Overview on Common Genes Involved in the Onset of Glioma and on the Role of Migraine as Risk Factor: Predictive Biomarkers or Therapeutic Targets?
Abstract
:Simple Summary
Abstract
1. Introduction
2. New Insight on Molecular Involvements of Gliomas
2.1. CLOCK Gene
2.2. BMAL1 Gene
2.3. CLOCK/BMAL1 Gene as Therapeutic Target
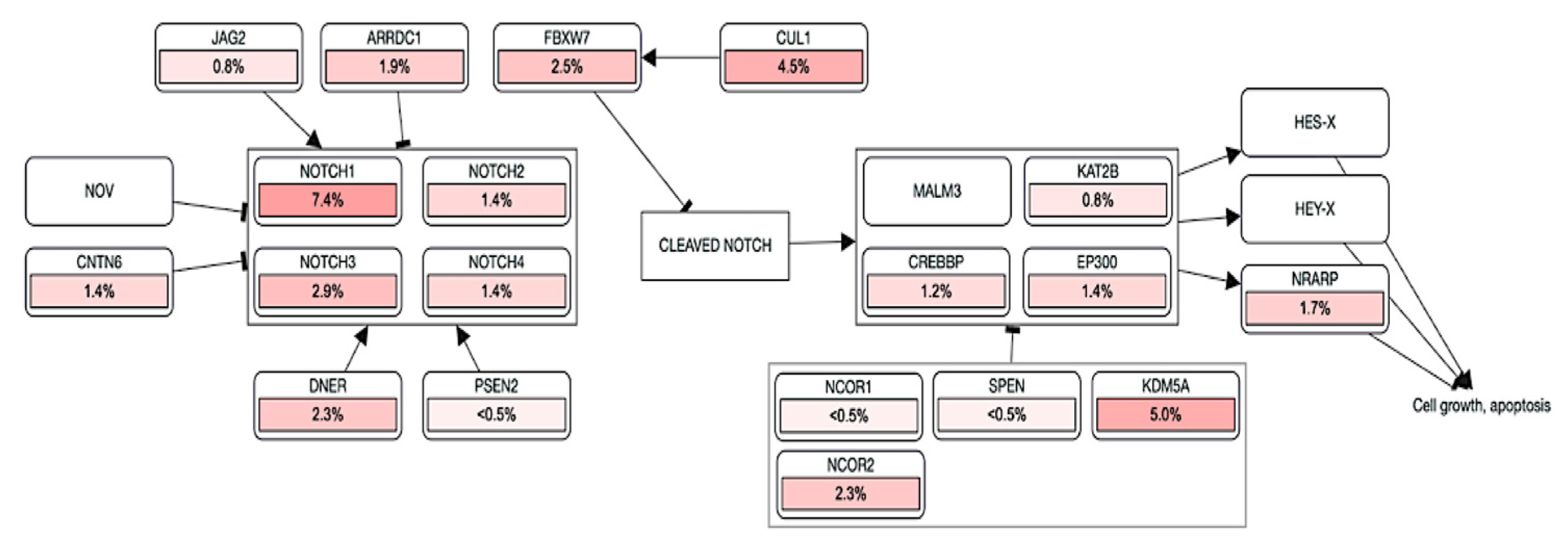
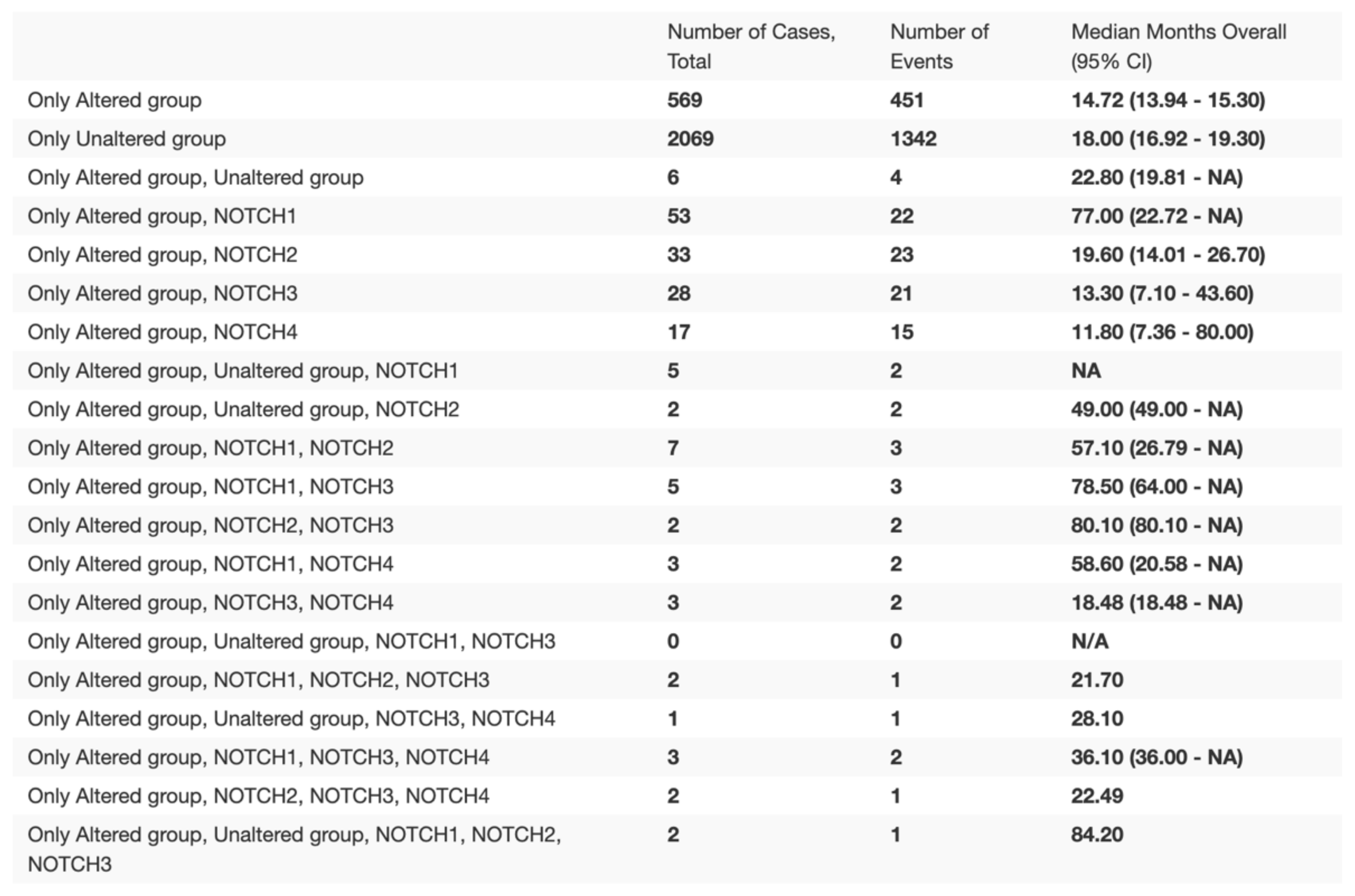
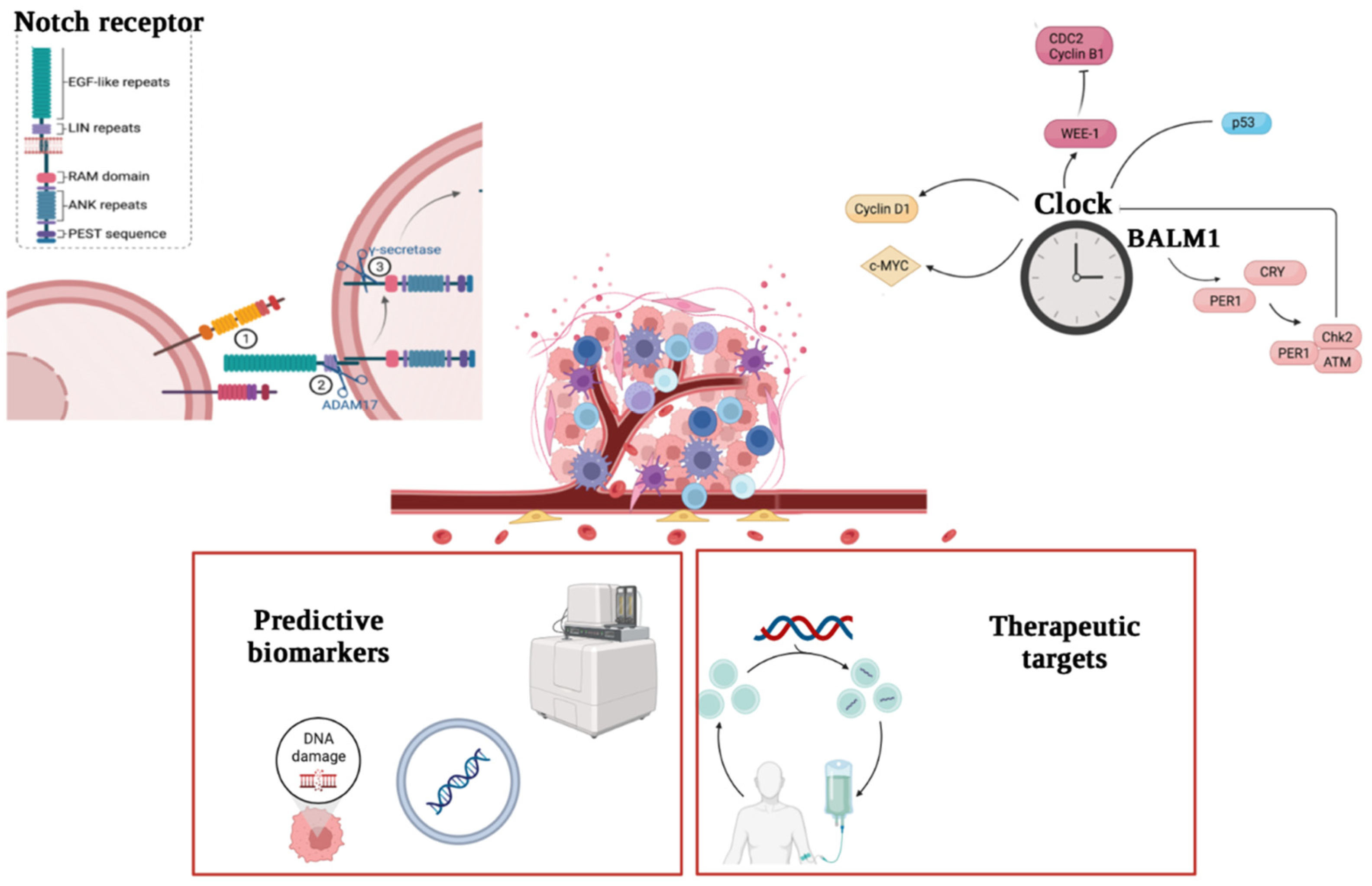
3. NOTCH Gene
NOTCH Gene as Therapeutic Target
4. Future Prospective
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The epidemiology of glioma in adults: A “state of the science” review. Neuro Oncol. 2014, 16, 896–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.-H.; Sheu, J.-J.; Lin, Y.-C.; Lin, H.-C. Association of migraines with brain tumors: A nationwide population-based study. J. Headache Pain 2018, 19, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Goffaux, P.; Fortin, D. Brain tumor headaches: From bedside to bench. Neurosurgery 2010, 67, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Lipton, R.B.; Bigal, M.E.; Diamond, M.; Freitag, F.; Reed, M.L.; Stewart, W.F.; On behalf of the AMPP Advisory Group. Migraine prevalence, disease burden, and the need for preventive therapy. Neurology 2007, 68, 343–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wakeman, T.P.; Lathia, J.D.; Hjelmeland, A.B.; Wang, X.-F.; White, R.R.; Rich, J.N.; Sullenger, B.A. Notch Promotes Radioresistance of Glioma Stem Cells. Stem Cells 2009, 28, 17–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haldbo-Classen, L.; Amidi, A.; Wu, L.; Lukacova, S.; Oettingen, G.; Lassen-Ramshad, Y.; Zachariae, R.; Kallehauge, J.; Høyer, M. Associations between patient-reported outcomes and radiation dose in patients treated with radiation therapy for primary brain tumours. Clin. Transl. Radiat. Oncol. 2021, 31, 86–92. [Google Scholar] [CrossRef]
- Liu, A.; Hou, C.; Chen, H.; Zong, X.; Zong, P. Genetics and Epigenetics of Glioblastoma: Applications and Overall Incidence of IDH1 Mutation. Front. Oncol. 2016, 6, 16. [Google Scholar] [CrossRef] [Green Version]
- Wagner, P.; Prucca, C.; Caputto, B.; Guido, M. Adjusting the Molecular Clock: The Importance of Circadian Rhythms in the Development of Glioblastomas and Its Intervention as a Therapeutic Strategy. Int. J. Mol. Sci. 2021, 22, 8289. [Google Scholar] [CrossRef]
- Jung, C.-H.; Kim, E.M.; Park, J.K.; Hwang, S.-G.; Moon, S.-K.; Kim, W.-J.; Um, H.-D. Bmal1 suppresses cancer cell invasion by blocking the phosphoinositide 3-kinase-Akt-MMP-2 signaling pathway. Oncol. Rep. 2013, 29, 2109–2113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.M. Notch signaling and Notch signaling modifiers. Int. J. Biochem. Cell Biol. 2011, 43, 1550–1562. [Google Scholar] [CrossRef]
