A Software Tool for Estimating Uncertainty of Bayesian Posterior Probability for Disease
Abstract
1. Introduction
1.1. Diagnosis in Medicine
1.1.1. Bayes’ Theorem in Medical Diagnostics
1.1.2. Challenges in Applying Bayesian Inference
Computational Complexity
Statistical Distributions in Diagnostics
Uncertainty of Bayesian Posterior Probabilities
1.1.3. Quantifying Uncertainty in Diagnostics
Combined Uncertainty
Measurement Uncertainty
Sampling Uncertainty
2. Methods
2.1. Computational Methods
2.1.1. Bayes’ Theorem
2.1.2. Parametric Distributions
- Normal distribution
- Lognormal distribution
- Gamma distribution.
2.1.3. Uncertainty Quantification
Measurement Uncertainty
Sampling Uncertainties of Means and Standard Deviations
Sampling Uncertainty of Prior Probability for Disease
Combined Uncertainty of Posterior Probability for Disease
2.1.4. Expanded Uncertainty of Posterior Probability for Disease
2.2. The Software
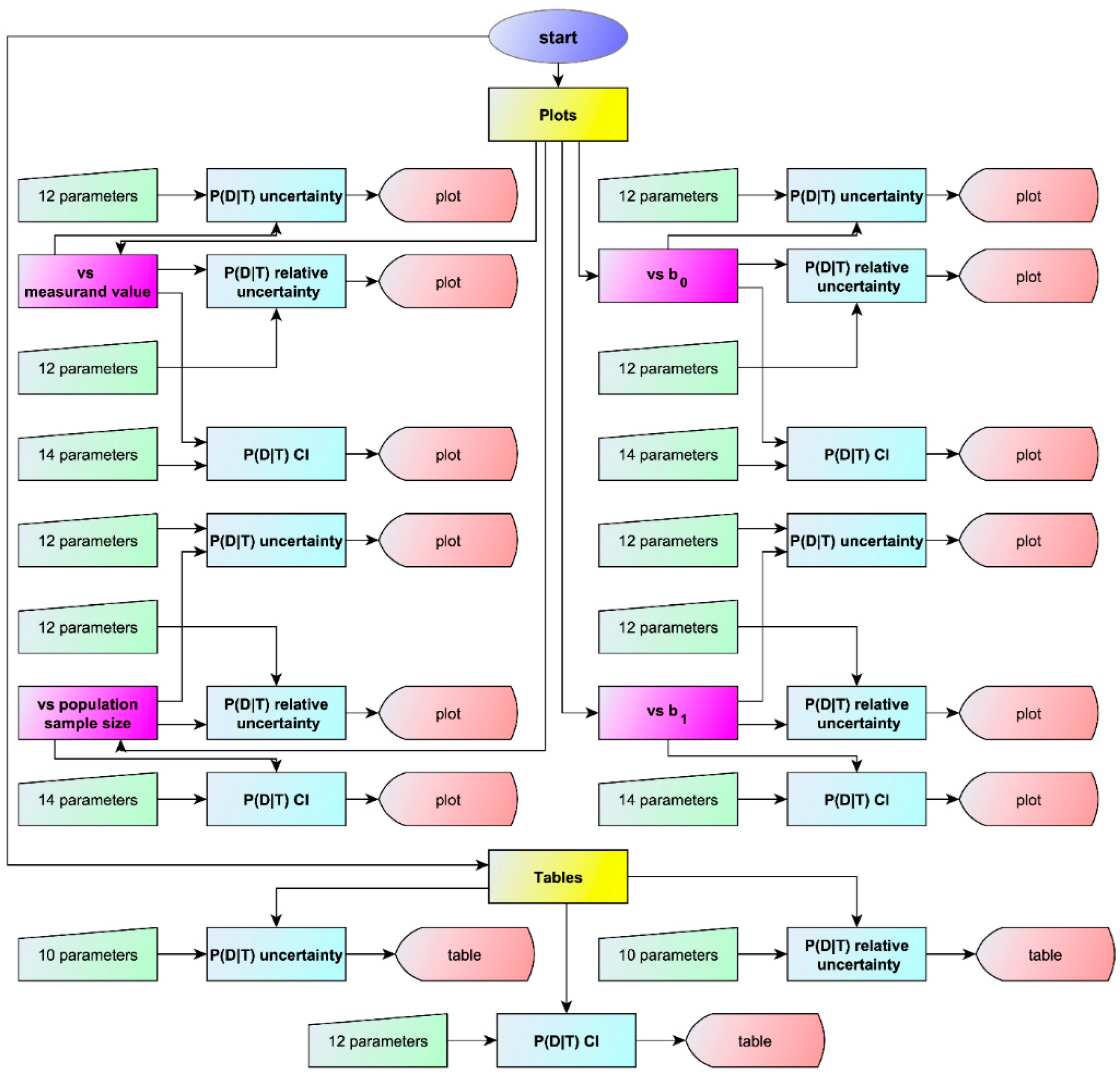
2.2.1. Program Overview
2.2.2. Input Parameters
- Normal distribution
- Lognormal distribution
- Gamma distribution.
