Multi-Layer Picture of Neurodegenerative Diseases: Lessons from the Use of Big Data through Artificial Intelligence
Abstract
1. Introduction
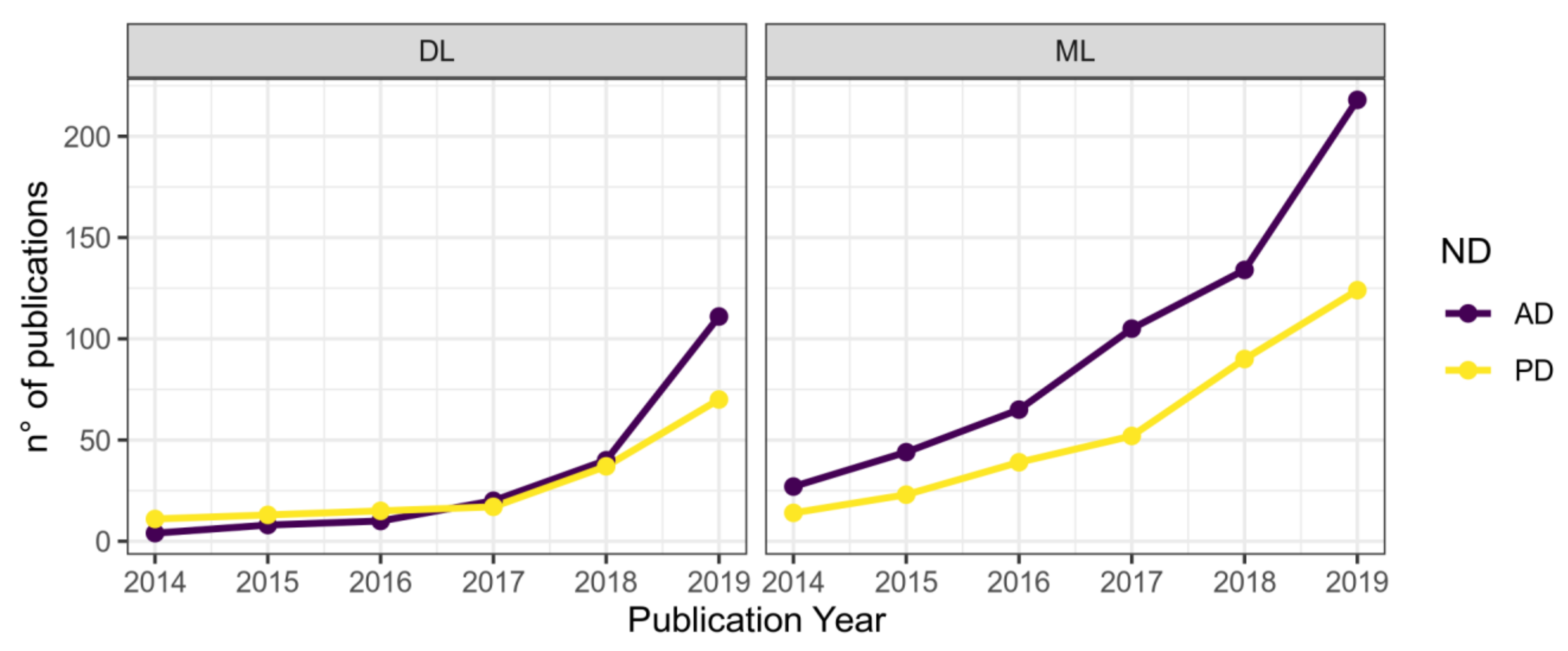
Literature Research
2. Basics of Machine Learning and Deep Learning
3. Artificial Intelligence in Neurology
3.1. Neuroimaging Classification and Segmentation
3.2. Clinical Records Investigation
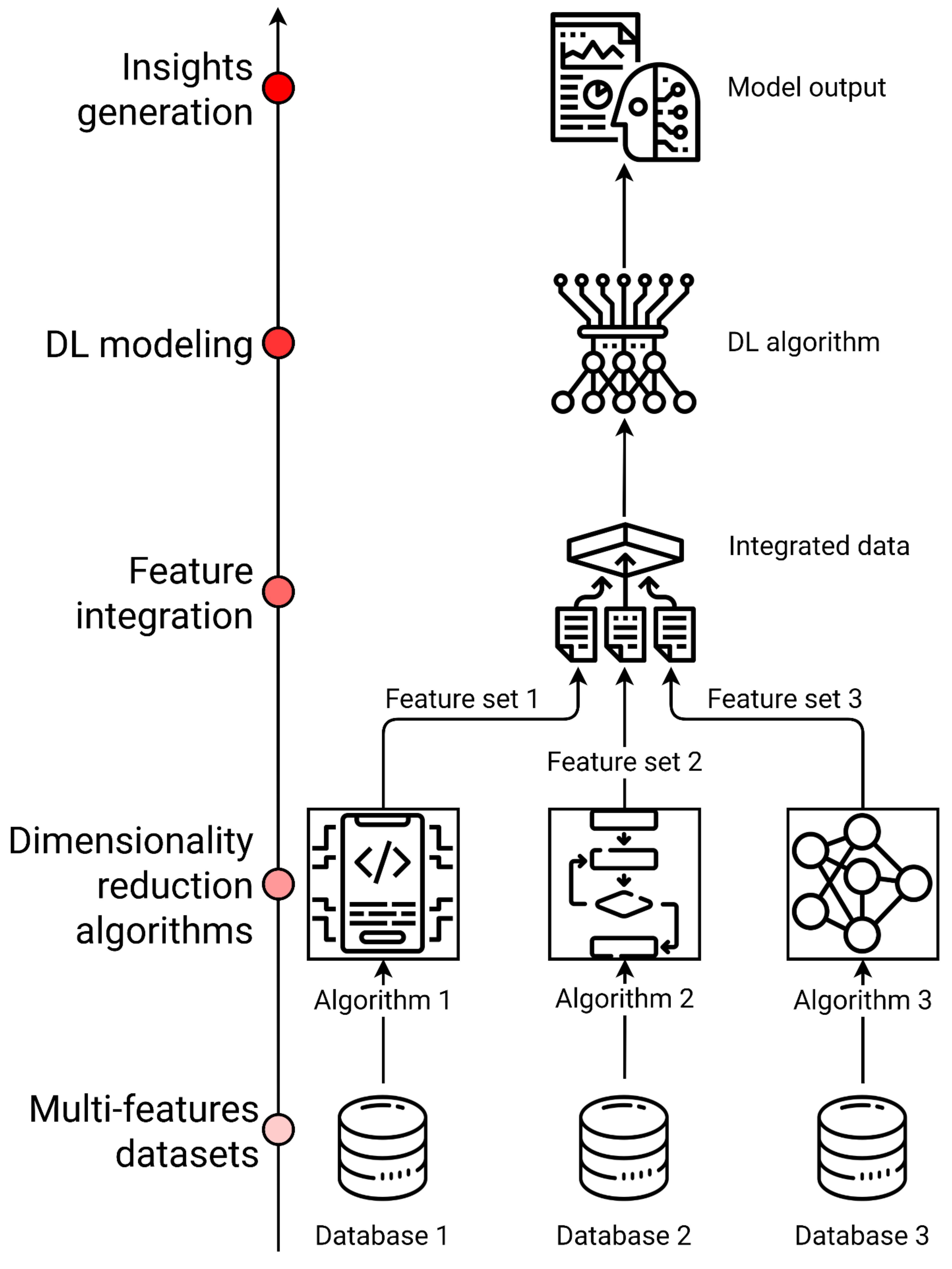
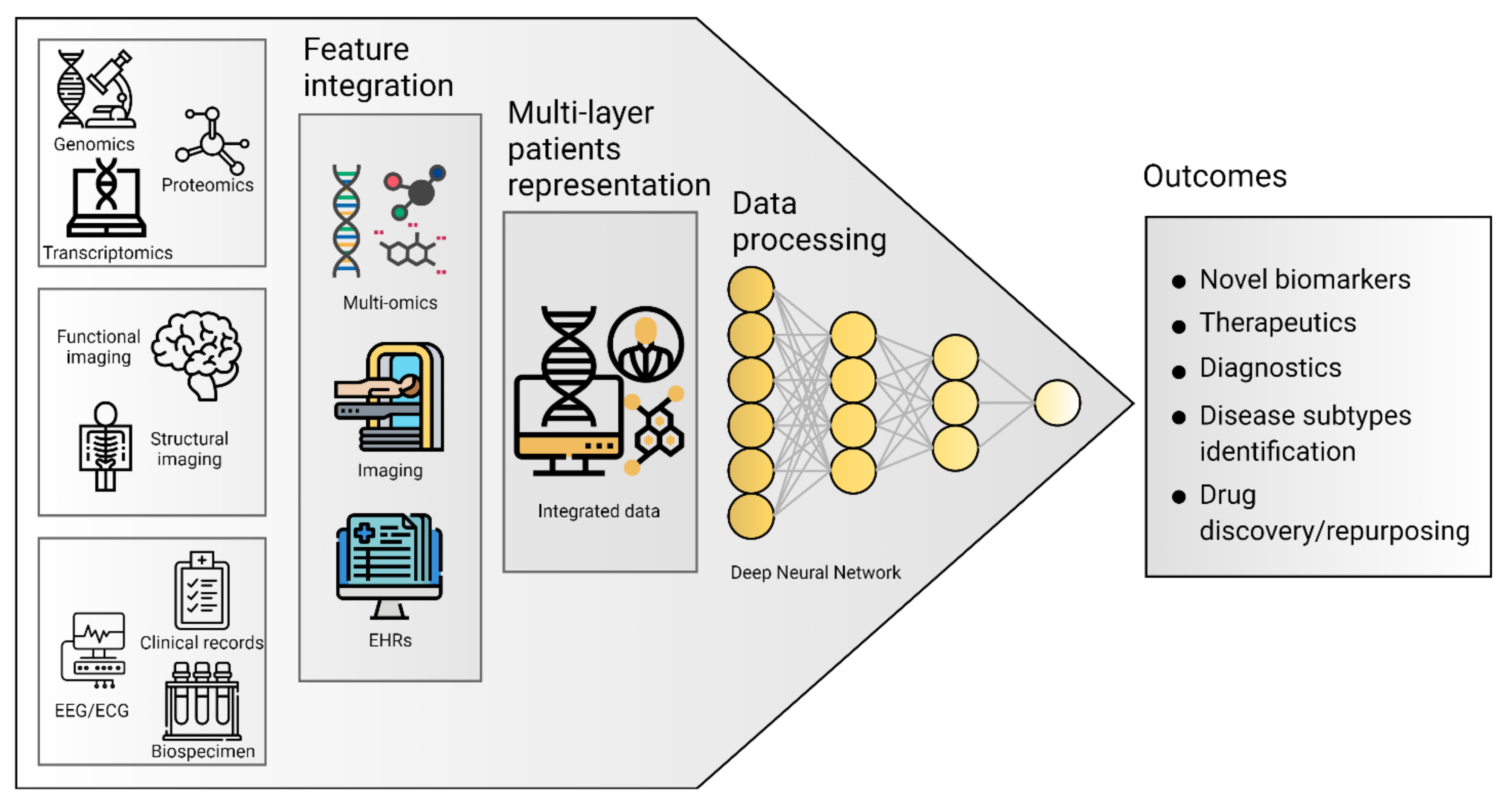
4. Big Data Integration
4.1. Multi-Omics
4.2. Electronic Health Records (EHRs)
4.3. Artificial Intelligence Applications on ND Multi-Omics and Clinical Data Integration
5. Databases
6. Challenges and Limitations for AI Techniques in ND Research
7. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Katsnelson, A.; De Strooper, B.; Zoghbi, H.Y. Neurodegeneration: From cellular concepts to clinical applications. Sci. Transl. Med. 2016, 8, 364ps18. [Google Scholar] [CrossRef] [PubMed]
- Bovolenta, T.M.; de Azevedo Silva, S.M.C.; Arb Saba, R.; Borges, V.; Ferraz, H.B.; Felicio, A.C. Systematic Review and Critical Analysis of Cost Studies Associated with Parkinson’s Disease. Park. Dis. 2017, 2017, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Erkkinen, M.G.; Kim, M.-O.; Geschwind, M.D. Clinical Neurology and Epidemiology of the Major Neurodegenerative Diseases. Cold Spring Harb. Perspect. Biol. 2018, 10, a033118. [Google Scholar] [CrossRef] [PubMed]
- Strafella, C.; Caputo, V.; Galota, M.R.; Zampatti, S.; Marella, G.; Mauriello, S.; Cascella, R.; Giardina, E. Application of Precision Medicine in Neurodegenerative Diseases. Front. Neurol. 2018, 9, 701. [Google Scholar] [CrossRef]
- Alexander, N.; Alexander, D.C.; Barkhof, F.; Denaxas, S. Using Unsupervised Learning to Identify Clinical Subtypes of Alzheimer’s Disease in Electronic Health Records. Stud. Health Technol. Inform. 2020, 270, 499–503. [Google Scholar]
- Dujardin, S.; Commins, C.; Lathuiliere, A.; Beerepoot, P.; Fernandes, A.R.; Kamath, T.V.; De Los Santos, M.B.; Klickstein, N.; Corjuc, D.L.; Corjuc, B.T.; et al. Tau molecular diversity contributes to clinical heterogeneity in Alzheimer’s disease. Nat. Med. 2020, 26, 1256–1263. [Google Scholar] [CrossRef]
- Maudsley, S.; Devanarayan, V.; Martin, B.; Geerts, H. Brain Health Modeling Initiative (BHMI) Intelligent and effective informatic deconvolution of “Big Data” and its future impact on the quantitative nature of neurodegenerative disease therapy. Alzheimers Dement. J. Alzheimers Assoc. 2018, 14, 961–975. [Google Scholar] [CrossRef]
- Young, A.L.; Marinescu, R.V.; Oxtoby, N.P.; Bocchetta, M.; Yong, K.; Firth, N.C.; Cash, D.M.; Thomas, D.L.; Dick, K.M.; Cardoso, J.; et al. Uncovering the heterogeneity and temporal complexity of neurodegenerative diseases with Subtype and Stage Inference. Nat. Commun. 2018, 9, 4273. [Google Scholar] [CrossRef]
- Manzoni, C.; Lewis, P.A.; Ferrari, R. Network Analysis for Complex Neurodegenerative Diseases. Curr. Genet. Med. Rep. 2020, 8, 17–25. [Google Scholar] [CrossRef]