- Ohgaki, H.; Kleihues, P. Population-Based Studies on Incidence, Survival Rates, and Genetic Alterations in Astrocytic and Oligodendroglial Gliomas. J. Neuropathol. Exp. Neurol. 2005, 64, 479–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohler, B.A.; Sherman, R.L.; Howlader, N.; Jemal, A.; Ryerson, A.B.; Henry, K.A.; Boscoe, F.P.; Cronin, K.A.; Lake, A.; Noone, A.-M.; et al. Annual Report to the Nation on the Status of Cancer, 1975–2011, Featuring Incidence of Breast Cancer Subtypes by Race/Ethnicity, Poverty, and State. J. Natl. Cancer Inst. 2015, 107, djv048, Erratum in J. Natl. Cancer Inst. 2015, 107, djv121. [Google Scholar] [CrossRef] [PubMed]
- WHO Classification of Tumours Editorial Board. WHO Classification of Tumours: Central Nervous System Tumours, 5th ed.; The new WHO classification of CNS tumours, the largest and most molecularly driven classification to date; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrom, Q.T.; Barnholtz-Sloan, J.S. Current State of Our Knowledge on Brain Tumor Epidemiology. Curr. Neurol. Neurosci. Rep. 2011, 11, 329–335. [Google Scholar] [CrossRef]
- Dico, A.L.; Salvatore, D.; Martelli, C.; Ronchi, D.; Diceglie, C.; Lucignani, G.; Ottobrini, L. Intracellular Redox-Balance Involvement in Temozolomide Resistance-Related Molecular Mechanisms in Glioblastoma. Cells 2019, 8, 1315. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Chen, G. Insights about circadian clock in glioma: From molecular pathways to therapeutic drugs. CNS Neurosci. Ther. 2022, 28, 1930–1941. [Google Scholar] [CrossRef]
- Grant, R. Overview: Brain tumour diagnosis and management/Royal College of Physicians guidelines. J. Neurol. Neurosurg. Psychiatry 2004, 75, ii18–ii23. [Google Scholar] [CrossRef] [Green Version]
- Chang, S.M.; Parney, I.; Huang, W.; Anderson, J.F.A.; Asher, A.L.; Bernstein, M.; Lillehei, K.O.; Brem, H.; Berger, M.S.; Laws, J.E.R.; et al. Patterns of Care for Adults with Newly Diagnosed Malignant Glioma. JAMA 2005, 293, 557–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanai, N.; Berger, M.S. Operative techniques for gliomas and the value of extent of resection. Neurotherapeutics 2009, 6, 478–486. [Google Scholar] [CrossRef] [Green Version]
- Keime-Guibert, F.; Chinot, O.; Taillandier, L.; Cartalat-Carel, S.; Frenay, M.; Kantor, G.; Guillamo, J.-S.; Jadaud, E.; Colin, P.; Bondiau, P.-Y.; et al. Radiotherapy for Glioblastoma in the Elderly. N. Engl. J. Med. 2007, 356, 1527–1535. [Google Scholar] [CrossRef]
