Patient-Specific Mathematical Model of the Clear Cell Renal Cell Carcinoma Microenvironment
Abstract
1. Introduction
2. Materials and Methods
2.1. Variables and Network
2.2. Mathematical Model
2.2.1. Cells
2.2.2. Molecules and Proteins
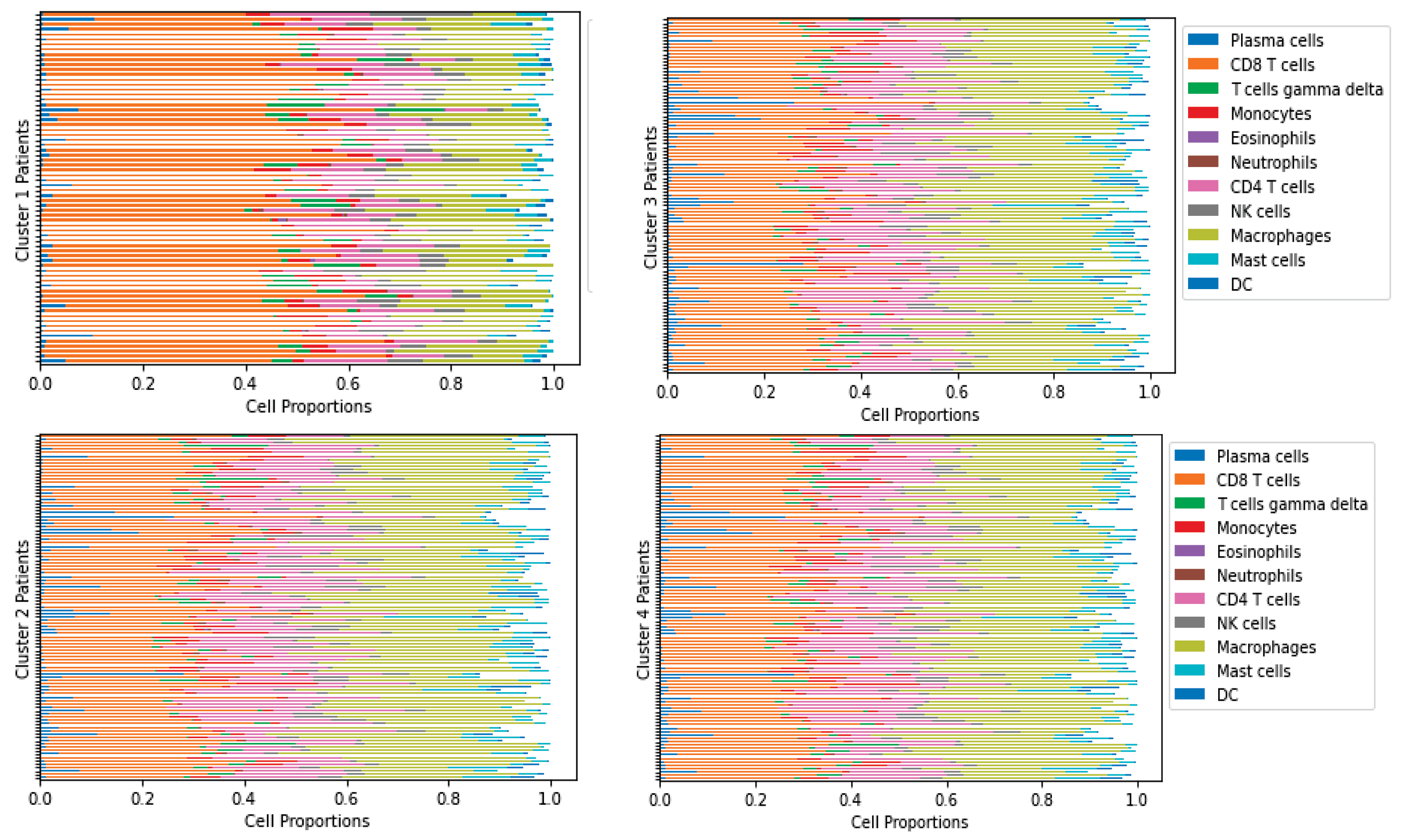
2.3. Data Preparation
2.4. Parameter Estimation
2.5. Sensitivity Analysis
3. Results
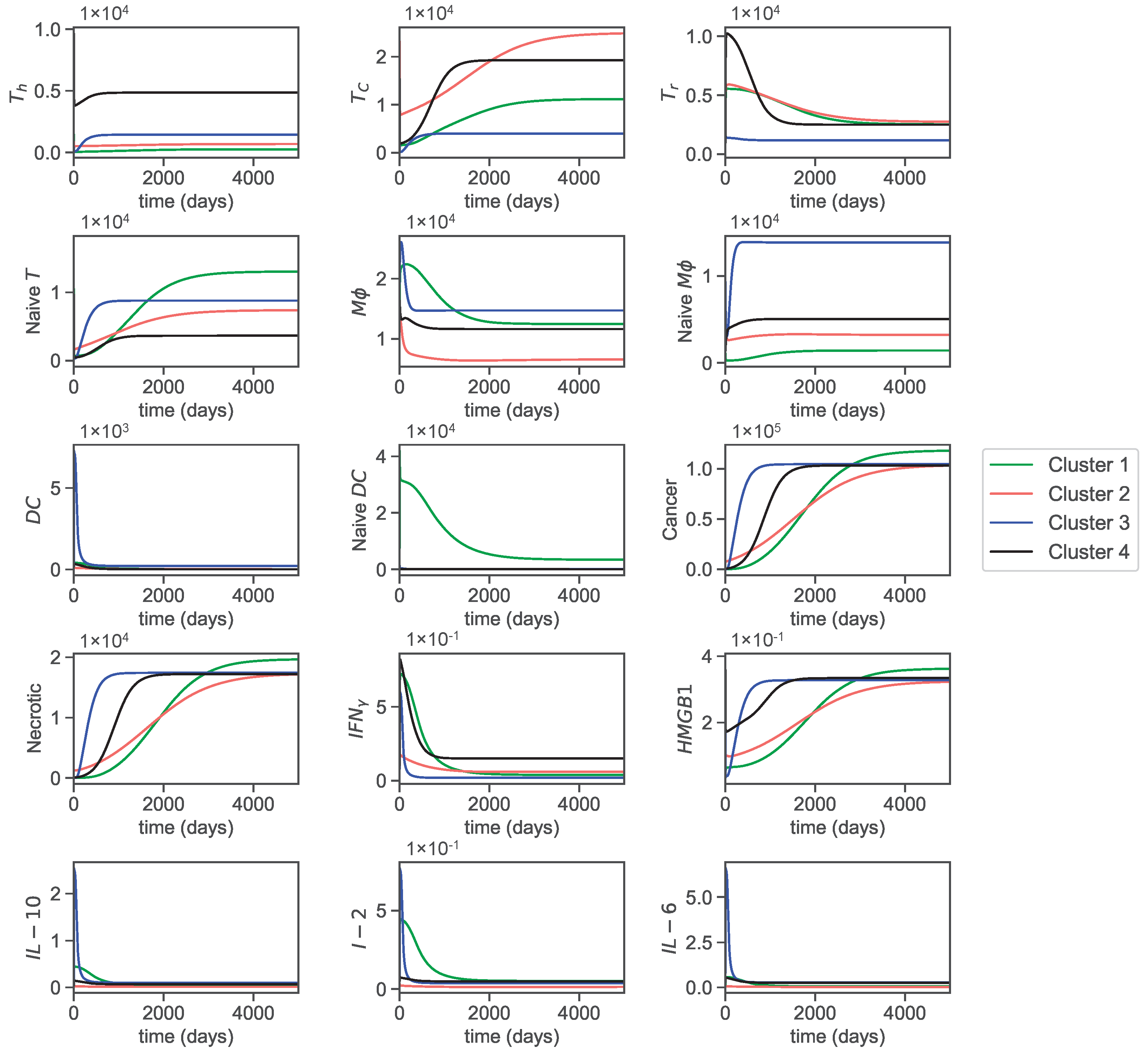
3.1. Dynamics
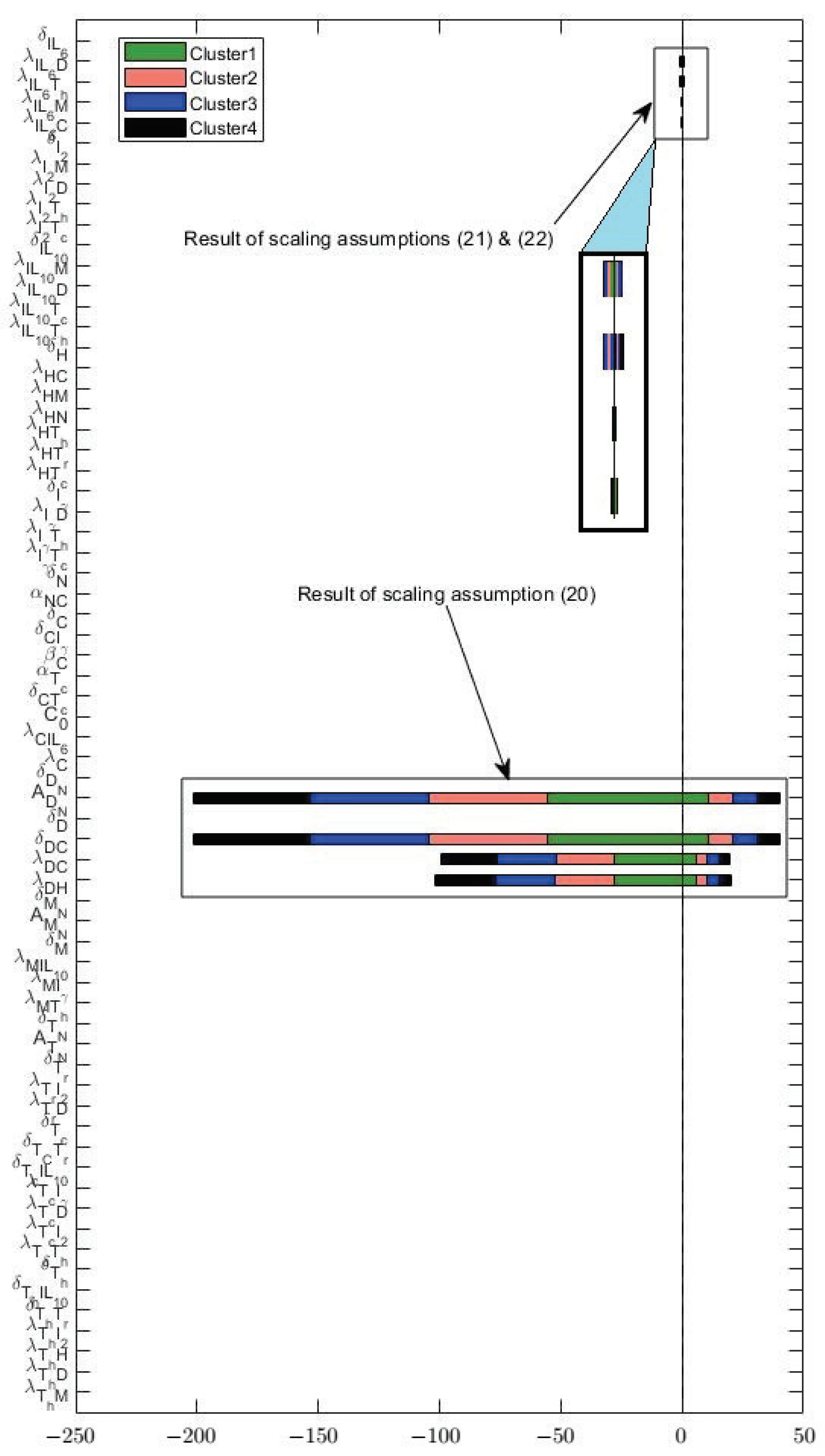
3.2. Sensitivity
3.3. Varying Dynamics of Cancer Cells with Scaled Assumptions
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ccRCC | clear cell renal cell carcinoma |
| ODE | ordinary differential equation |
| NK | natural killer |
| PD | programmed cell death |
| PD-L | programmed cell death ligand |
| IFN- | interferon- |
| DAMP | damage-associated molecular pattern |
| HMGB1 | high mobility group box-1 |
| VEGF | vascular endothelial growth factor |
| IL | interleukin |
| TCGA | The Cancer Genome Atlas |
| TCN | total cell number |
| TIC | total immune cell |
Appendix A. Derivation of Sample Parameters
Appendix A.1. Parameter Assumptions Based on Steady States
| 10 | ||||
Appendix A.2. Nondimensionalization of Parameters and Variables
| Parameter | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 |
|---|---|---|---|---|
References
- National Cancer Institute. Clear Cell Renal Cell Carcinoma. Available online: https://www.cancer.gov/pediatric-adult-rare-tumor/rare-tumors/rare-kidney-tumors/clear-cell-renal-cell-carcinoma (accessed on 12 August 2020).