- Dorsey, E.R.; Elbaz, A.; Nichols, E.; Abd-Allah, F.; Abdelalim, A.; Adsuar, J.C.; Ansha, M.G.; Brayne, C.; Choi, J.-Y.J.; Collado-Mateo, D.; et al. Global, regional, and national burden of Parkinson’s disease, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2018, 17, 939–953. [Google Scholar] [CrossRef]
- Nalls, M.A.; Blauwendraat, C.; Vallerga, C.L.; Heilbron, K.; Bandres-Ciga, S.; Chang, D.; Tan, M.; Kia, D.A.; Noyce, A.J.; Xue, A.; et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019, 18, 1091–1102. [Google Scholar] [CrossRef]
- De Jager, P.L.; Yang, H.-S.; Bennett, D.A. Deconstructing and targeting the genomic architecture of human neurodegeneration. Nat. Neurosci. 2018, 21, 1310–1317. [Google Scholar] [CrossRef]
- Docampo, E.; Giardina, E.; Riveira-Muñoz, E.; De Cid, R.; Escaramís, G.; Perricone, C.; Fernández-Sueiro, J.L.; Maymó, J.; González-Gay, M.A.; Blanco, F.J. Deletion of LCE3C and LCE3B is a susceptibility factor for psoriatic arthritis: A study in Spanish and Italian populations and meta-analysis. Arthritis Rheum. 2011, 63, 1860–1865. [Google Scholar] [CrossRef]
- Stocchi, L.; Cascella, R.; Zampatti, S.; Pirazzoli, A.; Novelli, G.; Giardina, E. The pharmacogenomic HLA biomarker associated to adverse abacavir reactions: Comparative analysis of different genotyping methods. Curr. Genom. 2012, 13, 314–320. [Google Scholar] [CrossRef]
- Arle, J.E.; Carlson, K.W. Medical diagnosis and treatment is NP-complete. J. Exp. Theor. Artif. Intell. 2021, 33, 297–312. [Google Scholar] [CrossRef]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-Generation Machine Learning for Biological Networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef]
- Perakakis, N.; Yazdani, A.; Karniadakis, G.E.; Mantzoros, C. Omics, big data and machine learning as tools to propel understanding of biological mechanisms and to discover novel diagnostics and therapeutics. Metabolism 2018, 87, A1–A9. [Google Scholar] [CrossRef]
- Valliani, A.A.-A.; Ranti, D.; Oermann, E.K. Deep Learning and Neurology: A Systematic Review. Neurol. Ther. 2019, 8, 351–365. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; ISBN 0-262-33737-1. [Google Scholar]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Hinton, G.E. A practical guide to training restricted Boltzmann machines. In Neural Networks: Tricks of the Trade; Springer: Berlin/Heidelberg, Germany, 2012; pp. 599–619. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Grapov, D.; Fahrmann, J.; Wanichthanarak, K.; Khoomrung, S. Rise of Deep Learning for Genomic, Proteomic, and Metabolomic Data Integration in Precision Medicine. Omics J. Integr. Biol. 2018, 22, 630–636. [Google Scholar] [CrossRef]
- Ramzan, F.; Khan, M.U.G.; Rehmat, A.; Iqbal, S.; Saba, T.; Rehman, A.; Mehmood, Z. A Deep Learning Approach for Automated Diagnosis and Multi-Class Classification of Alzheimer’s Disease Stages Using Resting-State fMRI and Residual Neural Networks. J. Med. Syst. 2019, 44, 37. [Google Scholar] [CrossRef]