Measurement Uncertainty
2.2.3. Output Specifications
Visualizations
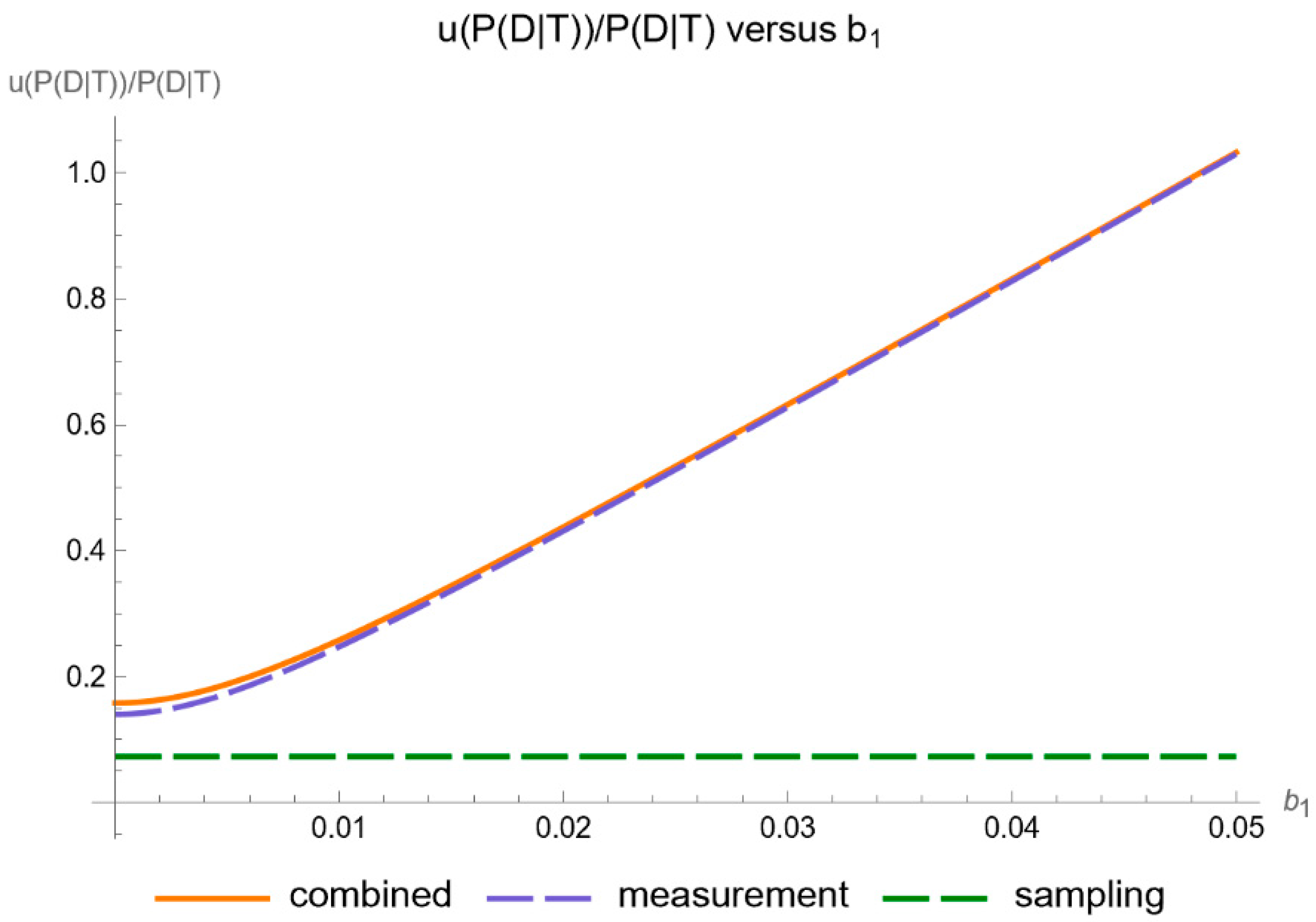
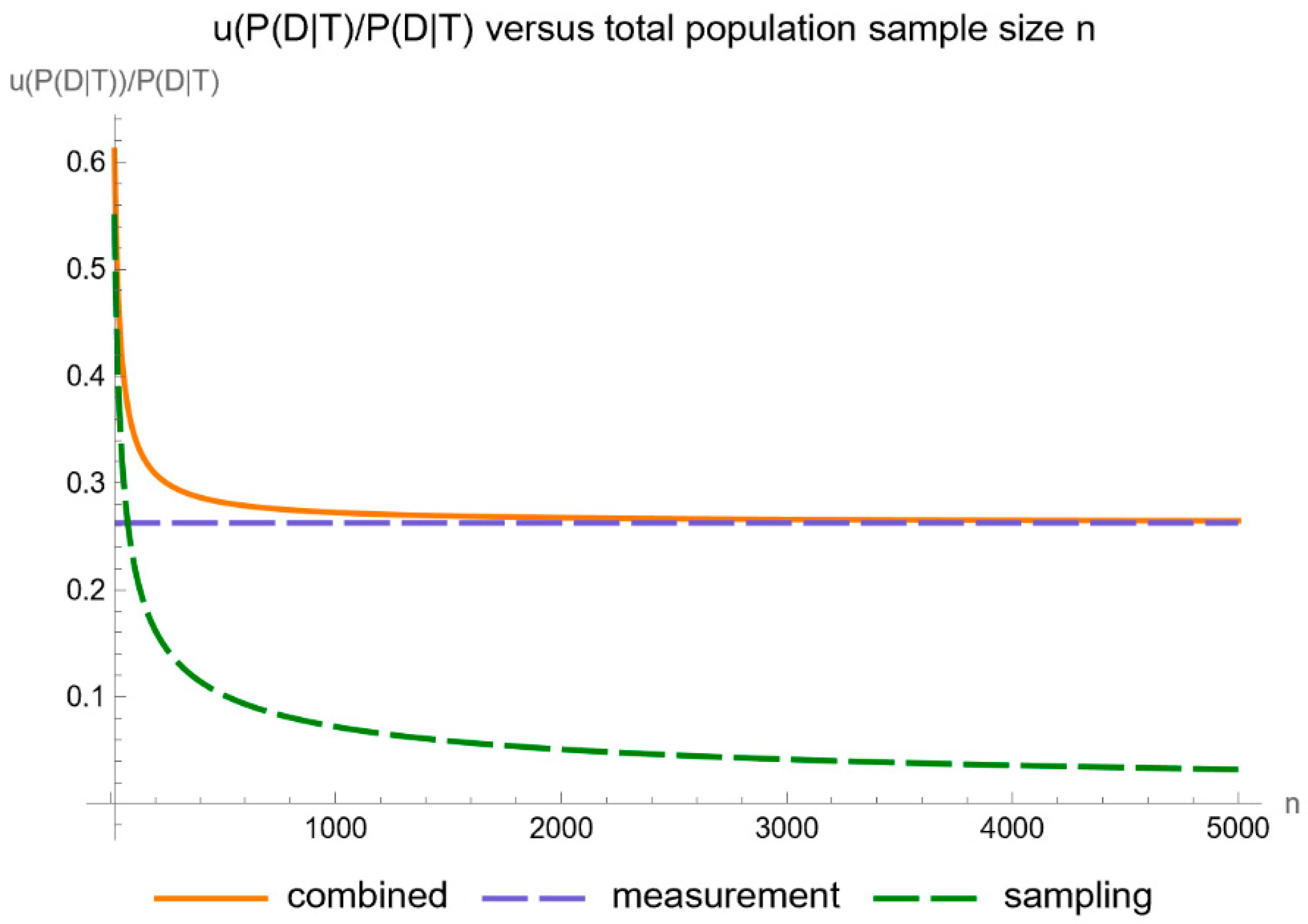
- Uncertainty of posterior probability for disease: Plots are generated to show the standard sampling, measurement, and combined uncertainty of the posterior probability for disease.