- Su, S.; Akbarinejad, S.; Shahriyari, L. Immune classification of clear cell renal cell carcinoma. Sci. Rep. 2021, 11, 4338. [Google Scholar] [CrossRef] [PubMed]
- Gkolfinopoulos, S.; Psyrri, A.; Bamias, A. Clear-cell renal cell carcinoma—A comprehensive review of agents used in the contemporary management of advanced/metastatic disease. Oncol. Rev. 2021, 15, 530. [Google Scholar] [CrossRef]
- Ganguly, R.; Puri, I.K. Mathematical model for chemotherapeutic drug efficacy in arresting tumour growth based on the cancer stem cell hypothesis. Cell Prolif. 2007, 40, 338–354. [Google Scholar] [CrossRef]
- Komarova, N.L.; Shahriyari, L.; Wodarz, D. Complex role of space in the crossing of fitness valleys by asexual populations. J. R. Soc. Interface 2014, 11, 20140014. [Google Scholar] [CrossRef] [PubMed]
- Shahriyari, L. Cell dynamics in tumour environment after treatments. J. R. Soc. Interface 2017, 14, 20160977. [Google Scholar] [CrossRef] [PubMed]
- Le, T.; Su, S.; Shahriyari, L. Investigating Optimal Chemotherapy Options for Osteosarcoma Patients through a Mathematical Model. Cells 2021, 10, 2009. [Google Scholar] [CrossRef]
- Shahriyari, L.; Komarova, N.L. The role of the bi-compartmental stem cell niche in delaying cancer. Phys. Biol. 2015, 12, 055001. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.; Hao, W. The Role of Exosomes in Pancreatic Cancer Microenvironment. Bull. Math. Biol. 2018, 80, 1111–1133. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Stiff, A.; Duggan, M.; Wesolowski, R.; Carson, W.E.; Friedman, A. Modeling combination therapy for breast cancer with BET and immune checkpoint inhibitors. Proc. Natl. Acad. Sci. USA 2018, 115, 5534–5539. [Google Scholar] [CrossRef]
- Rhodes, A.; Hillen, T. A mathematical model for the immune-mediated theory of metastasis. J. Theor. Biol. 2019, 482, 109999. [Google Scholar] [CrossRef] [PubMed]
- Budithi, A.; Su, S.; Kirshtein, A.; Shahriyari, L. Data Driven Mathematical Model of FOLFIRI Treatment for Colon Cancer. Cancers 2021, 13, 2632. [Google Scholar] [CrossRef] [PubMed]
- Mohammad Mirzaei, N.; Tatarova, Z.; Hao, W.; Changizi, N.; Asadpoure, A.; Zervantonakis, I.K.; Hu, Y.; Chang, Y.H.; Shahriyari, L. A PDE Model of Breast Tumor Progression in MMTV-PyMT Mice. J. Pers. Med. 2022, 12, 807. [Google Scholar] [CrossRef]
- Pillis, L.D.; Caldwell, T.; Sarapata, E.; Williams, H. Mathematical Modeling of the Regulatory T Cell Effects on Renal Cell Carcinoma Treatment. Discret. Contin. Dyn. Syst. Ser. B 2013, 18, 915–943. [Google Scholar] [CrossRef]
- Sharma, K.; Patidar, K.; Ali, M.A.; Patil, P.; Goud, H.; Hussain, T.; Nayarisseri, A.; Singh, S.K. Structure-Based Virtual Screening for the Identification of High Affinity Compounds as Potent VEGFR2 Inhibitors for the Treatment of Renal Cell Carcinoma. Curr. Top. Med. Chem. 2018, 18, 2174–2185. [Google Scholar] [CrossRef]
- Kirshtein, A.; Akbarinejad, S.; Hao, W.; Le, T.; Su, S.; Aronow, R.A.; Shahriyari, L. Data Driven Mathematical Model of Colon Cancer Progression. J. Clin. Med. 2020, 9, 3947. [Google Scholar] [CrossRef]
- Le, T.; Su, S.; Kirshtein, A.; Shahriyari, L. Data-Driven Mathematical Model of Osteosarcoma. Cancers 2021, 13, 2367. [Google Scholar] [CrossRef] [PubMed]