- Marek, K.; Jennings, D.; Lasch, S.; Siderowf, A.; Tanner, C.; Simuni, T.; Coffey, C.; Kieburtz, K.; Flagg, E.; Chowdhury, S.; et al. The Parkinson Progression Marker Initiative (PPMI). Prog. Neurobiol. 2011, 95, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Aich, S.; Kim, H.-C. Detection of Parkinson’s Disease from 3T T1 Weighted MRI Scans Using 3D Convolutional Neural Network. Diagnostics 2020, 10, 402. [Google Scholar] [CrossRef]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef]
- Basher, A.; Kim, B.C.; Lee, K.H.; Jung, H.Y. Automatic Localization and Discrete Volume Measurements of Hippocampi From MRI Data Using a Convolutional Neural Network. IEEE Access 2020, 8, 91725–91739. [Google Scholar] [CrossRef]
- Kushibar, K.; Valverde, S.; González-Villà, S.; Bernal, J.; Cabezas, M.; Oliver, A.; Lladó, X. Automated sub-cortical brain structure segmentation combining spatial and deep convolutional features. Med. Image Anal. 2018, 48, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Brusini, I.; Lindberg, O.; Muehlboeck, J.; Smedby, Ö.; Westman, E.; Wang, C. Shape information improves the cross-cohort performance of deep learning-based segmentation of the hippocampus. Front. Neurosci. 2020, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Liu, M.; Initiative, A.D.N. Alzheimer’s disease diagnosis based on multiple cluster dense convolutional networks. Comput. Med. Imaging Graph. 2018, 70, 101–110. [Google Scholar] [CrossRef]
- Jain, R.; Jain, N.; Aggarwal, A.; Hemanth, D.J. Convolutional neural network based Alzheimer’s disease classification from magnetic resonance brain images. Cogn. Syst. Res. 2019, 57, 147–159. [Google Scholar] [CrossRef]
- Pereira, C.R.; Pereira, D.R.; Rosa, G.H.; Albuquerque, V.H.C.; Weber, S.A.T.; Hook, C.; Papa, J.P. Handwritten dynamics assessment through convolutional neural networks: An application to Parkinson’s disease identification. Artif. Intell. Med. 2018, 87, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Drotár, P.; Mekyska, J.; Rektorová, I.; Masarová, L.; Smékal, Z.; Faundez-Zanuy, M. Decision Support Framework for Parkinson’s Disease Based on Novel Handwriting Markers. IEEE Trans. Neural Syst. Rehabil. Eng. 2015, 23, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.R.; Weber, S.A.T.; Hook, C.; Rosa, G.H.; Papa, J.P. Deep Learning-Aided Parkinson’s Disease Diagnosis from Handwritten Dynamics. In Proceedings of the 2016 29th SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI), Sao Paulo, Brazil, 4–7 October 2016; pp. 340–346. [Google Scholar]
- Berus, L.; Klancnik, S.; Brezocnik, M.; Ficko, M. Classifying Parkinson’s Disease Based on Acoustic Measures Using Artificial Neural Networks. Sensors 2018, 19, 16. [Google Scholar] [CrossRef] [PubMed]
- Al-Hameed, S.; Benaissa, M.; Christensen, H.; Mirheidari, B.; Blackburn, D.; Reuber, M. A new diagnostic approach for the identification of patients with neurodegenerative cognitive complaints. PLoS ONE 2019, 14, e0217388. [Google Scholar] [CrossRef]
