Comparison of Clonogenic Survival Data Obtained by Pre- and Post-Irradiation Methods
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Line and Cell Culture
2.2. Clonogenic Assays
2.3. Irradiation
2.4. Statistics
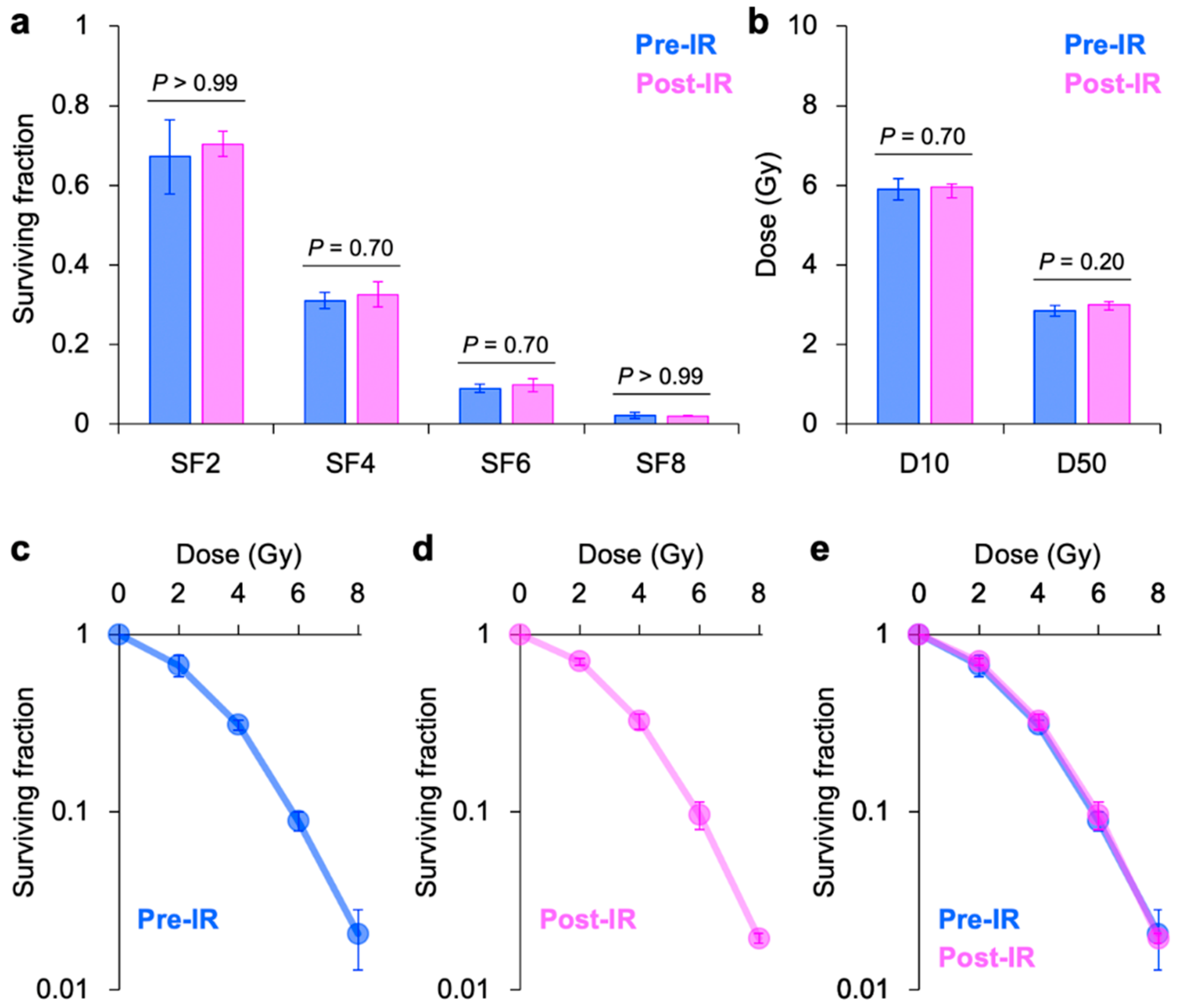
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Franken, N.A.P.; Rodermond, H.M.; Stap, J.; Haveman, J.; Van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Nuryadi, E.; Permata, T.B.M.; Komatsu, S.; Oike, T.; Nakano, T. Inter-assay precision of clonogenic assays for radiosensitivity in cancer cell line A549. Oncotarget 2018, 9, 13706–13712. [Google Scholar] [CrossRef] [PubMed]
- Kanai, T.; Endo, M.; Minohara, S.; Miyahara, N.; Koyama-Ito, H.; Tomura, H.; Matsufuji, N.; Futami, Y.; Fukumura, A.; Hiraoka, T.; et al. Biophysical characteristics of HIMAC clinical irradiation system for heavy-ion radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 1999, 44, 201–210. [Google Scholar] [CrossRef]
- Furusawa, Y.; Fukutsu, K.; Aoki, M.; Itsukaichi, H.; Eguchi-Kasai, K.; Ohara, H.; Yatagai, F.; Kanai, T.; Ando, K. Inactivation of aerobic and hypoxic cells from three different cell lines by accelerated (3)He-, (12)C- and (20)Ne-ion beams. Radiat. Res. 2000, 154, 485–496. [Google Scholar] [CrossRef]
- Kagawa, K.; Murakami, M.; Hishikawa, Y.; Abe, M.; Akagi, T.; Yanou, T.; Kagiya, G.; Furusawa, Y.; Ando, K.; Nojima, K.; et al. Preclinical biological assessment of proton and carbon ion beams at Hyogo Ion Beam Medical Center. Int. J. Radiat. Oncol. Biol. Phys. 2002, 54, 928–938. [Google Scholar] [CrossRef]
- COSMIC | Catalogue of Somatic Mutations in Cancer. Available online: https://cancer.sanger.ac.uk/cosmic (accessed on 2 August 2020).