- Mohammad Mirzaei, N.; Su, S.; Sofia, D.; Hegarty, M.; Abdel-Rahman, M.H.; Asadpoure, A.; Cebulla, C.M.; Chang, Y.H.; Hao, W.; Jackson, P.R.; et al. A Mathematical Model of Breast Tumor Progression Based on Immune Infiltration. J. Pers. Med. 2021, 11, 1031. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Falo, L.D.; You, Z. Knockdown of HMGB1 in tumor cells attenuates their ability to induce regulatory T cells and uncovers naturally acquired CD8 T cell-dependent antitumor immunity. J. Immunol. 2011, 187, 118–125. [Google Scholar] [CrossRef]
- Fyfe, G.; Fisher, R.I.; Rosenberg, S.A.; Sznol, M.; Parkinson, D.R.; Louie, A.C. Results of treatment of 255 patients with metastatic renal cell carcinoma who received high-dose recombinant interleukin-2 therapy. J. Clin. Oncol. 1995, 13, 688–696. [Google Scholar] [CrossRef]
- Escudier, B.; Pluzanska, A.; Koralewski, P.; Ravaud, A.; Bracarda, S.; Szczylik, C.; Chevreau, C.; Filipek, M.; Melichar, B.; Bajetta, E.; et al. Bevacizumab plus interferon alfa-2a for treatment of metastatic renal cell carcinoma: A randomised, double-blind phase III trial. Lancet 2007, 370, 2103–2111. [Google Scholar] [CrossRef]
- Rini, B.I.; Halabi, S.; Rosenberg, J.E.; Stadler, W.M.; Vaena, D.A.; Ou, S.S.; Archer, L.; Atkins, J.N.; Picus, J.; Czaykowski, P.; et al. Bevacizumab plus interferon alfa compared with interferon alfa monotherapy in patients with metastatic renal cell carcinoma: CALGB 90206. J. Clin. Oncol. 2008, 26, 5422. [Google Scholar] [CrossRef] [PubMed]
- Lalani, A.K.A.; McGregor, B.A.; Albiges, L.; Choueiri, T.K.; Motzer, R.; Powles, T.; Wood, C.; Bex, A. Systemic treatment of metastatic clear cell renal cell carcinoma in 2018: Current paradigms, use of immunotherapy, and future directions. Eur. Urol. 2019, 75, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Choueiri, T.K.; Larkin, J.; Oya, M.; Thistlethwaite, F.; Martignoni, M.; Nathan, P.; Powles, T.; McDermott, D.; Robbins, P.B.; Chism, D.D.; et al. Preliminary results for avelumab plus axitinib as first-line therapy in patients with advanced clear-cell renal-cell carcinoma (JAVELIN Renal 100): An open-label, dose-finding and dose-expansion, phase 1b trial. Lancet Oncol. 2018, 19, 451–460. [Google Scholar] [CrossRef]
- Peng, W.; Liu, C.; Xu, C.; Lou, Y.; Chen, J.; Yang, Y.; Yagita, H.; Overwijk, W.W.; Lizée, G.; Radvanyi, L.; et al. PD-1 Blockade Enhances T-cell Migration to Tumors by Elevating IFN-γ Inducible Chemokines. Cancer Res. 2012, 72, 5209–5218. [Google Scholar] [CrossRef]
- Han, Y.; Liu, D.; Li, L. PD-1/PD-L1 pathway: Current researches in cancer. Am. J. Cancer Res. 2020, 10, 727–742. [Google Scholar]
- Lai, X.; Hao, W.; Friedman, A. TNF-α inhibitor reduces drug-resistance to anti-PD-1: A mathematical model. PLoS ONE 2020, 15, e0231499. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hu, Y.; Wangkahart, E.; Liu, F.; Wang, A.; Zahran, E.; Maisey, K.R.; Liu, M.; Xu, Q.; Imarai, M.; et al. Interleukin (IL)-2 Is a Key Regulator of T Helper 1 and T Helper 2 Cytokine Expression in Fish: Functional Characterization of Two Divergent IL2 Paralogs in Salmonids. Front. Immunol. 2018, 9, 1683. [Google Scholar] [CrossRef]
- Xu, X.; Fu, X.Y.; Plate, J.; Chong, A.S. IFN-gamma induces cell growth inhibition by Fas-mediated apoptosis: Requirement of STAT1 protein for up-regulation of Fas and FasL expression. Cancer Res. 1998, 58, 2832–2837. [Google Scholar]
- Luo, Y.; Chihara, Y.; Fujimoto, K.; Sasahira, T.; Kuwada, M.; Fujiwara, R.; Fujii, K.; Ohmori, H.; Kuniyasu, H. High mobility group box 1 released from necrotic cells enhances regrowth and metastasis of cancer cells that have survived chemotherapy. Eur. J. Cancer 2013, 49, 741–751. [Google Scholar] [CrossRef]