- Camps, J.; Samà, A.; Martín, M.; Rodríguez-Martín, D.; Pérez-López, C.; Moreno Arostegui, J.M.; Cabestany, J.; Català, A.; Alcaine, S.; Mestre, B.; et al. Deep learning for freezing of gait detection in Parkinson’s disease patients in their homes using a waist-worn inertial measurement unit. Knowl. Based Syst. 2018, 139, 119–131. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Zorin, A.; Dass, G.; Vu, M.-T.; Xu, P.; Glont, M.; Vizcaíno, J.A.; Jarnuczak, A.F.; Petryszak, R.; Ping, P.; et al. Quantifying the impact of public omics data. Nat. Commun. 2019, 10, 3512. [Google Scholar] [CrossRef]
- Sonawane, A.R.; Weiss, S.T.; Glass, K.; Sharma, A. Network Medicine in the Age of Biomedical Big Data. Front. Genet. 2019, 10, 294. [Google Scholar] [CrossRef]
- Li, Y.; Wu, F.-X.; Ngom, A. A review on machine learning principles for multi-view biological data integration. Brief. Bioinform. 2018, 19, 325–340. [Google Scholar] [CrossRef]
- Espay, A.J.; Schwarzschild, M.A.; Tanner, C.M.; Fernandez, H.H.; Simon, D.K.; Leverenz, J.B.; Merola, A.; Chen-Plotkin, A.; Brundin, P.; Kauffman, M.A.; et al. Biomarker-driven phenotyping in Parkinson’s disease: A translational missing link in disease-modifying clinical trials. Mov. Disord. Off. J. Mov. Disord. Soc. 2017, 32, 319–324. [Google Scholar] [CrossRef]
- Ranchal, R.; Bastide, P.; Wang, X.; Gkoulalas-Divanis, A.; Mehra, M.; Bakthavachalam, S.; Lei, H.; Mohindra, A. Disrupting Healthcare Silos: Addressing Data Volume, Velocity and Variety with a Cloud-Native Healthcare Data Ingestion Service. IEEE J. Biomed. Health Inform. 2020, 24, 3182–3188. [Google Scholar] [CrossRef]
- Johnson, A.E.W.; Pollard, T.J.; Shen, L.; Lehman, L.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Anthony Celi, L.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef]
- Hernandez-Boussard, T.; Monda, K.L.; Crespo, B.C.; Riskin, D. Real world evidence in cardiovascular medicine: Ensuring data validity in electronic health record-based studies. J. Am. Med. Inform. Assoc. 2019, 26, 1189–1194. [Google Scholar] [CrossRef]
- Bender, D.; Sartipi, K. HL7 FHIR: An Agile and RESTful approach to healthcare information exchange. In Proceedings of the 26th IEEE international symposium on computer-based medical systems, Porto, Portugal, 20–22 June 2013; pp. 326–331.
- Kaur, J.; Mann, K.S. AI based healthcare platform for real time, predictive and prescriptive analytics using reactive programming. In Journal of Physics: Conference Series, Proceedings of the 10th International Conference on Computer and Electrical Engineering, Edmonton, AB, Canada, 11–13 October 2017; IOP Publishing: Bristol, UK, 2017; Volume 933, p. 012010. [Google Scholar]
- Lee, C.S.; Lee, A.Y. Clinical applications of continual learning machine learning. Lancet Digit. Health 2020, 2, e279–e281. [Google Scholar] [CrossRef]