- Prabhu, R.S.; Won, M.; Shaw, E.G.; Hu, C.; Brachman, D.G.; Buckner, J.C.; Stelzer, K.J.; Barger, G.R.; Brown, P.D.; Gilbert, M.R.; et al. Effect of the Addition of Chemotherapy to Radiotherapy on Cognitive Function in Patients With Low-Grade Glioma: Secondary Analysis of RTOG 98-02. J. Clin. Oncol. 2014, 32, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Hart, M.G.; Garside, R.; Rogers, G.; Somerville, M.; Stein, K.; Grant, R. Chemotherapy wafers for high grade glioma. Cochrane Database Syst. Rev. 2011, 2011, CD007294. [Google Scholar] [CrossRef] [PubMed]
- Lasorella, A.; Sanson, M.; Iavarone, A. FGFR-TACC gene fusions in human glioma. Neuro-Oncology 2016, 19, 475–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francisco, L.M.; Sage, P.T.; Sharpe, A.H. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev. 2010, 236, 219–242. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Cook, K.; Gee, H.E.; Hau, E. Hypoxia, metabolism, and the circadian clock: New links to overcome radiation resistance in high-grade gliomas. J. Exp. Clin. Cancer Res. 2020, 39, 129. [Google Scholar] [CrossRef]
- Li, A.; Lin, X.; Tan, X.; Yin, B.; Han, W.; Zhao, J.; Yuan, J.; Qiang, B.; Peng, X. Circadian gene Clock contributes to cell proliferation and migration of glioma and is directly regulated by tumor-suppressive miR-124. FEBS Lett. 2013, 587, 2455–2460. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Su, G.; Dai, Z.; Meng, M.; Zhang, H.; Fan, F.; Liu, Z.; Zhang, L.; Weygant, N.; He, F.; et al. Circadian clock genes promote glioma progression by affecting tumour immune infiltration and tumour cell proliferation. Cell Prolif. 2021, 54, e12988. [Google Scholar] [CrossRef]
- Xiang, S.; Mao, L.; Duplessis, T.; Yuan, L.; Dauchy, R.; Dauchy, E.; Blask, D.; Frasch, T.; Hill, S. Oscillation of Clock and Clock Controlled Genes Induced by Serum Shock in Human Breast Epithelial and Breast Cancer Cells: Regulation by Melatonin. Breast Cancer Basic Clin. Res. 2012, 6, 137–150. [Google Scholar] [CrossRef]
- Rana, S.; Mahmood, S. Circadian rhythm and its role in malignancy. J. Circadian Rhythm. 2010, 8, 3. [Google Scholar] [CrossRef] [Green Version]
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.R.; Huang, A.L.; Ptáček, L.J.; Fu, Y.-H. Genetic basis of human circadian rhythm disorders. Exp. Neurol. 2013, 243, 28–33. [Google Scholar] [CrossRef] [Green Version]
- Dunlap, J.C. Molecular Bases for Circadian Clocks. Cell 1999, 96, 271–290. [Google Scholar] [CrossRef] [Green Version]