- Relative uncertainty of posterior probability for disease: Plots are generated to show the relative standard sampling, measurement, and combined uncertainty of the posterior probability for disease.
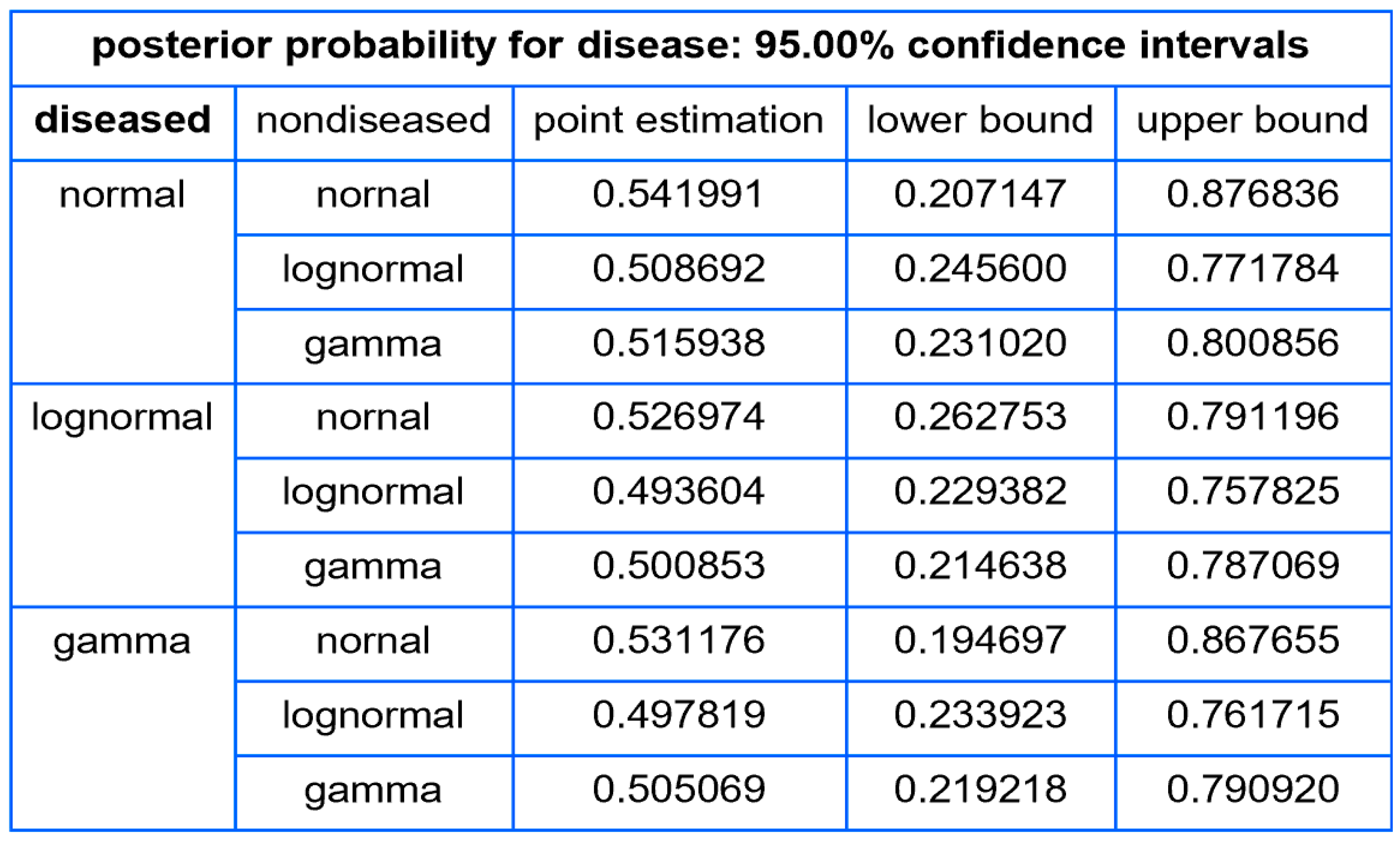
- Confidence intervals of posterior probability for disease: Plots are generated to show the confidence intervals of the posterior probability for disease, for a user defined confidence level.
Tables
- The standard sampling, measurement, and combined uncertainty of the posterior probability for disease.
- The relative standard sampling, measurement, and combined uncertainty of the posterior probability for disease.