- Broad Institute Cancer Cell Line Encyclopedia. Available online: https://portals.broadinstitute.org/ccle (accessed on 2 August 2020).
- Komatsu, S.; Oike, T.; Komatsu, Y.; Kubota, Y.; Sakai, M.; Matsui, T.; Nuryadi, E.; Permata, T.B.M.; Sato, H.; Kawamura, H.; et al. Deep learning-assisted literature mining for in vitro radiosensitivity data. Radiother. Oncol. 2019, 139, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Nuryadi, E.; Komatsu, S.; Hirota, Y.; Shibata, A.; Oike, T.; Nakano, T. Robustness of Clonogenic Assays as a Biomarker for Cancer Cell Radiosensitivity. Int. J. Mol. Sci. 2019, 20, 4148. [Google Scholar] [CrossRef] [PubMed]
- Anakura, M.; Nachankar, A.; Kobayashi, D.; Amornwichet, N.; Hirota, Y.; Shibata, A.; Oike, T.; Nakano, T. Radiosensitivity Differences between EGFR Mutant and Wild-Type Lung Cancer Cells are Larger at Lower Doses. Int. J. Mol. Sci. 2019, 20, 3635. [Google Scholar] [CrossRef] [PubMed]
- Amornwichet, N.; Oike, T.; Shibata, A.; Nirodi, C.S.; Ogiwara, H.; Makino, H.; Kimura, Y.; Hirota, Y.; Isono, M.; Yoshida, Y.; et al. The EGFR mutation status affects the relative biological effectiveness of carbon-ion beams in non-small cell lung carcinoma cells. Sci. Rep. 2015, 5, 11305. [Google Scholar] [CrossRef] [PubMed]
- Oike, T.; Ogiwara, H.; Torikai, K.; Nakano, T.; Yokota, J.; Kohno, T. Garcinol, a Histone Acetyltransferase Inhibitor, Radiosensitizes Cancer Cells by Inhibiting Non-Homologous End Joining. Int. J. Radiat. Oncol. Biol. Phys. 2012, 84, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Osu, N.; Kobayashi, D.; Shirai, K.; Musha, A.; Sato, H.; Hirota, Y.; Shibata, A.; Oike, T.; Ohno, T. Relative Biological Effectiveness of Carbon Ions for Head-and-Neck Squamous Cell Carcinomas According to Human Papillomavirus Status. J. Pers. Med. 2020, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Ichikawa, H.; Totoki, Y.; Yasuda, K.; Hiramoto, M.; Nammo, T.; Sakamoto, H.; Tsuta, K.; Furuta, K.; Shimada, Y.; et al. KIF5B-RET fusions in lung adenocarcinoma. Nat. Med. 2012, 18, 375–377. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oike, T.; Hirota, Y.; Dewi Maulany Darwis, N.; Shibata, A.; Ohno, T. Comparison of Clonogenic Survival Data Obtained by Pre- and Post-Irradiation Methods. J. Pers. Med. 2020, 10, 171. https://doi.org/10.3390/jpm10040171
Oike T, Hirota Y, Dewi Maulany Darwis N, Shibata A, Ohno T. Comparison of Clonogenic Survival Data Obtained by Pre- and Post-Irradiation Methods. Journal of Personalized Medicine. 2020; 10(4):171. https://doi.org/10.3390/jpm10040171
Chicago/Turabian StyleOike, Takahiro, Yuka Hirota, Narisa Dewi Maulany Darwis, Atsushi Shibata, and Tatsuya Ohno. 2020. "Comparison of Clonogenic Survival Data Obtained by Pre- and Post-Irradiation Methods" Journal of Personalized Medicine 10, no. 4: 171. https://doi.org/10.3390/jpm10040171
APA StyleOike, T., Hirota, Y., Dewi Maulany Darwis, N., Shibata, A., & Ohno, T. (2020). Comparison of Clonogenic Survival Data Obtained by Pre- and Post-Irradiation Methods. Journal of Personalized Medicine, 10(4), 171. https://doi.org/10.3390/jpm10040171