- Burish, M.J.; Chen, Z.; Yoo, S.-H. Emerging relevance of circadian rhythms in headaches and neuropathic pain. Acta Physiol. 2018, 225, e13161. [Google Scholar] [CrossRef]
- Burish, M.J.; Chen, Z.; Yoo, S.-H. Cluster Headache Is in Part a Disorder of the Circadian System. JAMA Neurol. 2018, 75, 783. [Google Scholar] [CrossRef]
- Bacchelli, E.; Cainazzo, M.M.; Cameli, C.; Guerzoni, S.; Martinelli, A.; Zoli, M.; Maestrini, E.; Pini, L.A. A genome-wide analysis in cluster headache points to neprilysin and PACAP receptor gene variants. J. Headache Pain 2016, 17, 114. [Google Scholar] [CrossRef] [Green Version]
- Van Oosterhout, F.; Michel, S.; Deboer, T.; Houben, T.; van de Ven, R.C.G.; Albus, H.; Westerhout, J.; Vansteensel, M.J.; Ferrari, M.D.; Maagdenberg, A.M.; et al. Enhanced circadian phase resetting in R192Q Cav2.1 calcium channel migraine mice. Ann. Neurol. 2008, 64, 315–324. [Google Scholar] [CrossRef]
- Janich, P.; Pascual, G.; Merlos-Suárez, A.; Batlle, E.; Ripperger, J.; Albrecht, U.; Cheng, H.-Y.M.; Obrietan, K.; Di Croce, L.; Benitah, S.A. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature 2011, 480, 209–214. [Google Scholar] [CrossRef] [Green Version]
- Fekry, B.; Ribas-Latre, A.; Baumgartner, C.; Deans, J.R.; Kwok, C.; Patel, P.; Fu, L.; Berdeaux, R.; Sun, K.; Kolonin, M.G.; et al. Incompatibility of the circadian protein BMAL1 and HNF4α in hepatocellular carcinoma. Nat. Commun. 2018, 9, 4349. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Lee, J.E.; Kahng, J.Y.; Kim, S.H.; Park, J.S.; Yoon, S.J.; Um, J.Y.; Kim, W.K.; Lee, J.-K.; Park, J.; et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 2018, 560, 243–247. [Google Scholar] [CrossRef]
- Pascual, G.; Dominguez, D.; Benitah, S.A. The contributions of cancer cell metabolism to metastasis. Dis. Model. Mech. 2018, 11, dmm032920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferns, G.A.; Abadi, M.S.S.; Raeisi, A.; Arjmand, M.-H. The Potential Role of Changes in the Glucose and Lipid Metabolic Pathways in Gastrointestinal Cancer Progression: Strategy in Cancer Therapy. Gastrointest. Tumors 2021, 8, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agnihotri, S.; Zadeh, G. Metabolic reprogramming in glioblastoma: The influence of cancer metabolism on epigenetics and unanswered questions. Neuro-Oncology 2015, 18, 160–172. [Google Scholar] [CrossRef] [Green Version]
- Marin-Valencia, I.; Cho, S.K.; Rakheja, D.; Hatanpaa, K.J.; Kapur, P.; Mashimo, T.; Jindal, A.; Vemireddy, V.; Good, L.B.; Raisanen, J.; et al. Glucose metabolism via the pentose phosphate pathway, glycolysis and Krebs cycle in an orthotopic mouse model of human brain tumors. NMR Biomed. 2012, 25, 1177–1186. [Google Scholar] [CrossRef] [Green Version]
