict the underlying distribution of cell types: A validation analysis. Epigenetics 2013, 8, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, A.E.; Irizarry, R.A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014, 15, R31. [Google Scholar] [CrossRef]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012, 13, 86. [Google Scholar] [CrossRef] [Green Version]
- Recommendations for standardized procedures for the on-line and off-line measurement of exhaled lower respiratory nitric oxide and nasal nitric oxide in adults and children-1999. This official statement of the American Thoracic Society was adopted by the ATS Board of Directors, July 1999. Am. J. Respir. Crit. Care Med. 1999, 160, 2104–2117. [CrossRef]
- Mukherjee, N.; Lockett, G.A.; Merid, S.K.; Melén, E.; Pershagen, G.; Holloway, J.W.; Arshad, S.H.; Ewart, S.; Zhang, H.; Karmaus, W. DNA methylation and genetic polymorphisms of the Leptin gene interact to influence lung function outcomes and asthma at 18 years of age. Int. J. Mol. Epidemiol. Genet. 2016, 7, 1–17. [Google Scholar]
- Boyd, A.; Golding, J.; Macleod, J.; Lawlor, D.A.; Fraser, A.; Henderson, J.; Molloy, L.; Ness, A.; Ring, S.; Davey Smith, G. Cohort Profile: The ‘children of the 90s’—The index offspring of the Avon Longitudinal Study of Parents and Children. Int. J. Epidemiol. 2013, 42, 111–127. [Google Scholar] [CrossRef] [Green Version]
- Fraser, A.; Macdonald-Wallis, C.; Tilling, K.; Boyd, A.; Golding, J.; Davey Smith, G.; Henderson, J.; Macleod, J.; Molloy, L.; Ness, A.; et al. Cohort Profile: The Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int. J. Epidemiol. 2013, 42, 97–110. [Google Scholar] [CrossRef] [Green Version]