- Goodstein, D. Defining the scientific method. Nat. Methods 2009, 6, 237. [Google Scholar]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.-M.; Zietz, M.; Hoffman, M.M.; et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 2018, 15, 20170387. [Google Scholar] [CrossRef]
- Zhavoronkov, A.; Mamoshina, P.; Vanhaelen, Q.; Scheibye-Knudsen, M.; Moskalev, A.; Aliper, A. Artificial intelligence for aging and longevity research: Recent advances and perspectives. Ageing Res. Rev. 2019, 49, 49–66. [Google Scholar] [CrossRef]
- Zhang, X.; Chou, J.; Liang, J.; Xiao, C.; Zhao, Y.; Sarva, H.; Henchcliffe, C.; Wang, F. Data-Driven Subtyping of Parkinson’s Disease Using Longitudinal Clinical Records: A Cohort Study. Sci. Rep. 2019, 9, 797. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Nho, K.; Kang, B.; Sohn, K.-A.; Kim, D. Predicting Alzheimer’s disease progression using multi-modal deep learning approach. Sci. Rep. 2019, 9, 1952. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Ha, J.; Park, S. Prediction of Alzheimer’s disease based on deep neural network by integrating gene expression and DNA methylation dataset. Expert Syst. Appl. 2020, 140, 112873. [Google Scholar] [CrossRef]
- Xu, J.; Wu, P.; Chen, Y.; Meng, Q.; Dawood, H.; Dawood, H. A hierarchical integration deep flexible neural forest framework for cancer subtype classification by integrating multi-omics data. BMC Bioinform. 2019, 20, 527. [Google Scholar] [CrossRef]
- Gligorijević, V.; Malod-Dognin, N.; Pržulj, N. Integrative methods for analyzing big data in precision medicine. Proteomics 2016, 16, 741–758. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, M.; da Veiga Leprevost, F.; Squizzato, S.; Park, Y.M.; Haug, K.; Carroll, A.J.; Spalding, D.; Paschall, J.; Wang, M.; et al. Discovering and linking public omics data sets using the Omics Discovery Index. Nat. Biotechnol. 2017, 35, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Kuzma, A.; Valladares, O.; Cweibel, R.; Greenfest-Allen, E.; Childress, D.M.; Malamon, J.; Gangadharan, P.; Zhao, Y.; Qu, L.; Leung, Y.Y.; et al. NIAGADS: The NIA Genetics of Alzheimer’s Disease Data Storage Site. Alzheimers Dement. 2016, 12, 1200–1203. [Google Scholar] [CrossRef]
- Toga, A.W.; Neu, S.C.; Bhatt, P.; Crawford, K.L.; Ashish, N. The Global Alzheimer’s Association Interactive Network. Alzheimers Dement. J. Alzheimers Assoc. 2016, 12, 49–54. [Google Scholar] [CrossRef]
- Mueller, S.G.; Weiner, M.W.; Thal, L.J.; Petersen, R.C.; Jack, C.; Jagust, W.; Trojanowski, J.Q.; Toga, A.W.; Beckett, L. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin. 2005, 15, 869–877. [Google Scholar] [CrossRef]
- Lill, C.M.; Roehr, J.T.; McQueen, M.B.; Kavvoura, F.K.; Bagade, S.; Schjeide, B.-M.M.; Schjeide, L.M.; Meissner, E.; Zauft, U.; Allen, N.C.; et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson’s disease genetics: The PDGene database. PLoS Genet. 2012, 8, e1002548. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLOS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef] [PubMed]