- Sulli, G.; Lam, M.T.Y.; Panda, S. Interplay between Circadian Clock and Cancer: New Frontiers for Cancer Treatment. Trends Cancer 2019, 5, 475–494. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [Green Version]
- De La Cruz Minyety, J.; Shuboni-Mulligan, D.D.; Briceno, N.; Young, D.; Gilbert, M.R., Jr.; Celiku, O.; Armstrong, T.S. Association of Circadian Clock Gene Expression with Glioma Tumor Microenvironment and Patient Survival. Cancers 2021, 13, 2756. [Google Scholar] [CrossRef]
- Xuan, W.; Hsu, W.H.; Khan, F.; Dunterman, M.; Pang, L.; Wainwright, D.A.; Ahmed, A.U.; Heimberger, A.B.; Lesniak, M.S.; Chen, P. Circadian Regulator CLOCK Drives Immunosuppression in Glioblastoma. Cancer Immunol. Res. 2022, 10, 770–784. [Google Scholar] [CrossRef]
- Ye, Y.; Xiang, Y.; Ozguc, F.M.; Kim, Y.; Liu, C.-J.; Park, P.K.; Hu, Q.; Diao, L.; Lou, Y.; Lin, C.; et al. The Genomic Landscape and Pharmacogenomic Interactions of Clock Genes in Cancer Chronotherapy. Cell Syst. 2018, 6, 314–328.e2. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, Y.; Zhang, N.; Yuan, H.; Wang, F.; Xu, H.; Yu, J.; Ma, J.; Hou, S.; Cao, X. IDH1 gene mutation activates Smad signaling molecules to regulate the expression levels of cell cycle and biological rhythm genes in human glioma U87-MG cells. Mol. Med. Rep. 2021, 23, 5. [Google Scholar] [CrossRef] [PubMed]
- Bunger, M.K.; Wilsbacher, L.D.; Moran, S.M.; Clendenin, C.; Radcliffe, L.A.; Hogenesch, J.B.; Simon, M.C.; Takahashi, J.S.; Bradfield, C.A. Mop3 Is an Essential Component of the Master Circadian Pacemaker in Mammals. Cell 2000, 103, 1009–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keesler, G.A.; Camacho, F.; Guo, Y.; Virshup, D.; Mondadori, C.; Yao, Z. Phosphorylation and destabilization of human period I clock protein by human casein kinase I epsilon. Neuroreport 2000, 11, 951–955. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Pelicano, H.; Liu, J.; Huang, P.; Lee, C.C. The circadian gene period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 2002, 111, 41–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Relógio, A.; Thomas, P.; Medina-Pérez, P.; Reischl, S.; Bervoets, S.; Gloc, E.; Riemer, P.; Mang-Fatehi, S.; Maier, B.; Schäfer, R.; et al. Ras-Mediated Deregulation of the Circadian Clock in Cancer. PLoS Genet. 2014, 10, e1004338. [Google Scholar] [CrossRef]
- Levi, F.; Schibler, U. Circadian Rhythms: Mechanisms and Therapeutic Implications. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 593–628. [Google Scholar] [CrossRef]