- Northstone, K.; Lewcock, M.; Groom, A.; Boyd, A.; Macleod, J.; Timpson, N.; Wells, N. The Avon Longitudinal Study of Parents and Children (ALSPAC): An update on the enrolled sample of index children in 2019. Wellcome Open Res. 2019, 4, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Relton, C.L.; Gaunt, T.; McArdle, W.; Ho, K.; Duggirala, A.; Shihab, H.; Woodward, G.; Lyttleton, O.; Evans, D.M.; Reik, W.; et al. Data Resource Profile: Accessible Resource for Integrated Epigenomic Studies (ARIES). Int. J. Epidemiol. 2015, 44, 1181–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Corcoran, C.; Kissoon, N.; Murphy, S.P.; Duckworth, L.J. Exhaled nitric oxide reflects asthma severity and asthma control. Pediatric Crit. Care Med. 2004, 5, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Mingotti, C.; Sarinho, J.; Stanigher, K.; Silva, J.; Roquette, E.; Marchi, E.; Ponte, E.V. Evaluating the FEV(1)/FVC ratio in the lower range of normality as a marker of worse clinical outcomes in asthmatic subjects without airway obstruction. Respir. Med. 2020, 162, 105880. [Google Scholar] [CrossRef] [PubMed]
- Peters, T.J.; Buckley, M.J.; Statham, A.L.; Pidsley, R.; Samaras, K.; R, V.L.; Clark, S.J.; Molloy, P.L. De novo identification of differentially methylated regions in the human genome. Epigenet. Chromatin 2015, 8, 6. [Google Scholar] [CrossRef] [Green Version]