- The confidence intervals of the posterior probability for disease for a user defined confidence level.
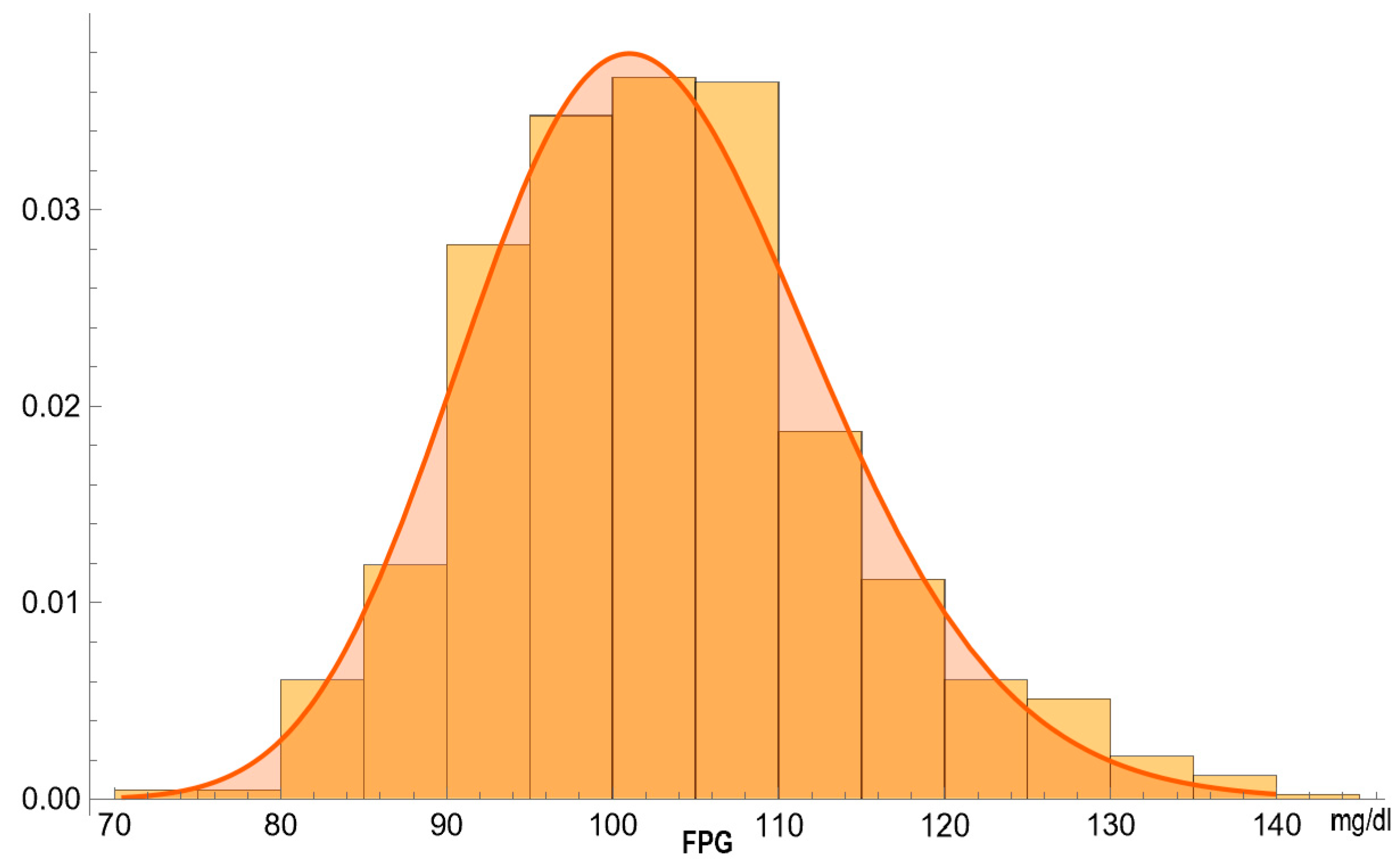
3. Illustrative Case Study
- Valid FPG and OGTT results (n = 13,836).
- A negative response to NHANES question DIQ010 regarding a diabetes diagnosis [34] (n = 13,465).
- Age 70–80 years (n = 976).