- Kiessling, S.; Beaulieu-Laroche, L.; Blum, I.D.; Landgraf, D.; Welsh, D.K.; Storch, K.-F.; Labrecque, N.; Cermakian, N. Enhancing circadian clock function in cancer cells inhibits tumor growth. BMC Biol. 2017, 15, 13. [Google Scholar] [CrossRef] [Green Version]
- Gorbacheva, V.Y.; Kondratov, R.V.; Zhang, R.; Cherukuri, S.; Gudkov, A.V.; Takahashi, J.S.; Antoch, M.P. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc. Natl. Acad. Sci. USA 2005, 102, 3407–3412. [Google Scholar] [CrossRef] [Green Version]
- Antoch, M.P.; Chernov, M.V. Pharmacological modulators of the circadian clock as potential therapeutic drugs. Mutat Res. 2009, 679, 17–23. [Google Scholar] [CrossRef] [Green Version]
- Ozturk, N.; Lee, J.H.; Gaddameedhi, S.; Sancar, A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc. Natl. Acad. Sci. USA 2009, 106, 2841–2846. [Google Scholar] [CrossRef]
- Lee, J.H.; Sancar, A. Circadian clock disruption improves the efficacy of chemotherapy through p73-mediated apoptosis. Proc. Natl. Acad. Sci. USA 2011, 108, 10668–10672. [Google Scholar] [CrossRef] [Green Version]
- Andersen, R.S.; Anand, A.; Harwood, D.S.L.; Kristensen, B.W. Tumor-Associated Microglia and Macrophages in the Glioblastoma Microenvironment and Their Implications for Therapy. Cancers 2021, 13, 4255. [Google Scholar] [CrossRef]
- Chen, P.; Hsu, W.-H.; Chang, A.; Tan, Z.; Lan, Z.; Zhou, A.; Spring, D.J.; Lang, F.F.; Wang, Y.A.; DePinho, R.A. Circadian Regulator CLOCK Recruits Immune-Suppressive Microglia into the GBM Tumor Microenvironment. Cancer Discov. 2020, 10, 371–381. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wang, J.; Zhang, X.; Tang, Q. New Insights into Cancer Chronotherapies. Front. Pharmacol. 2021, 12, 741295. [Google Scholar] [CrossRef]
- Slat, E.A.; Sponagel, J.; Marpegan, L.; Simon, T.; Kfoury, N.; Kim, A.; Binz, A.; Herzog, E.D.; Rubin, J.B. Cell-intrinsic, Bmal1-Dependent Circadian Regulation of Temozolomide Sensitivity in Glioblastoma. J. Biol. Rhythm. 2017, 32, 121–129. [Google Scholar] [CrossRef] [Green Version]
- Vlachostergios, P.; Hatzidaki, E.; Stathakis, N.E.; Koukoulis, G.K.; Papandreou, C.N. Bortezomib Downregulates MGMT Expression in T98G Glioblastoma Cells. Cell. Mol. Neurobiol. 2013, 33, 313–318. [Google Scholar] [CrossRef]
- Goldsmith, C.S.; Kim, S.M.; Karunarathna, N.; Neuendorff, N.; Toussaint, L.G.; Earnest, D.J.; Bell-Pedersen, D. Inhibition of p38 MAPK activity leads to cell type-specific effects on the molecular circadian clock and time-dependent reduction of glioma cell invasiveness. BMC Cancer 2018, 18, 43. [Google Scholar] [CrossRef] [Green Version]