- Geeleher, P.; Hartnett, L.; Egan, L.J.; Golden, A.; Raja Ali, R.A.; Seoighe, C. Gene-set analysis is severely biased when applied to genome-wide methylation data. Bioinformatics 2013, 29, 1851–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leynaert, B.; Sunyer, J.; Garcia-Esteban, R.; Svanes, C.; Jarvis, D.; Cerveri, I.; Dratva, J.; Gislason, T.; Heinrich, J.; Janson, C.; et al. Gender differences in prevalence, diagnosis and incidence of allergic and non-allergic asthma: A population-based cohort. Thorax 2012, 67, 625. [Google Scholar] [CrossRef] [Green Version]
- Saik, O.V.; Demenkov, P.S.; Ivanisenko, T.V.; Bragina, E.Y.; Freidin, M.B.; Goncharova, I.A.; Dosenko, V.E.; Zolotareva, O.I.; Hofestaedt, R.; Lavrik, I.N.; et al. Novel candidate genes important for asthma and hypertension comorbidity revealed from associative gene networks. BMC Med. Genom. 2018, 11, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huo, Y.; Zhang, H.Y. Genetic Mechanisms of Asthma and the Implications for Drug Repositioning. Genes 2018, 9, 237. [Google Scholar] [CrossRef] [Green Version]
- Qi, S.; Liu, G.; Dong, X.; Huang, N.; Li, W.; Chen, H. Microarray data analysis to identify differentially expressed genes and biological pathways associated with asthma. Exp. Ther. Med. 2018, 16, 1613–1620. [Google Scholar] [CrossRef]
- Reeder, K.M.; Dunaway, C.W.; Blackburn, J.P.; Yu, Z.; Matalon, S.; Hastie, A.T.; Ampleford, E.J.; Meyers, D.A.; Steele, C. The common γ-chain cytokine IL-7 promotes immunopathogenesis during fungal asthma. Mucosal Immunol. 2018, 11, 1352–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, E.A.; Koziol-White, C.J.; Clay, K.J.; Liu, L.Y.; Bates, M.E.; Bertics, P.J.; Jarjour, N.N. Potential contribution of IL-7 to allergen-induced eosinophilic airway inflammation in asthma. J. Immunol. 2009, 182, 1404–1410. [Google Scholar] [CrossRef] [Green Version]