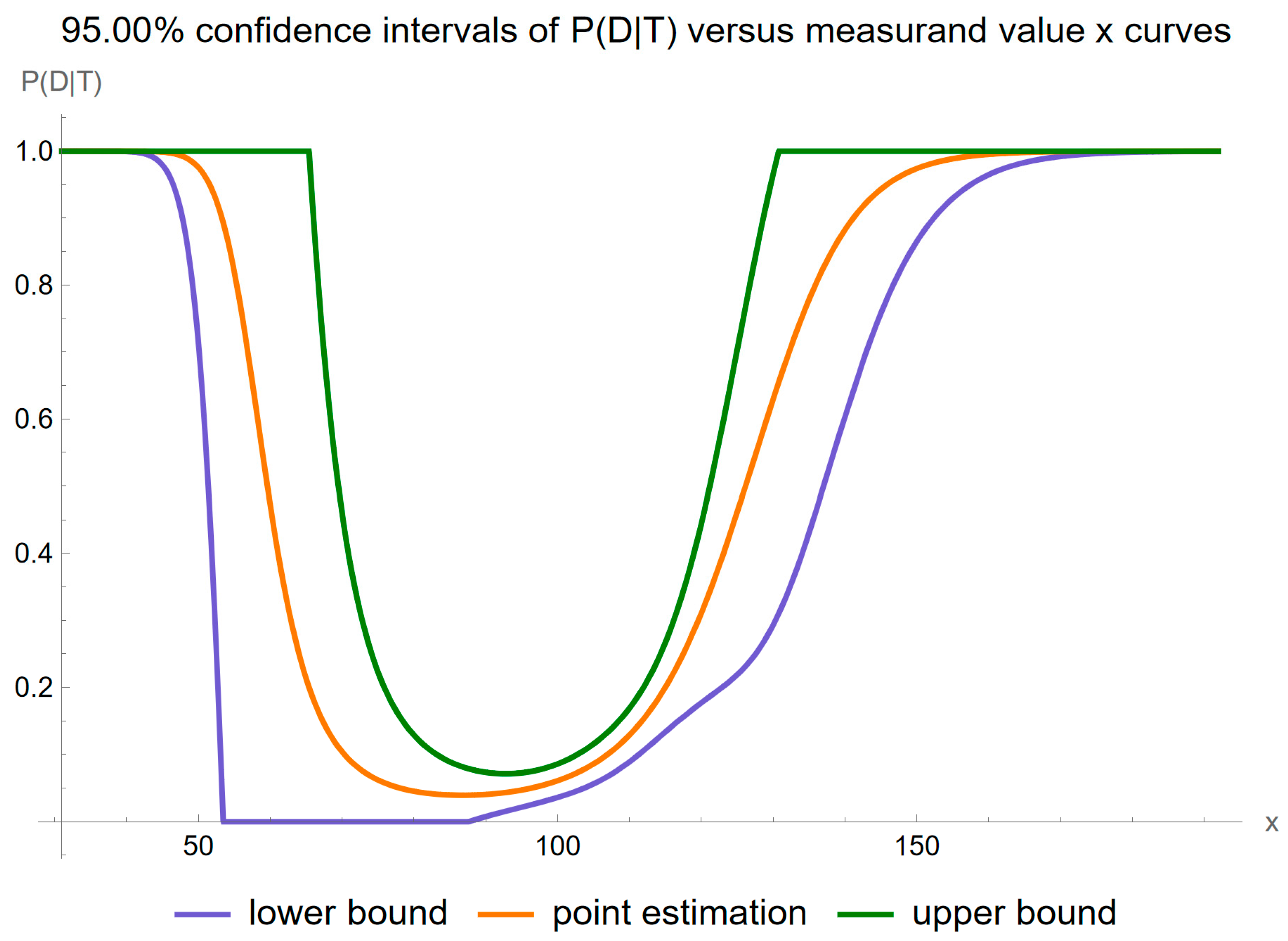
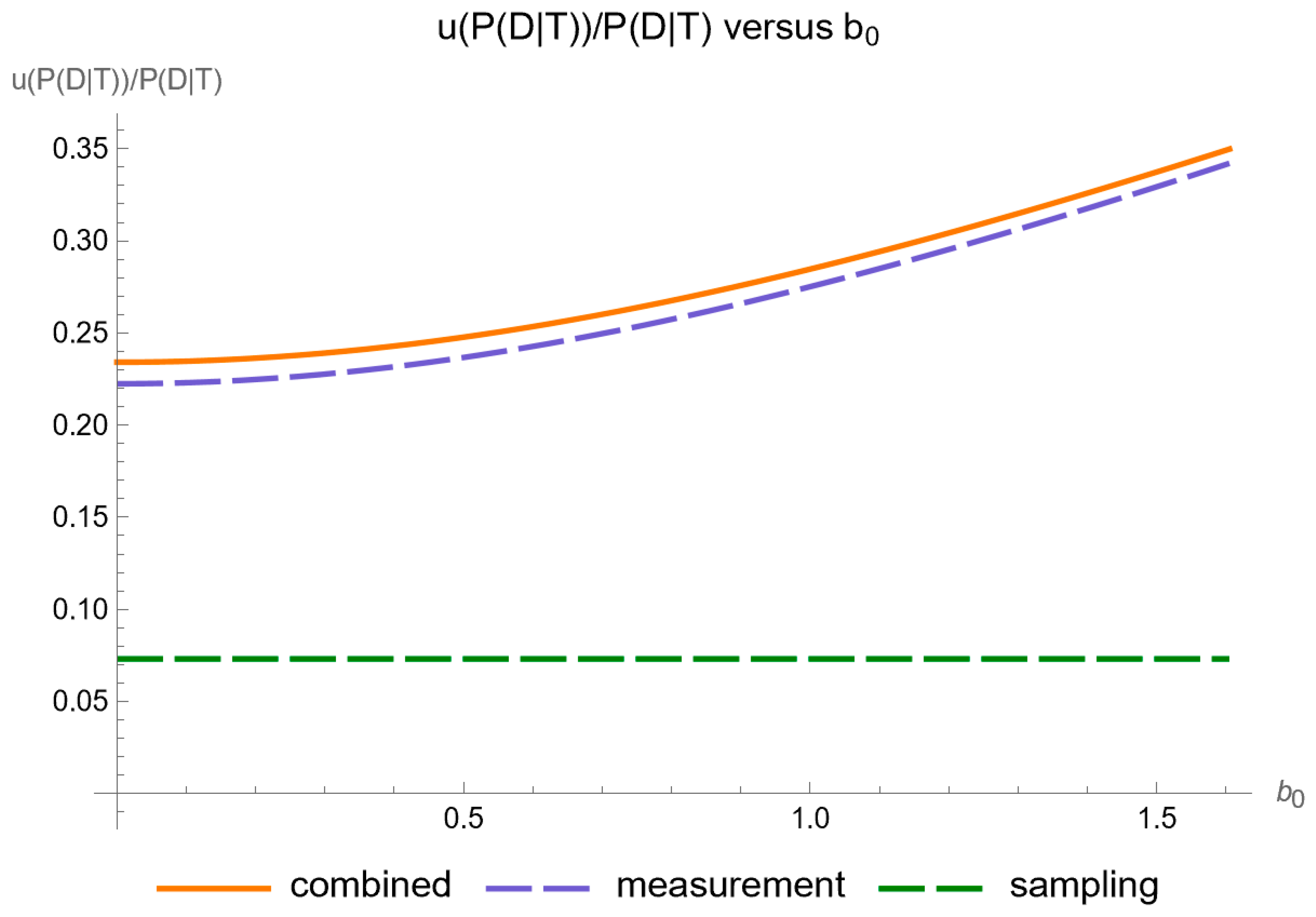
4. Results
- It is substantially affected by measurement uncertainty of FPG.