- Kopan, R.; Ilagan, M.X.G. The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef] [Green Version]
- Pierfelice, T.; Alberi, L.; Gaiano, N. Notch in the Vertebrate Nervous System: An Old Dog with New Tricks. Neuron 2011, 69, 840–855. [Google Scholar] [CrossRef] [Green Version]
- Sutherland, H.G.; Albury, C.L.; Griffiths, L.R. Advances in genetics of migraine. J. Headache Pain 2019, 20, 72. [Google Scholar] [CrossRef]
- Wang, W.; Prince, C.Z.; Mou, Y.; Pollman, M.J. Notch3 Signaling in Vascular Smooth Muscle Cells Induces c-FLIP Expression via ERK/MAPK Activation. J. Biol. Chem. 2002, 277, 21723–21729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joutel, A.; Corpechot, C.; Ducros, A.; Vahedi, K.; Chabriat, H.; Mouton, P.; Alamowitch, S.; Domenga, V.; Cécillion, M.; Maréchal, E.; et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 1996, 383, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Rusanescu, G.; Mao, J. Notch3 is necessary for neuronal differentiation and maturation in the adult spinal cord. J. Cell Mol. Med. 2014, 18, 2103–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siebel, C.; Lendahl, U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowell, C.S.; Radtke, F. Notch as a tumour suppressor. Nat. Rev. Cancer 2017, 17, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Simpson, M.A.; Irving, M.D.; Asilmaz, E.; Gray, M.J.; Dafou, D.; Elmslie, F.V.; Mansour, S.; Holder, S.E.; Brain, C.E.; Burton, B.K.; et al. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat. Genet. 2011, 43, 303–305. [Google Scholar] [CrossRef]
- Miao, K.; Lei, J.H.; Valecha, M.V.; Zhang, A.; Xu, J.; Wang, L.; Lyu, X.; Chen, S.; Miao, Z.; Zhang, X.; et al. NOTCH1 activation compensates BRCA1 deficiency and promotes triple-negative breast cancer formation. Nat. Commun. 2020, 11, 3256. [Google Scholar] [CrossRef]
- Parmigiani, E.; Taylor, V.; Giachino, C. Oncogenic and Tumor-Suppressive Functions of NOTCH Signaling in Glioma. Cells 2020, 9, 2304. [Google Scholar] [CrossRef]
- Goruganthu, M.U.L.; Shanker, A.; Dikov, M.M.; Carbone, D.P. Specific Targeting of Notch Ligand-Receptor Interactions to Modulate Immune Responses: A Review of Clinical and Preclinical Findings. Front. Immunol. 2020, 11, 1958. [Google Scholar] [CrossRef]
- Abravanel, D.L.; Belka, G.K.; Pan, T.C.; Pant, D.K.; Collins, M.A.; Sterner, C.J.; Chodosh, L.A. Notch promotes recurrence of dormant tumor cells following HER2/neu-targeted therapy. J. Clin. Investig. 2015, 125, 2484–2496. [Google Scholar] [CrossRef]