- Kostova, N.; Zlateva, S.; Ugrinova, I.; Pasheva, E. The expression of HMGB1 protein and its receptor RAGE in human malignant tumors. Mol. Cell Biochem. 2010, 337, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Apetoh, L.; Ghiringhelli, F.; Tesniere, A.; Criollo, A.; Ortiz, C.; Lidereau, R.; Mariette, C.; Chaput, N.; Mira, J.P.; Delaloge, S.; et al. The interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radiotherapy. Immunol. Rev. 2007, 220, 47–59. [Google Scholar] [CrossRef]
- Scaffidi, P.; Misteli, T.; Bianchi, M.E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002, 418, 191–195. [Google Scholar] [CrossRef]
- Lotze, M.T.; Tracey, K.J. High-mobility group box 1 protein (HMGB1): Nuclear weapon in the immune arsenal. Nat. Rev. Immunol. 2005, 5, 331–342. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Dieu-Nosjean, M.C.; Sautès-Fridman, C.; Fridman, W.H. Cancer immune contexture and immunotherapy. Curr. Opin. Immunol. 2016, 39, 7–13. [Google Scholar] [CrossRef]
- Vuong, L.; Kotecha, R.R.; Voss, M.H.; Hakimi, A.A. Tumor microenvironment dynamics in clear-cell renal cell carcinoma. Cancer Discov. 2019, 9, 1349–1357. [Google Scholar] [CrossRef]
- Vesely, M.D.; Kershaw, M.H.; Schreiber, R.D.; Smyth, M.J. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011, 29, 235–271. [Google Scholar] [CrossRef]
- Hakimi, A.A.; Voss, M.H.; Kuo, F.; Sanchez, A.; Liu, M.; Nixon, B.G.; Vuong, L.; Ostrovnaya, I.; Chen, Y.B.; Reuter, V.; et al. Transcriptomic Profiling of the Tumor Microenvironment Reveals Distinct Subgroups of Clear Cell Renal Cell Cancer: Data from a Randomized Phase III TrialGenomic Predictors of TKI Response in the COMPARZ Trial. Cancer Discov. 2019, 9, 510–525. [Google Scholar] [CrossRef]
- Beuselinck, B.; Job, S.; Becht, E.; Karadimou, A.; Verkarre, V.; Couchy, G.; Giraldo, N.; Rioux-Leclercq, N.; Molinié, V.; Sibony, M.; et al. Molecular Subtypes of Clear Cell Renal Cell Carcinoma Are Associated with Sunitinib Response in the Metastatic SettingTranscriptomic Predictor of Sunitinib Response in RCC. Clin. Cancer Res. 2015, 21, 1329–1339. [Google Scholar] [CrossRef]
- Ju, S.A.; Park, S.M.; Joe, Y.; Chung, H.T.; An, W.G.; Kim, B.S. T cells in renal cell carcinoma. Oncol. Lett. 2022, 23, 43. [Google Scholar] [CrossRef]
- Okeke, E.B.; Uzonna, J.E. The Pivotal Role of Regulatory T Cells in the Regulation of Innate Immune Cells. Front. Immunol. 2019, 10, 680. [Google Scholar] [CrossRef]
- Teh, B.T.; Farber, L.J.; Furge, K. Molecular Characterization of Renal Cell Carcinoma. In Renal Cell Carcinoma; Figlin, R.A., Rathmell, W.K., Rini, B.I., Eds.; Springer: Boston, MA, USA, 2012; pp. 91–111. [Google Scholar] [CrossRef]
- Wang, Y.; Chaudhri, G.; Jackson, R.J.; Karupiah, G. IL-12p40 and IL-18 play pivotal roles in orchestrating the cell-mediated immune response to a poxvirus infection. J. Immunol. 2009, 183, 3324–3331. [Google Scholar] [CrossRef]
- Aras, S.; Zaidi, M.R. TAMeless traitors: Macrophages in cancer progression and metastasis. Br. J. Cancer 2017, 117, 1583–1591. [Google Scholar] [CrossRef]
- Iyer, S.S.; Cheng, G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol. 2012, 32, 23–63. [Google Scholar] [CrossRef]
- Ng, T.H.; Britton, G.J.; Hill, E.V.; Verhagen, J.; Burton, B.R.; Wraith, D.C. Regulation of adaptive immunity; the role of interleukin-10. Front. Immunol. 2013, 4, 129. [Google Scholar] [CrossRef]