- Giardina, E.; Caltagirone, C. The IRCCS Network of Neuroscience and Neurorehabilitation: The Italian Platform for Care and Research about Neurodegenerative Disorders. Eur. J. Neurol. 2018, 25, 209. [Google Scholar]
- Beekly, D.L.; Ramos, E.M.; Lee, W.W.; Deitrich, W.D.; Jacka, M.E.; Wu, J.; Hubbard, J.L.; Koepsell, T.D.; Morris, J.C.; Kukull, W.A.; et al. The National Alzheimer’s Coordinating Center (NACC) Database: The Uniform Data Set. Alzheimer Dis. Assoc. Disord. 2007, 21, 249–258. [Google Scholar] [CrossRef]
- Leff, D.R.; Yang, G.-Z. Big Data for Precision Medicine. Engineering 2015, 1, 277–279. [Google Scholar] [CrossRef]
- Khoury, M.J.; Iademarco, M.F.; Riley, W.T. Precision Public Health for the Era of Precision Medicine. Am. J. Prev. Med. 2016, 50, 398–401. [Google Scholar] [CrossRef]
- Zhou, L.; Verstreken, P. Reprogramming neurodegeneration in the big data era. Curr. Opin. Neurobiol. 2018, 48, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Barabási, A.-L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef]
- Huang, S.; Chaudhary, K.; Garmire, L.X. More is better: Recent progress in multi-omics data integration methods. Front. Genet. 2017, 8, 84. [Google Scholar] [CrossRef] [PubMed]
- Sethi, A.; Sankaran, A.; Panwar, N.; Khare, S.; Mani, S. DLPaper2Code: Auto-generation of Code from Deep Learning Research Papers. arXiv 2017, arXiv:1711.03543. [Google Scholar]
- Sammut, C.; Webb, G.I. Encyclopedia of Machine Learning; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2011; ISBN 0-387-30768-0. [Google Scholar]
- Lever, J.; Krzywinski, M.; Altman, N. Regularization; Nature Publishing Group: Berlin/Heidelberg, Germany, 2016; ISBN 1548-7105. [Google Scholar]
- Molina, L.C.; Belanche, L.; Nebot, À. Feature selection algorithms: A survey and experimental evaluation. In Proceedings of the 2002 IEEE International Conference on Data Mining, 2002. Proceedings, Maebashi City, Japan, 9–12 December 2002; pp. 306–313. [Google Scholar]
- Gibney, E. Google AI algorithm masters ancient game of Go. Nat. News 2016, 529, 445. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Singh, A.; Sengupta, S.; Lakshminarayanan, V. Explainable Deep Learning Models in Medical Image Analysis. J. Imaging 2020, 6, 52. [Google Scholar] [CrossRef]
- Sharma, A.; Shukla, D.; Goel, T.; Mandal, P.K. BHARAT: An Integrated Big Data Analytic Model for Early Diagnostic Biomarker of Alzheimer’s Disease. Front. Neurol. 2019, 10, 9. [Google Scholar] [CrossRef]
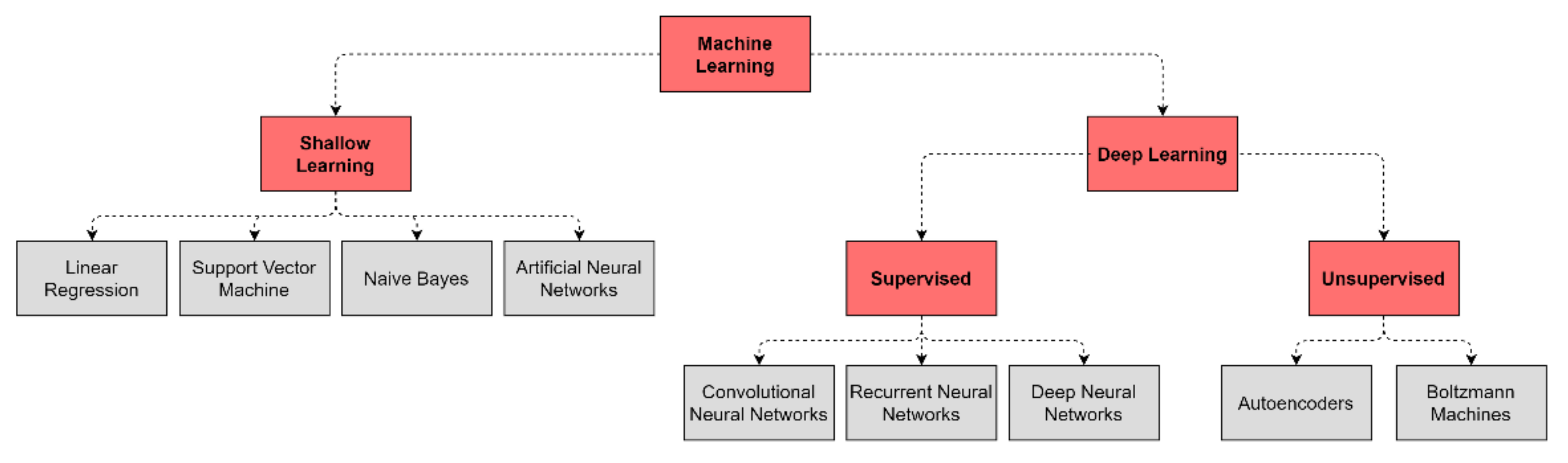
| Architecture | Description | Graph |
|---|---|---|
| Deep Neural Network (DNN) | The basic network is made of multiple hidden layers. It is capable of modeling complex non-linear relationships by learning input data representation to be matched with a specific output [19]. | 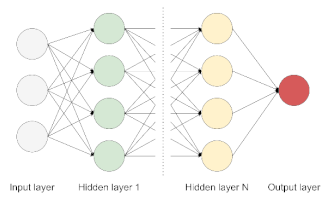 |