- Michaeloudes, C.; Bhavsar, P.K.; Mumby, S.; Xu, B.; Hui, C.K.M.; Chung, K.F.; Adcock, I.M. Role of Metabolic Reprogramming in Pulmonary Innate Immunity and Its Impact on Lung Diseases. J. Innate Immun. 2020, 12, 31–46. [Google Scholar] [CrossRef]
- Rodriguez-Coira, J.; Villaseñor, A.; Izquierdo, E.; Huang, M.; Barker-Tejeda, T.C.; Radzikowska, U.; Sokolowska, M.; Barber, D. The Importance of Metabolism for Immune Homeostasis in Allergic Diseases. Front. Immunol. 2021, 12, 692004. [Google Scholar] [CrossRef]
- Rodriguez-Perez, N.; Schiavi, E.; Frei, R.; Ferstl, R.; Wawrzyniak, P.; Smolinska, S.; Sokolowska, M.; Sievi, N.A.; Kohler, M.; Schmid-Grendelmeier, P.; et al. Altered fatty acid metabolism and reduced stearoyl-coenzyme a desaturase activity in asthma. Allergy 2017, 72, 1744–1752. [Google Scholar] [CrossRef] [Green Version]
- Kuo, C.S.; Pavlidis, S.; Loza, M.; Baribaud, F.; Rowe, A.; Pandis, I.; Sousa, A.; Corfield, J.; Djukanovic, R.; Lutter, R.; et al. T-helper cell type 2 (Th2) and non-Th2 molecular phenotypes of asthma using sputum transcriptomics in U-BIOPRED. Eur. Respir. J. 2017, 49, 1602135. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.T.; Sikdar, S.; Xu, C.J.; Lee, M.K.; Cardwell, J.; Forno, E.; Imboden, M.; Jeong, A.; Madore, A.M.; Qi, C.; et al. Epigenome-wide association study of DNA methylation and adult asthma in the Agricultural Lung Health Study. Eur. Respir. J. 2020, 56, 2000217. [Google Scholar] [CrossRef] [PubMed]