- Two local maxima are observed, corresponding to the regions near the steepest segments of the posterior probability curve, which exhibits an approximately double sigmoidal configuration. These maxima are quantitatively defined as following:
- 2.1.
- At an FPG value of 58.7 mg/dL, the posterior probability for disease is equal to 0.585, while the combined standard uncertainty is equal to 0.893.
- 2.2.
- At an FPG value of 133.2 mg/dL, the posterior probability for disease is equal to 0.725, while the combined standard uncertainty is equal to 0.182.
- At an FPG value of 64.1 mg/dL, the posterior probability for disease is equal to 0.257, while the relative combined standard uncertainty is equal to 2.044.
- At an FPG value of 128.1 mg/dL, the posterior probability for disease is equal to 0.561, while the relative combined standard uncertainty is equal to 0.278.
5. Discussion
5.1. Reevaluation of Traditional Diagnostic Methods
- Cardiac troponin for diagnosing myocardial injury and infarction [40];
- Natriuretic peptides for the diagnosis of heart failure [41];
- D-dimer for diagnosing thromboembolic events [42];
- FPG, OGTT, and glycated hemoglobin (HbA1c) for diagnosing diabetes [31];
- OGTT for the diagnosis of gestational diabetes [43];
- Thyroid stimulating hormone (TSH), free serum triiodothyronine (T3), and free serum thyroxine (T4) for diagnosing thyroid dysfunction [44];
- Protein-to-creatinine ratio for the diagnosis of preeclampsia [45];
- Creatinine or cystatin C derived glomerular filtration rate (GFR), and albuminuria for diagnosing chronic kidney disease [46].
5.2. Limitations of the Program
- Underlying assumptions:
- 1.1.
- 1.2.
- The hypothesis of parametric distribution of measurements or their transformations. However, existing literature underlines the robustness of nonparametric techniques in capturing complex data distributions [55].
- 1.3.
- 2.
- The use of first-order Taylor series approximations in uncertainty propagation calculations, where higher-order approximations may provide more accurate estimations [15].
- 3.
- The approximation of the uncertainty of the prior probability for disease using the Agresti–Coull-adjusted Waldo interval, despite more accurate methods being available [58].
- 4.
- 5.
5.3. Limitations of the Case Study
5.4. Challenges in Bayesian Analysis for Disease Diagnosis
5.5. Implications of Incomplete Data
- Over-reliance on prior probabilities: Limited empirical data may cause an overdependence on prior probabilities, leading to distorted posterior probabilities and potentially flawed clinical decisions [73].
- Increased uncertainty: Insufficient data amplifies the uncertainty of computed posterior probabilities, which in turn could exacerbate clinical indecision [74].
- Bias risks: Unrepresentative datasets could introduce systemic bias, increasing the uncertainty in Bayesian computations [5].
5.6. Analysis of the Double Sigmoidal Curve in Posterior Probability Estimation and Its Impact on Uncertainty
5.7. Software Comparison
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix A.1. Formalisms and Notation
- Acronyms
- Notation
- Bayes’ Theorem
Appendix A.1.1. Parametric Distributions
- The domain of a random variable X following a normal distribution is the set of all real numbers, denoting .
- The domain of a random variable X following a lognormal distribution is the set of all positive real numbers, denoting .
- The domain of a random variable X following a gamma distribution is the set of all positive real numbers, denoting .
Appendix A.1.2. Calculations of the Posterior Probability for Disease and Its Uncertainty
Appendix A.2. Software Availability and Requirements
Appendix A.3. A Note about the Program
- About the Program Controls
- Range of input parameters
References
- Weiner, E.S.C.; Simpson, J.A. The Oxford English Dictionary; Oxford Univeristy Press: Oxford, UK, 1989; ISBN 9780198611868. [Google Scholar]
- Chatzimichail, T.; Hatjimihail, A.T. A Bayesian Inference Based Computational Tool for Parametric and Nonparametric Medical Diagnosis. Diagnostics 2023, 13, 3135. [Google Scholar] [CrossRef]
- Choi, Y.-K.; Johnson, W.O.; Thurmond, M.C. Diagnosis Using Predictive Probabilities without Cut-Offs. Stat. Med. 2006, 25, 699–717. [Google Scholar] [CrossRef] [PubMed]
- Bours, M.J. Bayes’ Rule in Diagnosis. J. Clin. Epidemiol. 2021, 131, 158–160. [Google Scholar] [CrossRef] [PubMed]
- Gelman, A.; Carlin, J.B.; Stern, H.S.; Dunson, D.B.; Vehtari, A.; Rubin, D.B. Bayesian Data Analysis; CRC Press: Boca Raton, FL, USA, 2013; ISBN 9781439898208. [Google Scholar]
- van de Schoot, R.; Depaoli, S.; King, R.; Kramer, B.; Märtens, K.; Tadesse, M.G.; Vannucci, M.; Gelman, A.; Veen, D.; Willemsen, J.; et al. Bayesian Statistics and Modelling. Nat. Rev. Methods Primers 2021, 1, 1. [Google Scholar] [CrossRef]
- Viana, M.A.G.; Ramakrishnan, V. Bayesian Estimates of Predictive Value and Related Parameters of a Diagnostic Test. Can. J. Stat. 1992, 20, 311–321. [Google Scholar] [CrossRef]