- Zitvogel, L.; Kroemer, G. Targeting PD-1/PD-L1 interactions for cancer immunotherapy. Oncoimmunology 2012, 1, 1223–1225. [Google Scholar] [CrossRef]
- Gudbrandsdottir, G.; Aarstad, H.H.; Hjelle, K.M.; Førde, K.; Reisæter, L.; Bostad, L.; Aarstad, H.J.; Beisland, C. The levels of IL-6 and soluble IL-33R are increased in the renal vein during surgery for clear cell renal cell carcinoma. Cytokine 2021, 144, 155586. [Google Scholar] [CrossRef]
- Grivennikov, S.; Karin, E.; Terzic, J.; Mucida, D.; Yu, G.Y.; Vallabhapurapu, S.; Scheller, J.; Rose-John, S.; Cheroutre, H.; Eckmann, L.; et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 2009, 15, 103–113. [Google Scholar] [CrossRef]
- Negrier, S.; Perol, D.; Menetrier-Caux, C.; Escudier, B.; Pallardy, M.; Ravaud, A.; Douillard, J.Y.; Chevreau, C.; Lasset, C.; Blay, J.Y. Interleukin-6, Interleukin-10, and Vascular Endothelial Growth Factor in Metastatic Renal Cell Carcinoma: Prognostic Value of Interleukin-6—From the Groupe Français d’Immunothérapie. JCO 2004, 22, 2371–2378. [Google Scholar] [CrossRef]
- Palucka, K.; Banchereau, J. Cancer immunotherapy via dendritic cells. Nat. Rev. Cancer 2012, 12, 265–277. [Google Scholar] [CrossRef]
- Süren, D.; Yıldırım, M.; Demirpençe, Ö.; Kaya, V.; Alikanoğlu, A.S.; Bülbüller, N.; Yıldız, M.; Sezer, C. The role of high mobility group box 1 (HMGB1) in colorectal cancer. Med. Sci. Monit. 2014, 20, 530–537. [Google Scholar]
- Galluzzi, L.; Kepp, O.; Kroemer, G. Immunogenic cell death in radiation therapy. Oncoimmunology 2013, 2, e26536. [Google Scholar] [CrossRef]
- Albert, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Boyman, O.; Sprent, J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 2012, 12, 180–190. [Google Scholar] [CrossRef]
- Chen, D.; Mellman, I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef]
- Levy, E.M.; Roberti, M.P.; Mordoh, J. Natural Killer Cells in Human Cancer: From Biological Functions to Clinical Applications. J. Biomed. Biotechnol. 2011, 2011, 676198 . [Google Scholar] [CrossRef]
- Pili, R.; Quinn, D.I.; Hammers, H.J.; Monk, P.; George, S.; Dorff, T.B.; Olencki, T.; Shen, L.; Orillion, A.; Lamonica, D.; et al. Immunomodulation by Entinostat in Renal Cell Carcinoma Patients Receiving High-Dose Interleukin 2: A Multicenter, Single-Arm, Phase I/II Trial (NCI-CTEP#7870). Clin. Cancer Res. 2017, 23, 7199–7208. [Google Scholar] [CrossRef]
- Schreiner, B.; Mitsdoerffer, M.; Kieseier, B.C.; Chen, L.; Hartung, H.P.; Weller, M.; Wiendl, H. Interferon-beta enhances monocyte and dendritic cell expression of B7-H1 (PD-L1), a strong inhibitor of autologous T-cell activation: Relevance for the immune modulatory effect in multiple sclerosis. J. Neuroimmunol. 2004, 155, 172–182. [Google Scholar] [CrossRef]
- Canderan, G.; Dellabona, P. T helper 17 T cells do good for cancer immunotherapy. Immunotherapy 2010, 2, 21–24. [Google Scholar] [CrossRef]
- Ni, L.; Lu, J. Interferon gamma in cancer immunotherapy. Cancer Med. 2018, 7, 4509–4516. [Google Scholar] [CrossRef]
- Whiteside, T.L. The role of regulatory T cells in cancer immunology. Immunotargets Ther. 2015, 4, 159–171. [Google Scholar] [CrossRef]
- Tomar, M.S.; Kumar, S.; Kumar, S.; Gautam, P.K.; Singh, R.K.; Verma, P.K.; Singh, S.P.; Acharya, A. NK Cell Effector Functions Regulation by Modulating nTreg Cell Population During Progressive Growth of Dalton’s Lymphoma in Mice. Immunol. Invest. 2018, 47, 40–56. [Google Scholar] [CrossRef]