- Himes, B.E.; Klanderman, B.; Ziniti, J.; Senter-Sylvia, J.; Soto-Quiros, M.E.; Avila, L.; Celedón, J.C.; Lange, C.; Mariani, T.J.; Lasky-Su, J.; et al. Association of SERPINE2 with asthma. Chest 2011, 140, 667–674. [Google Scholar] [CrossRef] [Green Version]
- Nahm, D.H.; Lee, K.H.; Shin, J.Y.; Ye, Y.M.; Kang, Y.; Park, H.S. Identification of alpha-enolase as an autoantigen associated with severe asthma. J. Allergy Clin. Immunol. 2006, 118, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, N.; Arathimos, R.; Chen, S.; Kheirkhah Rahimabad, P.; Han, L.; Zhang, H.; Holloway, J.W.; Relton, C.; Henderson, A.J.; Arshad, S.H.; et al. DNA methylation at birth is associated with lung function development until age 26 years. Eur. Respir. J. 2021, 57, 2003505. [Google Scholar] [CrossRef]
- Sunny, S.K.; Zhang, H.; Mzayek, F.; Relton, C.L.; Ring, S.; Henderson, A.J.; Ewart, S.; Holloway, J.W.; Arshad, S.H. Pre-adolescence DNA methylation is associated with lung function trajectories from pre-adolescence to adulthood. Clin. Epigenet. 2021, 13, 5. [Google Scholar] [CrossRef]
- Sunny, S.K.; Zhang, H.; Relton, C.L.; Ring, S.; Kadalayil, L.; Mzayek, F.; Ewart, S.; Holloway, J.W.; Arshad, S.H. Sex-specific longitudinal association of DNA methylation with lung function. ERJ Open Res. 2021, 7, 00127-2021. [Google Scholar] [CrossRef]
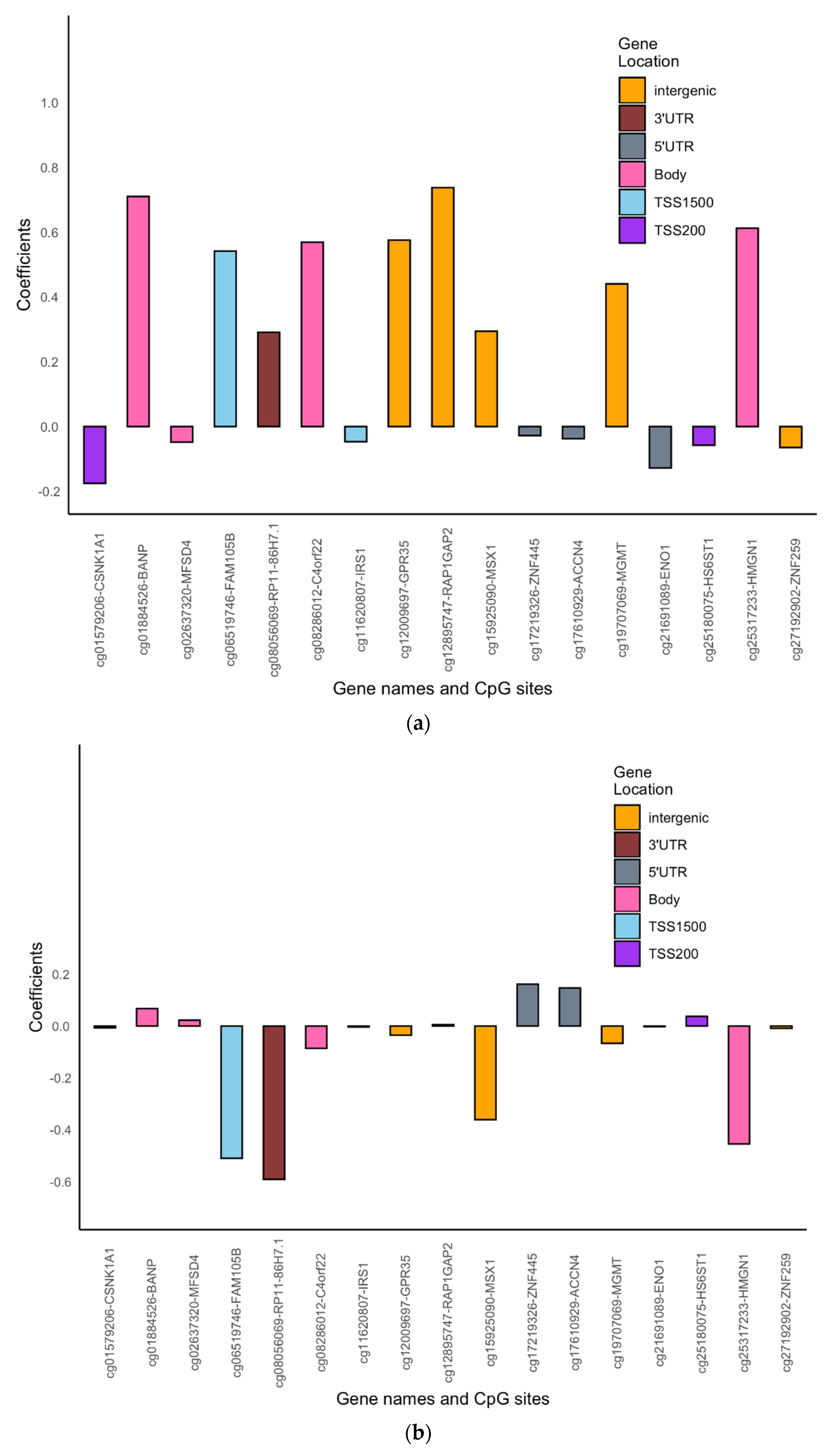
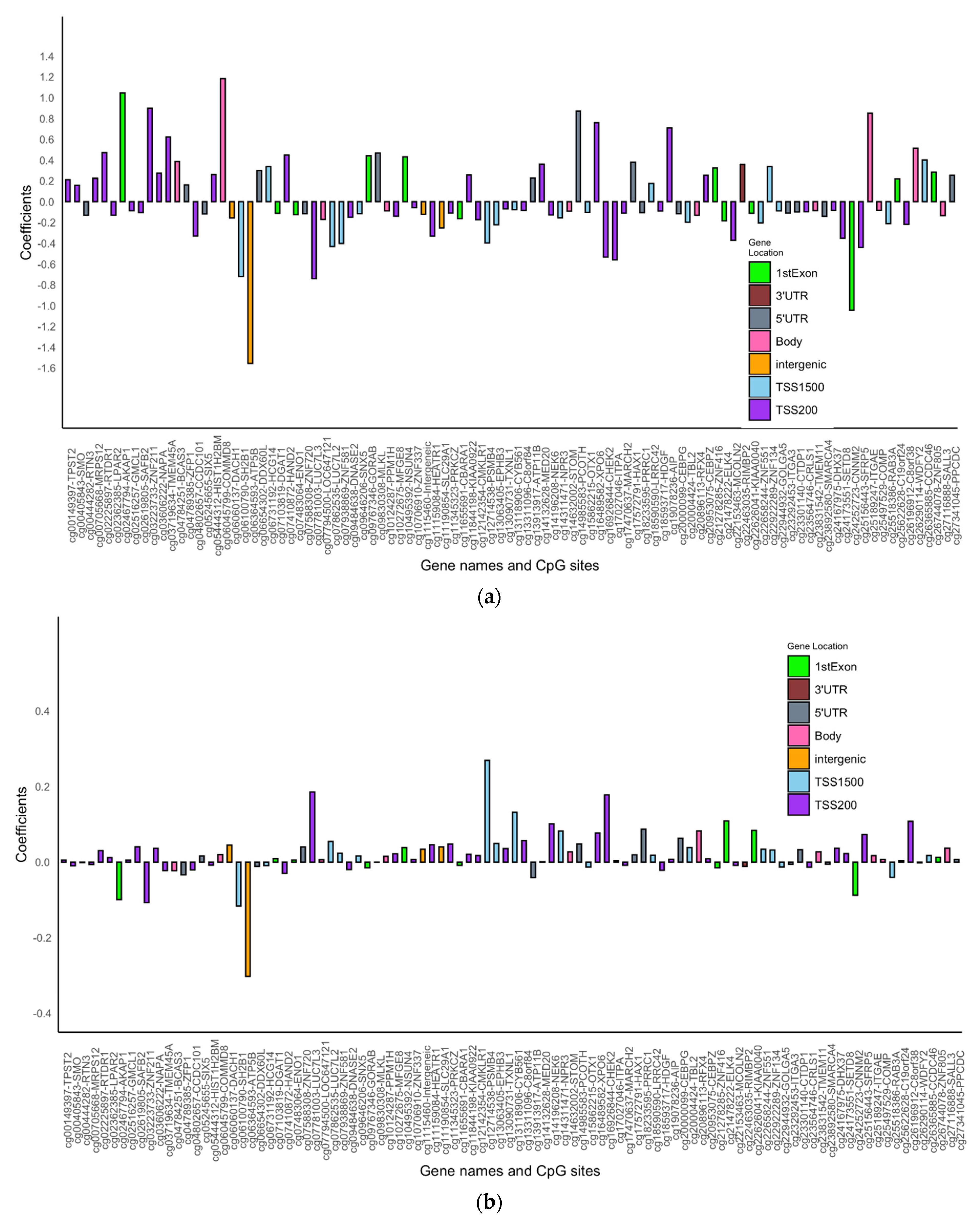
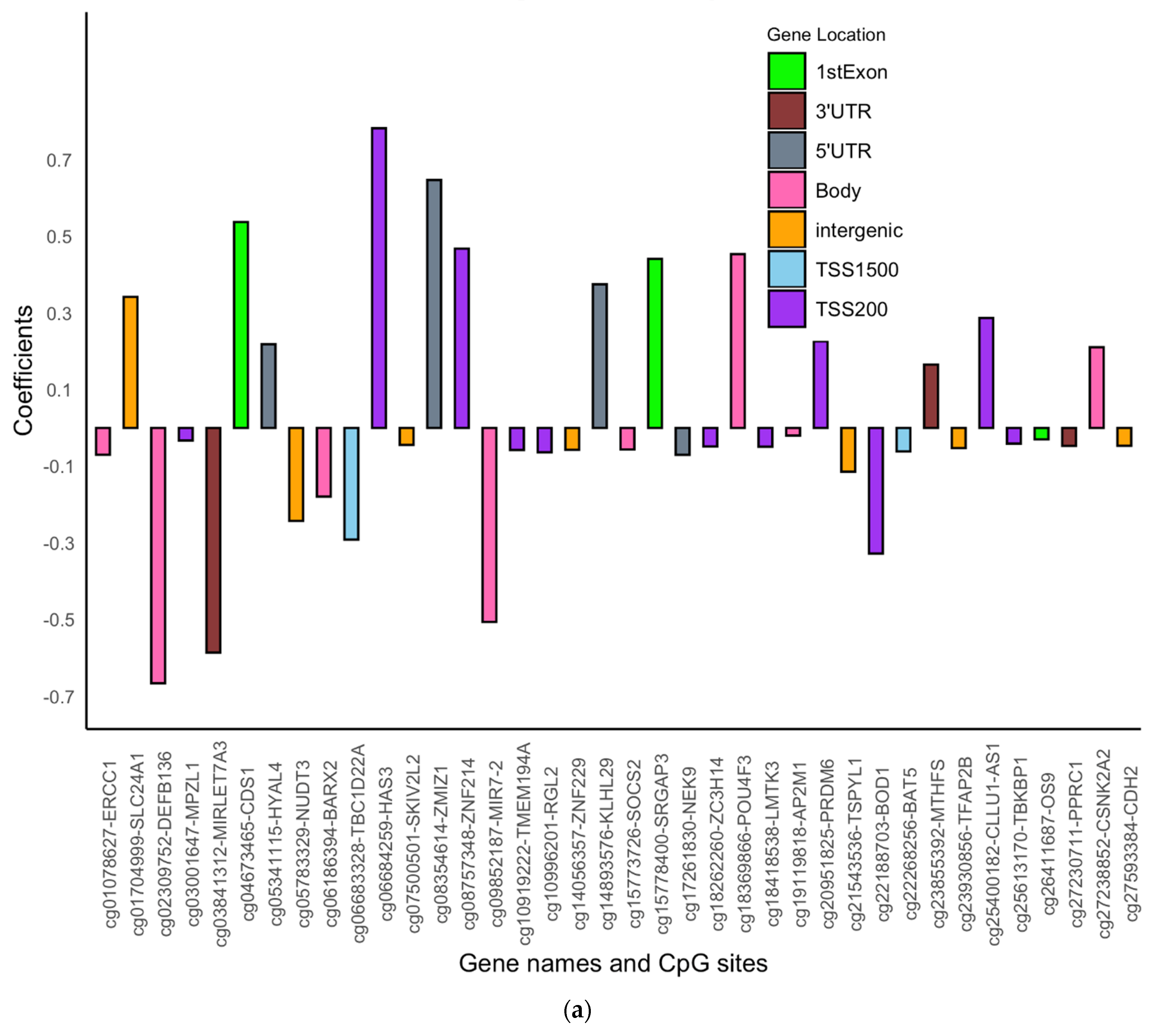
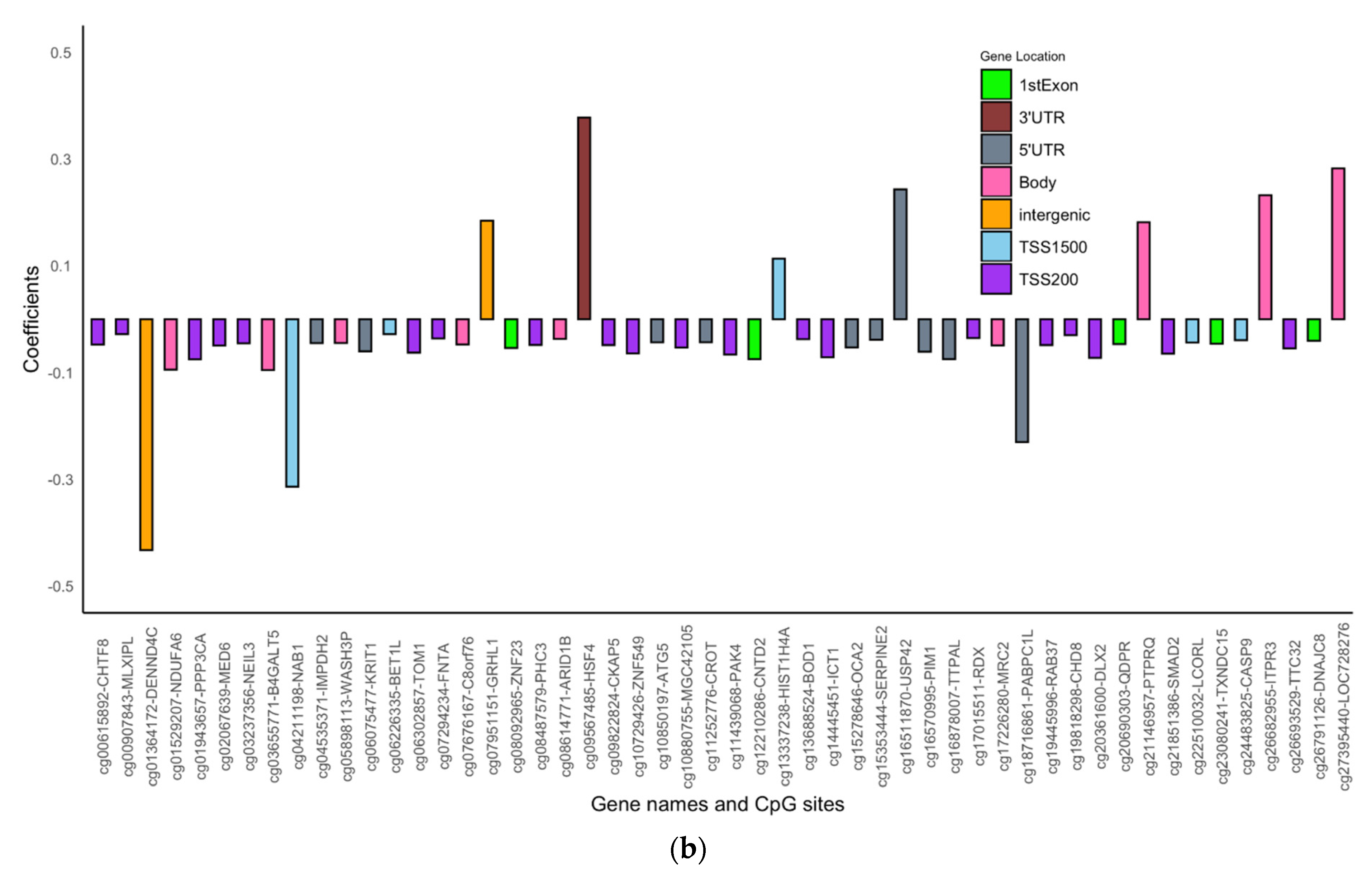
| Variables N (%) | Females | Males | |||||
|---|---|---|---|---|---|---|---|
| Subsample N = 102; n (%) | Complete Cohort N = 431; n (%) | p-Value | Subsample N = 133; n (%) | Complete Cohort N = 402; n (%) | p-Value | ||