- Topol, E.J. Individualized Medicine from Prewomb to Tomb. Cell 2014, 157, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Joyce, J. Bayes’ Theorem. In The Stanford Encyclopedia of Philosophy; Stanford University: Stanford, CA, USA, 2021. [Google Scholar]
- Lehmann, E.L.; Romano, J.P. Testing Statistical Hypotheses; Springer: New York, NY, USA, 2008; ISBN 9780387988641. [Google Scholar]
- Box, G.E.P.; Cox, D.R. An Analysis of Transformations. J. R. Stat. Soc. Series B Stat. Methodol. 1964, 26, 211–243. [Google Scholar] [CrossRef]
- D’Agostino, R.; Pearson, E.S. Tests for Departure from Normality. Empirical Results for the Distributions of b2 and √b1. Biometrika 1973, 60, 613–622. [Google Scholar] [CrossRef]
- Srinivasan, P.; Westover, M.B.; Bianchi, M.T. Propagation of Uncertainty in Bayesian Diagnostic Test Interpretation. South Med. J. 2012, 105, 452–459. [Google Scholar] [CrossRef]
- Ayyub, B.M.; Klir, G.J. Uncertainty Modeling and Analysis in Engineering and the Sciences; Chapman and Hall/CRC: Boca Raton, FL, USA, 2006. [Google Scholar]
- Joint Committee for Guides in Metrology. Evaluation of Measurement Data—Supplement 2 to the “Guide to the Expression of Uncertainty in Measurement”—Extension to Any Number of Output Quantities; BIPM: Sèvres, France, 2011. [Google Scholar]
- Kallner, A.; Boyd, J.C.; Duewer, D.L.; Giroud, C.; Hatjimihail, A.T.; Klee, G.G.; Lo, S.F.; Pennello, G.; Sogin, D.; Tholen, D.W.; et al. Expression of Measurement Uncertainty in Laboratory Medicine; Approved Guideline; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Smith, A.F.; Shinkins, B.; Hall, P.S.; Hulme, C.T.; Messenger, M.P. Toward a Framework for Outcome-Based Analytical Performance Specifications: A Methodology Review of Indirect Methods for Evaluating the Impact of Measurement Uncertainty on Clinical Outcomes. Clin. Chem. 2019, 65, 1363–1374. [Google Scholar] [CrossRef]
- Ceriotti, F.; Fernandez-Calle, P.; Klee, G.G.; Nordin, G.; Sandberg, S.; Streichert, T.; Vives-Corrons, J.-L.; Panteghini, M. Criteria for Assigning Laboratory Measurands to Models for Analytical Performance Specifications Defined in the 1st EFLM Strategic Conference. Clin. Chem. Lab. Med. 2017, 55, 189–194. [Google Scholar] [CrossRef]
- Chatzimichail, T.; Hatjimihail, A.T. A Software Tool for Calculating the Uncertainty of Diagnostic Accuracy Measures. Diagnostics 2021, 11, 406. [Google Scholar] [CrossRef]
- Rostron, P.D.; Fearn, T.; Ramsey, M.H. Confidence Intervals for Robust Estimates of Measurement Uncertainty. Accredit. Qual. Assur. 2020, 25, 107–119. [Google Scholar] [CrossRef]
- Geisser, S.; Johnson, W.O. Modes of Parametric Statistical Inference; John Wiley & Sons: Hoboken, NJ, USA, 2006; ISBN 9780471743125. [Google Scholar]
- White, G.H. Basics of Estimating Measurement Uncertainty. Clin. Biochem. Rev. 2008, 29 (Suppl. S1), S53–S60. [Google Scholar]
- Ellison, S.L.R.; Williams, A. Quantifying Uncertainty in Analytical Measurement, 3rd ed.; EURACHEM/CITAC: Teddington, UK, 2012. [Google Scholar]
- Agresti, A.; Franklin, C.; Klingenberg, B. Statistics: The Art and Science of Learning from Data, Global Edition, 4th ed.; Pearson Education: London, UK, 2023; ISBN 9781292442464. [Google Scholar]
- Miller, J.; Miller, J.C. Statistics and Chemometrics for Analytical Chemistry, 7th ed.; Pearson Education: London, UK, 2018; ISBN 9781292186719. [Google Scholar]
- Aitchison, J.; Brown, J.A.C. The Lognormal Distribution with Special Reference to Its Uses in Econometrics; Cambridge University Press: Cambridge, UK, 1957. [Google Scholar]
- Agresti, A.; Coull, B.A. Approximate Is Better than “Exact” for Interval Estimation of Binomial Proportions. Am. Stat. 1998, 52, 119–126. [Google Scholar] [CrossRef]
- Wilson, B.M.; Smith, B.L. Taylor-Series and Monte-Carlo-Method Uncertainty Estimation of the Width of a Probability Distribution Based on Varying Bias and Random Error. Meas. Sci. Technol. 2013, 24, 035301. [Google Scholar] [CrossRef]
- Welch, B.L. The Generalization of ‘Student’s’ Problem When Several Different Population Variances Are Involved. Biometrika 1947, 34, 28–35. [Google Scholar] [CrossRef]
- Satterthwaite, F.E. An Approximate Distribution of Estimates of Variance Components. Biometrics 1946, 2, 110–114. [Google Scholar] [CrossRef]
- ElSayed, N.A.; Aleppo, G.; Aroda, V.R.; Bannuru, R.R.; Brown, F.M.; Bruemmer, D.; Collins, B.S.; Hilliard, M.E.; Isaacs, D.; Johnson, E.L.; et al. 2. Classification and Diagnosis of Diabetes: Standards of Care in Diabetes—2023. Diabetes Care 2023, 46, S19–S40. [Google Scholar] [CrossRef]
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.N.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, Regional and Country-Level Diabetes Prevalence Estimates for 2021 and Projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef]
- National Center for Health Statistics. National Health and Nutrition Examination Survey Data. Available online: https://wwwn.cdc.gov/nchs/nhanes/default.aspx (accessed on 4 September 2023).