| Autoencoder (AE) | It allows detecting patterns in the data in an unsupervised fashion. The model is made of an encoder and a decoder, transforming input data to generate its own representation, aiming to minimize the difference between the input and its output representation [20]. |  |
| Restricted Boltzmann Machine (RBM) | This model is made of two layers, where nodes are bidirectionally connected but there are no connections within one layer. It is trained to learn a probability distribution for the input data and can be used as a building block for deep probabilistic models, where multiple RBMs can be stacked to build a deeper network [21]. | 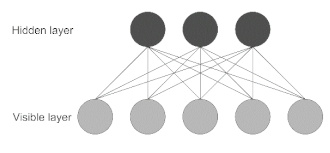 |
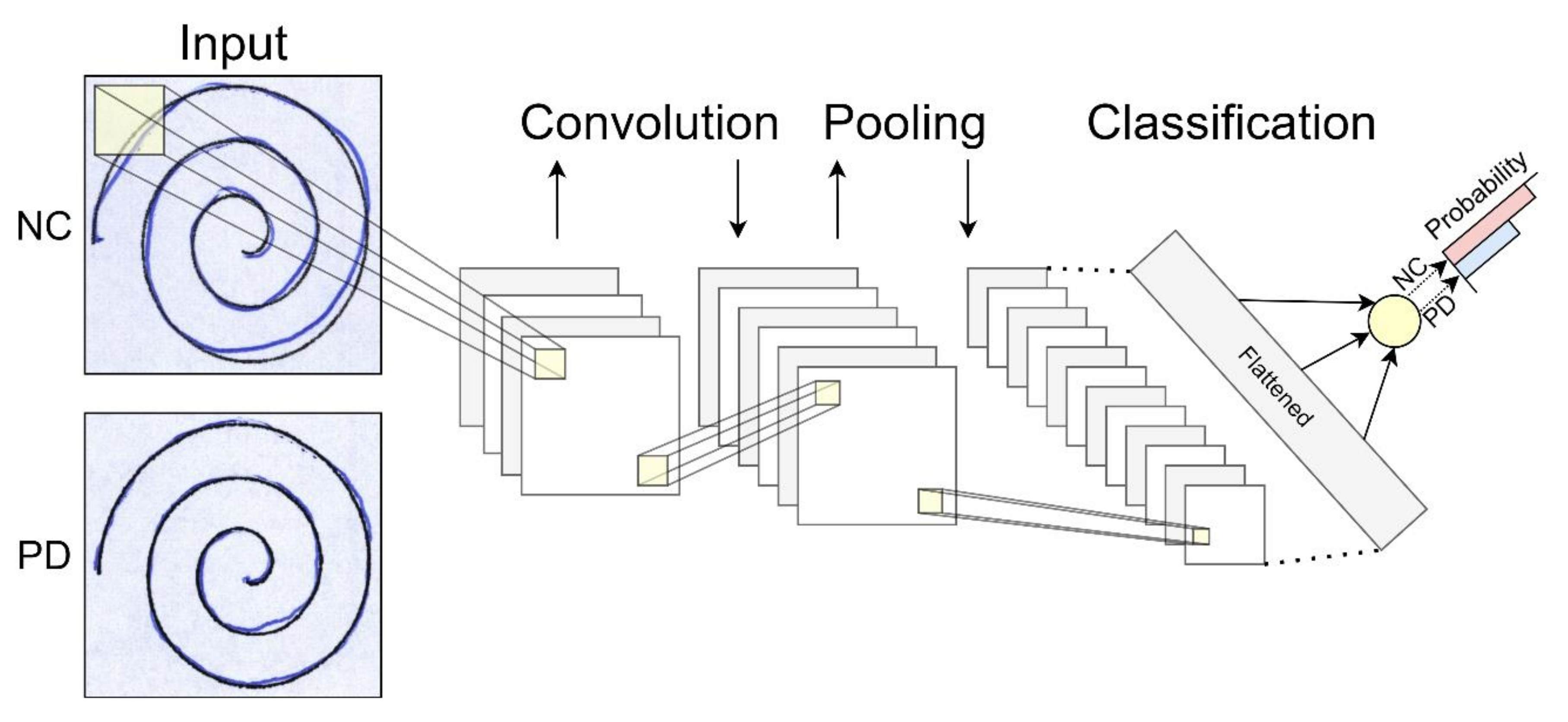
| Convolutional Neural Network (CNN) | Most used for image processing in computer vision applications. The network uses convolution and pooling operations to extract relevant features from data, useful for image classification. This architecture is inspired by the organization of the visual cortex [22]. | 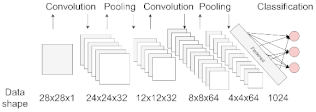 |
| Recurrent Neural Network (RNN) | Best suited to process sequential data and used to predict the future from the past. The network can give an output for every timestep and takes the previous inputs into account to determine the output. Long-Short Term Memory (LSTM) and Gated Recurrent Units (GRUs) are RNN architectures [19]. | 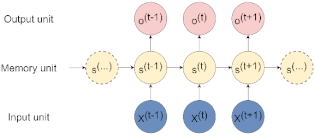 |
| Database Name | ND | URL | Data Type | Description |
|---|---|---|---|---|
| PDGene | PD | http://www.pdgene.org, accessed on 19 February 2021 | Omics | PDGene is a database providing results for potential risk loci in PD [61]. |
| PPMI | PD | https://www.ppmi-info.org, accessed on 19 February 2021 | Mixed | The Parkinson’s Progression Markers Initiative holds a comprehensive set of clinical, imaging, and biosample data to define biomarkers of PD progression [25]. |
| NIAGADS | AD | https://www.niagads.org, accessed on 19 February 2021 | Omics | The National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site is a repository that collects and shares genotypic data for the study of AD and related dementias [58]. |
| ADNI | AD | http://adni.loni.usc.edu, accessed on 19 February 2021 | Mixed | The Alzheimer’s Disease Neuroimaging Initiative is a multisite study for the prevention and treatment of AD. Its database stores a collection of validated study data to define the progression of AD, including mild cognitive impairment subjects and elderly controls [60]. |
| NACC | AD | https://www.alz.washington.edu, accessed on 19 February 2021 | Clinical | The National Alzheimer’s Coordinating Center holds a large relational database of standardized clinical and neuropathological research data for both exploratory and explanatory AD research [65]. |
| LAADC | AD | https://www.ohsu.edu/brain-institute/clinical-data-resources, accessed on 19 February 2021 | Clinical | Longitudinal relational database from the Layton Aging and Alzheimer’s Disease Center holding clinical data for over 4000 research subjects. |