| Asthma transition | Acquisition | 7 (6.86) | 41 (9.51) | 0.40 | 11 (8.27) | 26 (6.47) | 0.98 |
| Never Asthma | 95 (93.14) | 390 (90.49) | 122 (91.73) | 376 (93.53) | |||
| Active smoking | Yes | 25 (23.36) | 120 (25.86) | 0.59 | 33 (21.85) | 105 (22.98) | 0.48 |
| No | 82 (76.64) | 344 (74.14) | 118 (78.15) | 352 (77.02) | |||
| Second-hand smoking | Yes | 54 (50) | 212 (44.92) | 0.34 | 66 (43.42) | 204 (43.59) | 0.78 |
| No | 54 (50) | 260 (55.08) | 86 (56.58) | 264 (56.41) | |||
| Atopy | Yes | 14 (12.96) | 76 (19.1) | 0.14 | 42 (27.81) | 108 (27.91) | 0.97 |
| No | 94 (87.04) | 322 (80.9) | 109 (72.19) | 279 (72.09) | |||
| Variables N (%) | Females | Males | |||||
|---|---|---|---|---|---|---|---|
| Subsample N = 156; n (%) | Complete Cohort N = 330; n (%) | p-Value | Subsample N = 121; n (%) | Complete Cohort N = 286; n (%) | p-Value | ||
| Asthma transition | Acquisition | 5 (3.21) | 12 (3.64) | 0.81 | 3 (2.48) | 10 (3.5) | 0.59 |
| Never Asthma | 151 (96.79) | 318 (96.36) | 118 (97.52) | 276 (96.5) | |||
| Active smoking | Yes | 46 (27.54) | 107 (26.95) | 0.89 | 46 (31.94) | 99 (28.95) | 0.51 |
| No | 121 (72.46) | 290 (73.05) | 98 (68.06) | 243 (71.05) | |||
| Second-hand smoking | Yes | 35 (20.96) | 98 (24.75) | 0.33 | 27 (18.75) | 80 (23.39) | 0.26 |
| No | 132 (79.04) | 298 (75.25) | 117 (81.25) | 262 (76.61) | |||
| Atopy | Yes | 55 (29.73) | 115 (32.63) | 0.36 | 73 (42.2) | 146 (46.65) | 0.35 |
| No | 130(70.27) | 227 (66.37) | 100 (57.8) | 167 (53.35) | |||
| GO term | Biological Processes | p-Value | No. of Genes |
|---|---|---|---|
| GO:1901575 | organic substance catabolic process | 0.0005 | 12 |