- Ruytinx, P.; Proost, P.; Van Damme, J.; Struyf, S. Chemokine-Induced Macrophage Polarization in Inflammatory Conditions. Front. Immunol. 2018, 9, 1930. [Google Scholar] [CrossRef]
- Beutler, B.; Greenwald, D.; Hulmes, J.D.; Chang, M.; Pan, Y.C.; Mathison, J.; Ulevitch, R.; Cerami, A. Identity of tumour necrosis factor and the macrophage-secreted factor cachectin. Nature 1985, 316, 552–554. [Google Scholar] [CrossRef]
- Bogdan, C.; Stenger, S.; Röllinghoff, M.; Solbach, W. Cytokine interactions in experimental cutaneous leishmaniasis. Interleukin 4 synergizes with interferon-gamma to activate murine macrophages for killing of Leishmania major amastigotes. Eur. J. Immunol. 1991, 21, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C.F.; Murray, H.W.; Wiebe, M.E.; Rubin, B.Y. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med. 1983, 158, 670–689. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Ang, B.; Xu, X.; Huang, X.; Wu, Y.; Sun, Y.; Wang, W.; Li, N.; Cao, X.; Wan, T. TLR4 is essential for dendritic cell activation and anti-tumor T-cell response enhancement by DAMPs released from chemically stressed cancer cells. Cell Mol. Immunol. 2014, 11, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Lutz, M.B.; Schuler, G. Immature, semi-mature and fully mature dendritic cells: Which signals induce tolerance or immunity? Trends Immunol. 2002, 23, 445–449. [Google Scholar] [CrossRef]
- Dudek, A.M.; Martin, S.; Garg, A.D.; Agostinis, P. Immature, Semi-Mature, and Fully Mature Dendritic Cells: Toward a DC-Cancer Cells Interface That Augments Anticancer Immunity. Front. Immunol. 2013, 4, 438. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, Inflammation, and Cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef]
- Achkar, T.; Arjunan, A.; Wang, H.; Saul, M.; Davar, D.; Appleman, L.J.; Friedland, D.; Parikh, R.A. High-dose interleukin 2 in patients with metastatic renal cell carcinoma with sarcomatoid features. PLoS ONE 2017, 12, e0190084. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Aronow, R.A.; Akbarinejad, S.; Le, T.; Su, S.; Shahriyari, L. TumorDecon: A digital cytometry software. SoftwareX 2022, 18, 101072. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Friedman, A. Interaction of tumor with its micro-environment: A mathematical model. Bull Math. Biol. 2010, 72, 1029–1068. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S.A. IL-2: The first effective immunotherapy for human cancer. J. Immunol. 2014, 192, 5451–5458. [Google Scholar] [CrossRef]
- Komohara, Y.; Hasita, H.; Ohnishi, K.; Fujiwara, Y.; Suzu, S.; Eto, M.; Takeya, M. Macrophage infiltration and its prognostic relevance in clear cell renal cell carcinoma. Cancer Sci. 2011, 102, 1424–1431. [Google Scholar] [CrossRef] [PubMed]
- Dannenmann, S.R.; Thielicke, J.; Stöckli, M.; Matter, C.; Von Boehmer, L.; Cecconi, V.; Hermanns, T.; Hefermehl, L.; Schraml, P.; Moch, H.; et al. Tumor-associated macrophages subvert T-cell function and correlate with reduced survival in clear cell renal cell carcinoma. Oncoimmunology 2013, 2, e23562. [Google Scholar] [CrossRef]
- Thiounn, N.; Pages, F.; Flam, T.; Tartour, E.; Mosseri, V.; Zerbib, M.; Beuzeboc, P.; Deneux, L.; Fridman, W.H.; Debré, B. IL-6 is a survival prognostic factor in renal cell carcinoma. Immunol. Lett. 1997, 58, 121–124. [Google Scholar] [CrossRef]