| GEO | Mixed | http://www.ncbi.nlm.nih.gov/geo, accessed on 19 February 2021 | Omics | Gene Expression Omnibus is a public functional genomics data repository of array-and sequence-based data [62]. |
| UK Biobank | Mixed | https://www.ukbiobank.ac.uk, accessed on 19 February 2021 | Omics | UK Biobank contains data from a large prospective study with over 500,000 participants and it aims to improve the prevention, diagnosis, and treatment of various illnesses, including dementia [63]. |
| OmicsDI | Mixed | https://www.omicsdi.org/, accessed on 19 February 2021 | Omics | Omics Discovery Index facilitates access to omics datasets from multiple studies through an integrated and open-source platform [57]. |
| JPND | Mixed | https://www.neurodegenerationresearch.eu, accessed on 19 February 2021 | Mixed | The Joint Programme Neurodegenerative Disease Research Database contains data from research related to neurodegenerative diseases from 27 member countries. |
| GAAIN | Mixed | http://www.gaaindata.org, accessed on 19 February 2021 | Mixed | The Global Alzheimer’s Association Interactive Network is an online integrated research platform affiliated with partners all over the world, providing resources and data enabling comparative data analysis and cohort discovery [59]. |
| Bio FINDER | Mixed | https://biofinder.se/, accessed on 19 February 2021 | Mixed | The Swedish Biomarkers for Identifying Neurodegenerative Disorders Early and Reliably study aims to develop early diagnostic tests to identify novel treatment targets and understand the links between different ND and clinical symptoms. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Termine, A.; Fabrizio, C.; Strafella, C.; Caputo, V.; Petrosini, L.; Caltagirone, C.; Giardina, E.; Cascella, R. Multi-Layer Picture of Neurodegenerative Diseases: Lessons from the Use of Big Data through Artificial Intelligence. J. Pers. Med. 2021, 11, 280. https://doi.org/10.3390/jpm11040280
Termine A, Fabrizio C, Strafella C, Caputo V, Petrosini L, Caltagirone C, Giardina E, Cascella R. Multi-Layer Picture of Neurodegenerative Diseases: Lessons from the Use of Big Data through Artificial Intelligence. Journal of Personalized Medicine. 2021; 11(4):280. https://doi.org/10.3390/jpm11040280
Chicago/Turabian StyleTermine, Andrea, Carlo Fabrizio, Claudia Strafella, Valerio Caputo, Laura Petrosini, Carlo Caltagirone, Emiliano Giardina, and Raffaella Cascella. 2021. "Multi-Layer Picture of Neurodegenerative Diseases: Lessons from the Use of Big Data through Artificial Intelligence" Journal of Personalized Medicine 11, no. 4: 280. https://doi.org/10.3390/jpm11040280
APA StyleTermine, A., Fabrizio, C., Strafella, C., Caputo, V., Petrosini, L., Caltagirone, C., Giardina, E., & Cascella, R. (2021). Multi-Layer Picture of Neurodegenerative Diseases: Lessons from the Use of Big Data through Artificial Intelligence. Journal of Personalized Medicine, 11(4), 280. https://doi.org/10.3390/jpm11040280