| GO:0046657 | folic acid catabolic process | 0.001 | 1 |
| GO:0042219 | cellular modified amino acid catabolic process | 0.002 | 1 |
| GO:0038111 | interleukin-7-mediated signaling pathway | 0.002 | 2 |
| GO:1990261 | pre-mRNA catabolic process | 0.002 | 1 |
| GO:0071544 | diphosphoinositol polyphosphate catabolic process | 0.002 | 1 |
| GO:0042365 | water-soluble vitamin catabolic process | 0.003 | 1 |
| GO:0009056 | catabolic process | 0.003 | 13 |
| GO:0098760 | response to interleukin-7 | 0.003 | 2 |
| GO:0098761 | cellular response to interleukin-7 | 0.003 | 2 |
| GO term | Biological Processes | p-Value | No. of Genes |
|---|---|---|---|
| GO:0019438 | aromatic compound biosynthetic process | 0.0004 | 57 |
| GO:0032774 | RNA biosynthetic process | 0.0007 | 49 |
| GO:0008589 | regulation of smoothened signaling pathway | 0.0007 | 5 |
| GO:0018130 | heterocycle biosynthetic process | 0.0008 | 56 |
| GO:1903506 | regulation of nucleic acid-templated transcription | 0.0008 | 49 |
| GO:2001141 | regulation of RNA biosynthetic process | 0.0008 | 49 |
| GO:1901362 | organic cyclic compound biosynthetic process | 0.001 | 57 |
| GO:0034654 | nucleobase-containing compound biosynthetic process | 0.001 | 55 |
| GO:0006355 | regulation of transcription, DNA-templated | 0.001 | 48 |
| GO:0009757 | hexose-mediated signaling | 0.001 | 2 |
| Sex | Chr. $ | Start # | End & | Gene £ | CpGs ¥ | p-Value |
|---|---|---|---|---|---|---|
| M | 3 | 196065106 | 196065569 | TM4SF19 | cg05556202, cg05445326 | 4.89 × 10−206 |
| 12 | 57472396 | 57472611 | TMEM194A | cg10919222, cg09934365 | 3.03 × 10−150 | |