- Gordon, W.; Ulu, D.V.; Histen, G.; Sanchez-Irizarry, C.; Aster, J.C.; Blacklow, S.C. Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 2007, 14, 295–300. [Google Scholar] [CrossRef]
- Yahyanejad, S.; King, H.; Iglesias, V.S.; Granton, P.V.; Barbeau, L.M.; van Hoof, S.J.; Groot, A.J.; Habets, R.; Prickaerts, J.; Chalmers, A.J.; et al. NOTCH blockade combined with radiation therapy and temozolomide prolongs survival of orthotopic glioblastoma. Oncotarget 2016, 7, 41251–41264. [Google Scholar] [CrossRef] [Green Version]
- Krop, I.; Demuth, T.; Guthrie, T.; Wen, P.Y.; Mason, W.P.; Chinnaiyan, P.; Butowski, N.; Groves, M.D.; Kesari, S.; Freedman, S.J.; et al. Phase I Pharmacologic and Pharmacodynamic Study of the Gamma Secretase (Notch) Inhibitor MK-0752 in Adult Patients with Advanced Solid Tumors. J. Clin. Oncol. 2012, 30, 2307–2313. [Google Scholar] [CrossRef]
- Espinoza, I.; Miele, L. Notch inhibitors for cancer treatment. Pharmacol. Ther. 2013, 139, 95–110. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casili, G.; Lanza, M.; Filippone, A.; Caffo, M.; Paterniti, I.; Campolo, M.; Colarossi, L.; Sciacca, D.; Lombardo, S.P.; Cuzzocrea, S.; et al. Overview on Common Genes Involved in the Onset of Glioma and on the Role of Migraine as Risk Factor: Predictive Biomarkers or Therapeutic Targets? J. Pers. Med. 2022, 12, 1969. https://doi.org/10.3390/jpm12121969
Casili G, Lanza M, Filippone A, Caffo M, Paterniti I, Campolo M, Colarossi L, Sciacca D, Lombardo SP, Cuzzocrea S, et al. Overview on Common Genes Involved in the Onset of Glioma and on the Role of Migraine as Risk Factor: Predictive Biomarkers or Therapeutic Targets? Journal of Personalized Medicine. 2022; 12(12):1969. https://doi.org/10.3390/jpm12121969
Chicago/Turabian StyleCasili, Giovanna, Marika Lanza, Alessia Filippone, Maria Caffo, Irene Paterniti, Michela Campolo, Lorenzo Colarossi, Dorotea Sciacca, Sofia Paola Lombardo, Salvatore Cuzzocrea, and et al. 2022. "Overview on Common Genes Involved in the Onset of Glioma and on the Role of Migraine as Risk Factor: Predictive Biomarkers or Therapeutic Targets?" Journal of Personalized Medicine 12, no. 12: 1969. https://doi.org/10.3390/jpm12121969
APA StyleCasili, G., Lanza, M., Filippone, A., Caffo, M., Paterniti, I., Campolo, M., Colarossi, L., Sciacca, D., Lombardo, S. P., Cuzzocrea, S., & Esposito, E. (2022). Overview on Common Genes Involved in the Onset of Glioma and on the Role of Migraine as Risk Factor: Predictive Biomarkers or Therapeutic Targets? Journal of Personalized Medicine, 12(12), 1969. https://doi.org/10.3390/jpm12121969