- Gudbrandsdottir, G.; Aarstad, H.H.; Bostad, L.; Hjelle, K.M.; Aarstad, H.J.; Bruserud, Ø.; Tvedt, T.H.A.; Beisland, C. Serum levels of the IL-6 family of cytokines predict prognosis in renal cell carcinoma (RCC). Cancer Immunol. Immunother. 2021, 70, 19–30. [Google Scholar] [CrossRef]
- Walther, M.M.; Johnson, B.; Culley, D.; Shah, R.; Weber, J.; Venzon, D.; Yang, J.C.; Linehan, W.M.; Rosenberg, S.A. Serum interleukin-6 levels in metastatic renal cell carcinoma before treatment with interleukin-2 correlates with paraneoplastic syndromes but not patient survival. J. Urol. 1998, 159, 718–722. [Google Scholar] [CrossRef]
- Wang, Y.; Fu, D.; Chen, Y.; Su, J.; Wang, Y.; Li, X.; Zhai, W.; Niu, Y.; Yue, D.; Geng, H. G3BP1 promotes tumor progression and metastasis through IL-6/G3BP1/STAT3 signaling axis in renal cell carcinomas. Cell Death Dis. 2018, 9, 501. [Google Scholar] [CrossRef]
- Nerli, R.; Devaraju, S.; Hiremath, M.B.; Guntaka, A.K.; Patne, P.; Dixit, N. Tumor doubling time of renal cell carcinoma measured by CT. Indian J. Urol. 2014, 30, 153–157. [Google Scholar] [CrossRef]
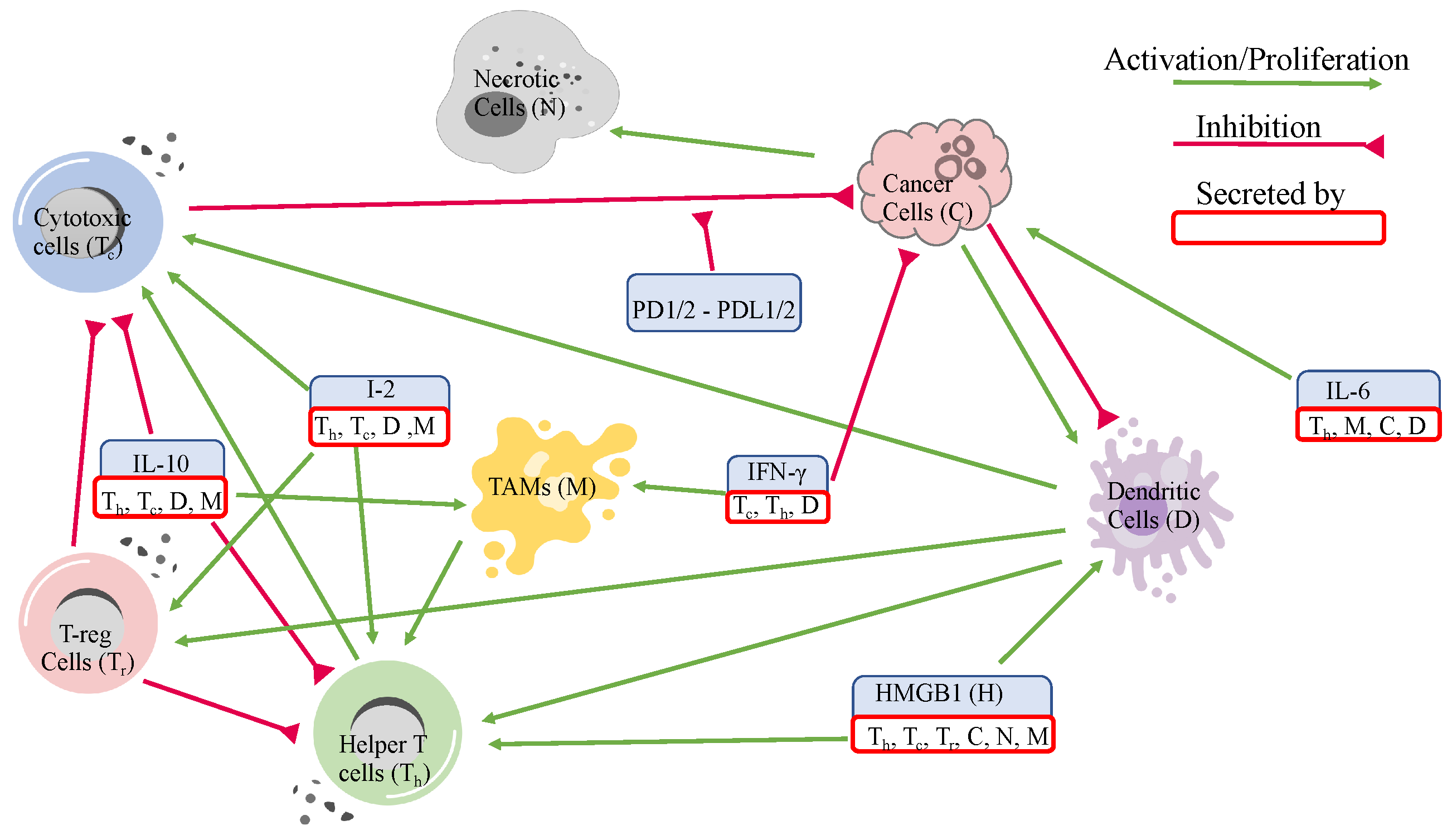
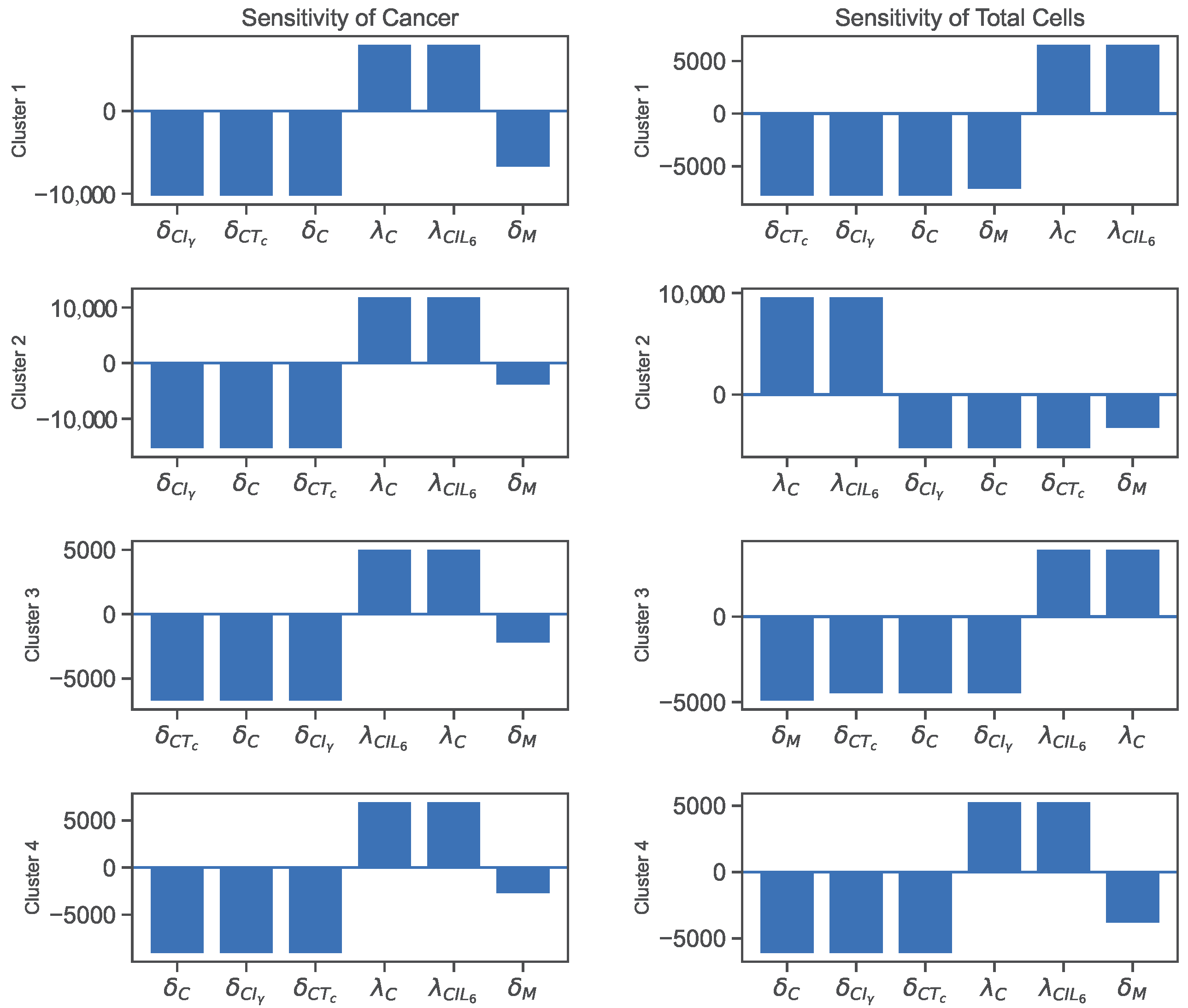
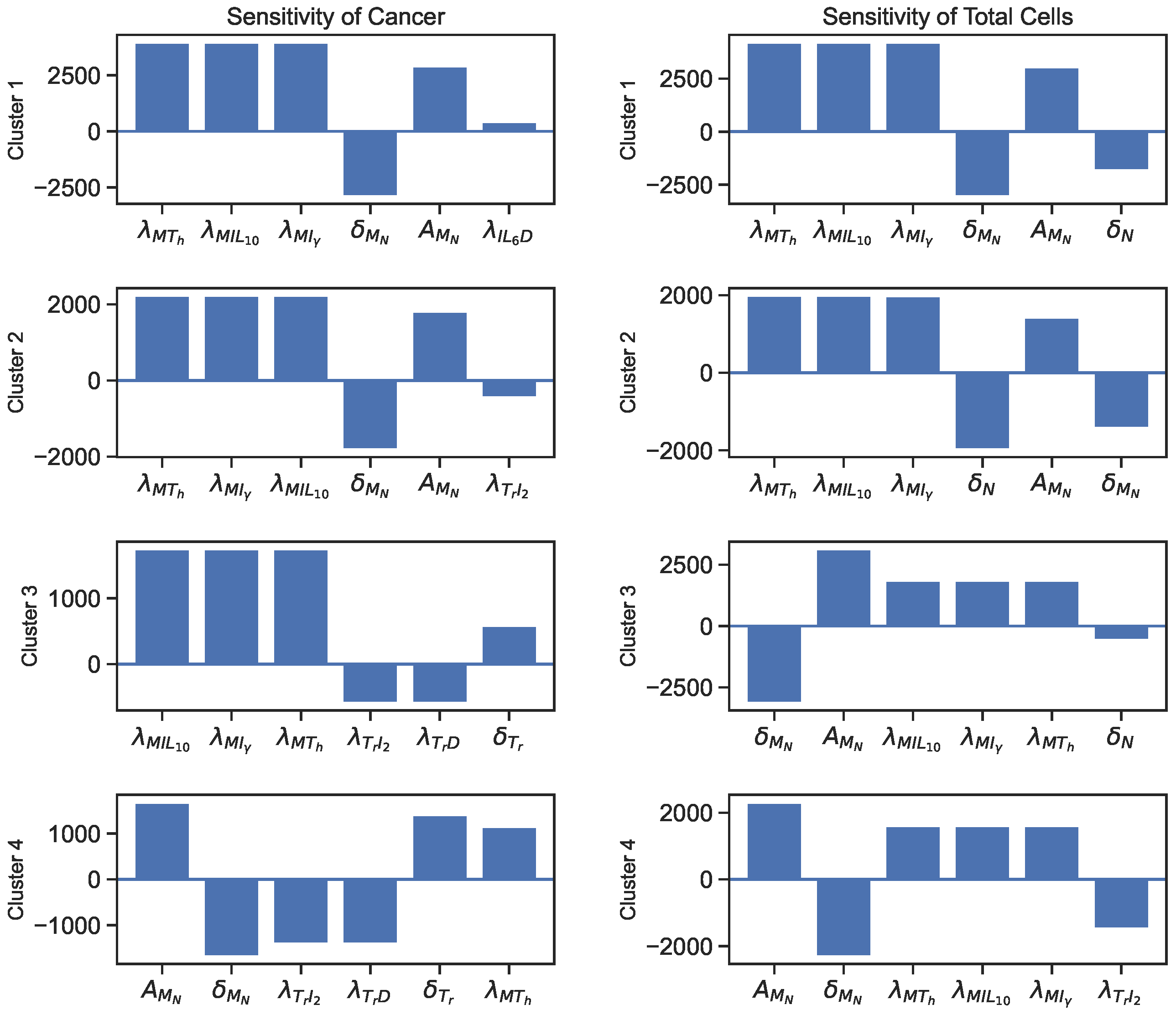
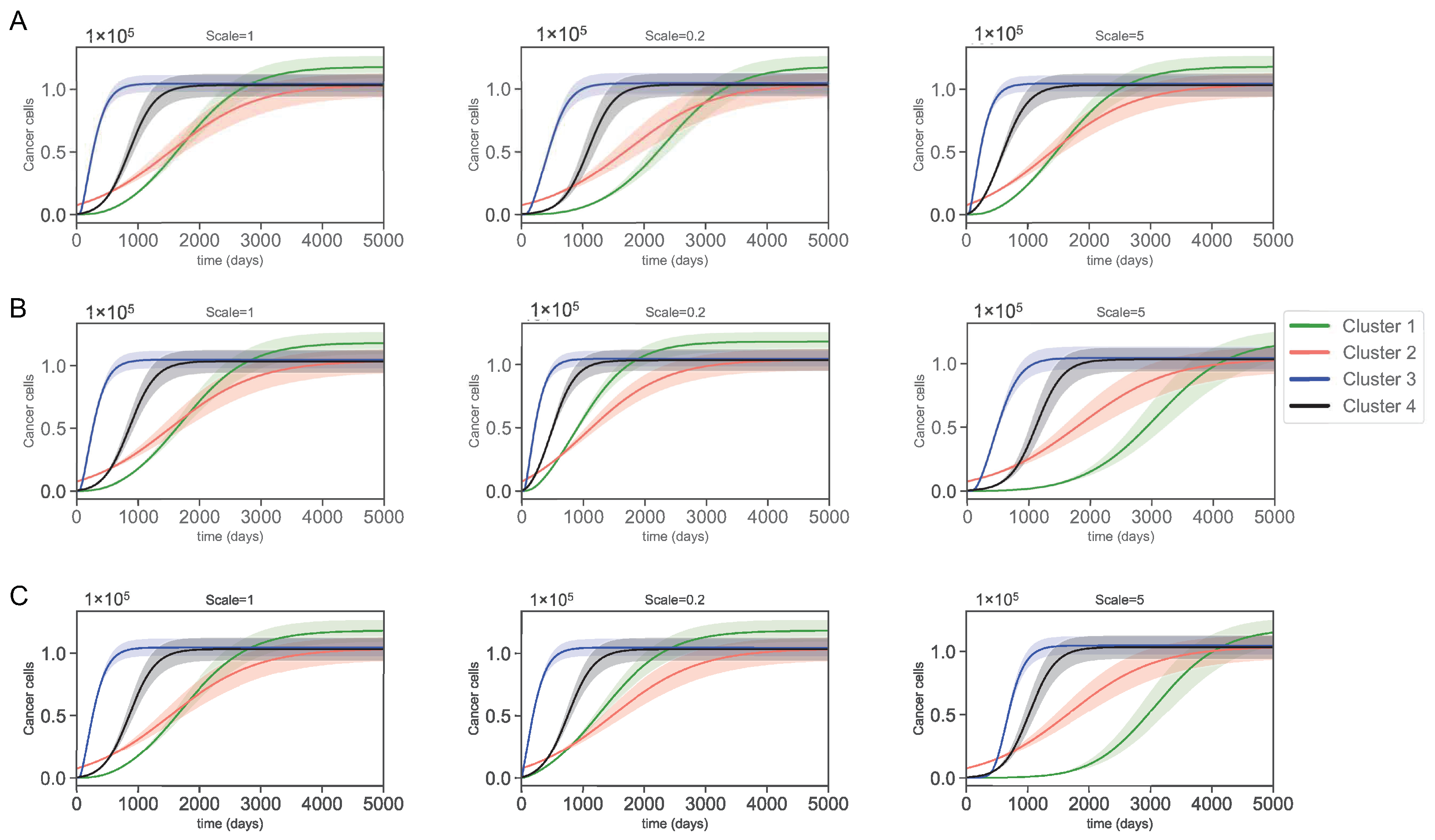
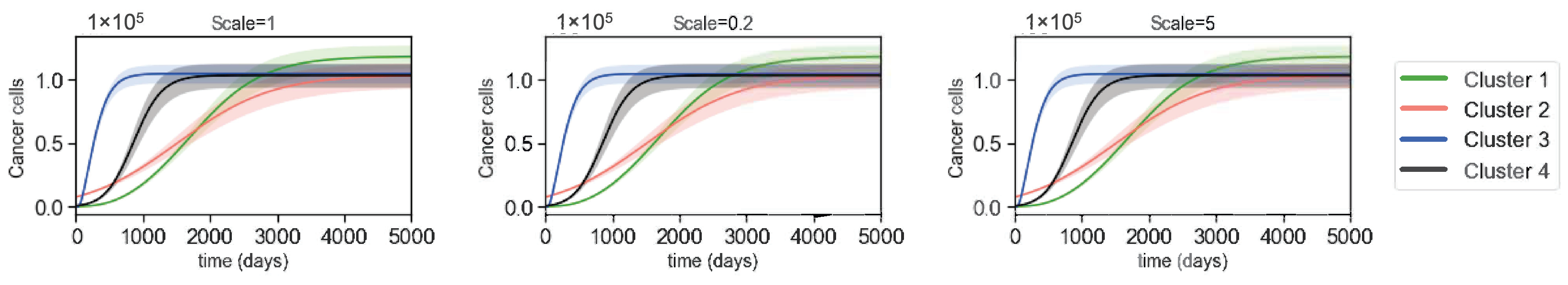
| Variables | Names | Combinations from Data |
|---|---|---|
| Helper T-cells | Activated memory CD4 T-cells and follicular helper T-cells | |
| Cytotoxic cells | CD8 T-cells and activated NK cells | |
| Regulatory T-cells | Regulatory T-cells | |
| Naive T-cells | Naive CD4 T-cells, memory resting CD4 T-cells | |
| and resting natural killer (NK) cells | ||
| Naive dendritic cells | Naive dendritic cells | |
| D | Dendritic cells | Mature dendritic cells |
| Naive Macrophages | M0 macrophages and monocytes | |
| M | Macrophages | M1 and M2 macrophages |
| C | Cancer cells | Estimated from the data |
| N | Necrotic cells | Estimated from the data |
| IFN- | Interferon- from gene expression data | |
| H | HMGB1 | HMGB1 from gene expression data |
| IL-10 | IL10 from gene expression data | |
| IL-2 and IL-12 | IL2 and IL12 from gene expression data | |
| IL-6 | IL6 from gene expression data |
| Cells and Cytokines | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 |
|---|---|---|---|---|
| 1 | ||||
| 1 | ||||
| Cells and Cytokines | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 |
|---|---|---|---|---|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sofia, D.; Mohammad Mirzaei, N.; Shahriyari, L. Patient-Specific Mathematical Model of the Clear Cell Renal Cell Carcinoma Microenvironment. J. Pers. Med. 2022, 12, 1681. https://doi.org/10.3390/jpm12101681
Sofia D, Mohammad Mirzaei N, Shahriyari L. Patient-Specific Mathematical Model of the Clear Cell Renal Cell Carcinoma Microenvironment. Journal of Personalized Medicine. 2022; 12(10):1681. https://doi.org/10.3390/jpm12101681
Chicago/Turabian StyleSofia, Dilruba, Navid Mohammad Mirzaei, and Leili Shahriyari. 2022. "Patient-Specific Mathematical Model of the Clear Cell Renal Cell Carcinoma Microenvironment" Journal of Personalized Medicine 12, no. 10: 1681. https://doi.org/10.3390/jpm12101681
APA StyleSofia, D., Mohammad Mirzaei, N., & Shahriyari, L. (2022). Patient-Specific Mathematical Model of the Clear Cell Renal Cell Carcinoma Microenvironment. Journal of Personalized Medicine, 12(10), 1681. https://doi.org/10.3390/jpm12101681