| 17 | 27899874 | 27899966 | TP53I13 | cg05877788, cg04498198 | 2.69 × 10−53 | |
| F | 2 | 224903369 | 224903487 | SERPINE2 | cg15353444, cg11719885 | 5.84 × 10−177 |
| 1 | 156721844 | 156722068 | HDGF | cg04402095, cg18593717 | 5.35 × 10−162 | |
| 17 | 79633496 | 79633565 | CCDC137, C17orf90 | cg199963747, cg11820993 | 1.70 × 10−160 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rathod, A.; Zhang, H.; Arshad, S.H.; Ewart, S.; Relton, C.L.; Karmaus, W.; Holloway, J.W. DNA Methylation and Asthma Acquisition during Adolescence and Post-Adolescence, an Epigenome-Wide Longitudinal Study. J. Pers. Med. 2022, 12, 202. https://doi.org/10.3390/jpm12020202
Rathod A, Zhang H, Arshad SH, Ewart S, Relton CL, Karmaus W, Holloway JW. DNA Methylation and Asthma Acquisition during Adolescence and Post-Adolescence, an Epigenome-Wide Longitudinal Study. Journal of Personalized Medicine. 2022; 12(2):202. https://doi.org/10.3390/jpm12020202
Chicago/Turabian StyleRathod, Aniruddha, Hongmei Zhang, Syed Hasan Arshad, Susan Ewart, Caroline L. Relton, Wilfried Karmaus, and John W. Holloway. 2022. "DNA Methylation and Asthma Acquisition during Adolescence and Post-Adolescence, an Epigenome-Wide Longitudinal Study" Journal of Personalized Medicine 12, no. 2: 202. https://doi.org/10.3390/jpm12020202
APA StyleRathod, A., Zhang, H., Arshad, S. H., Ewart, S., Relton, C. L., Karmaus, W., & Holloway, J. W. (2022). DNA Methylation and Asthma Acquisition during Adolescence and Post-Adolescence, an Epigenome-Wide Longitudinal Study. Journal of Personalized Medicine, 12(2), 202. https://doi.org/10.3390/jpm12020202