- National Center for Health Statistics. National Health and Nutrition Examination Survey Questionnaire. Available online: https://wwwn.cdc.gov/nchs/nhanes/Search/variablelist.aspx?Component=Questionnaire (accessed on 4 September 2023).
- Myung, I.J. Tutorial on Maximum Likelihood Estimation. J. Math. Psychol. 2003, 47, 90–100. [Google Scholar] [CrossRef]
- Nielsen, A.A. Least Squares Adjustment: Linear and Nonlinear Weighted Regression Analysis; Technical University of Denmark: Kongens Lyngby, Denmark, 2007. [Google Scholar]
- Darling, D.A. The Kolmogorov-Smirnov, Cramer-von Mises Tests. Ann. Math. Stat. 1957, 28, 823–838. [Google Scholar] [CrossRef]
- Tucker, L.A. Limited Agreement between Classifications of Diabetes and Prediabetes Resulting from the OGTT, Hemoglobin A1c, and Fasting Glucose Tests in 7412 U.S. Adults. J. Clin. Med. Res. 2020, 9, 2207. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Emanuel, E.J. Predicting the Future—Big Data, Machine Learning, and Clinical Medicine. N. Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef]
- Wereski, R.; Kimenai, D.M.; Taggart, C.; Doudesis, D.; Lee, K.K.; Lowry, M.T.H.; Bularga, A.; Lowe, D.J.; Fujisawa, T.; Apple, F.S.; et al. Cardiac Troponin Thresholds and Kinetics to Differentiate Myocardial Injury and Myocardial Infarction. Circulation 2021, 144, 528–538. [Google Scholar] [CrossRef]
- Roberts, E.; Ludman, A.J.; Dworzynski, K.; Al-Mohammad, A.; Cowie, M.R.; McMurray, J.J.V.; Mant, J.; NICE Guideline Development Group for Acute Heart Failure. The Diagnostic Accuracy of the Natriuretic Peptides in Heart Failure: Systematic Review and Diagnostic Meta-Analysis in the Acute Care Setting. BMJ 2015, 350, h910. [Google Scholar] [CrossRef]
- Freund, Y.; Chauvin, A.; Jimenez, S.; Philippon, A.-L.; Curac, S.; Fémy, F.; Gorlicki, J.; Chouihed, T.; Goulet, H.; Montassier, E.; et al. Effect of a Diagnostic Strategy Using an Elevated and Age-Adjusted D-Dimer Threshold on Thromboembolic Events in Emergency Department Patients with Suspected Pulmonary Embolism: A Randomized Clinical Trial. JAMA 2021, 326, 2141–2149. [Google Scholar] [CrossRef]
- Rani, P.R.; Begum, J. Screening and Diagnosis of Gestational Diabetes Mellitus, Where Do We Stand. J. Clin. Diagn. Res. 2016, 10, QE01–QE04. [Google Scholar] [CrossRef]
- Reyes Domingo, F.; Avey, M.T.; Doull, M. Screening for Thyroid Dysfunction and Treatment of Screen-Detected Thyroid Dysfunction in Asymptomatic, Community-Dwelling Adults: A Systematic Review. Syst. Rev. 2019, 8, 260. [Google Scholar] [CrossRef]
- Rodriguez-Thompson, D.; Lieberman, E.S. Use of a Random Urinary Protein-to-Creatinine Ratio for the Diagnosis of Significant Proteinuria during Pregnancy. Am. J. Obstet. Gynecol. 2001, 185, 808–811. [Google Scholar] [CrossRef]
- Moynihan, R.; Glassock, R.; Doust, J. Chronic Kidney Disease Controversy: How Expanding Definitions Are Unnecessarily Labelling Many People as Diseased. BMJ 2013, 347, f4298. [Google Scholar] [CrossRef] [PubMed]
- Haeckel, R.; Wosniok, W.; Gurr, E.; Peil, B.; on behalf of Arbeitsgruppe Richtwer. Supplements to a Recent Proposal for Permissible Uncertainty of Measurements in Laboratory Medicine. LaboratoriumsMedizin 2016, 40, 141–145. [Google Scholar] [CrossRef]
- Knottnerus, J.A.; Buntinx, F. (Eds.) The Evidence Base of Clinical Diagnosis. In Evidence-Based Medicine, 2nd ed.; BMJ Books: London, UK, 2011; ISBN 9781444360639. [Google Scholar]
- Hajian-Tilaki, K. Sample Size Estimation in Diagnostic Test Studies of Biomedical Informatics. J. Biomed. Inform. 2014, 48, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Baron, J.A. Uncertainty in Bayes. Med. Decis. Mak. 1994, 14, 46–51. [Google Scholar] [CrossRef]
- Ashby, D.; Smith, A.F. Evidence-Based Medicine as Bayesian Decision-Making. Stat. Med. 2000, 19, 3291–3305. [Google Scholar] [CrossRef]
- Knottnerus, J.A.; Dinant, G.J. Medicine Based Evidence, a Prerequisite for Evidence Based Medicine. BMJ 1997, 315, 1109–1110. [Google Scholar] [CrossRef]
- Pfeiffer, R.M.; Castle, P.E. With or without a Gold Standard. Epidemiology 2005, 16, 595–597. [Google Scholar] [CrossRef]
- van Smeden, M.; Naaktgeboren, C.A.; Reitsma, J.B.; Moons, K.G.M.; de Groot, J.A.H. Latent Class Models in Diagnostic Studies When There Is No Reference Standard—A Systematic Review. Am. J. Epidemiol. 2014, 179, 423–431. [Google Scholar] [CrossRef]
- Wasserman, L. All of Nonparametric Statistics; Springer: Berlin/Heidelberg, Germany, 2006; ISBN 9780387306230. [Google Scholar]
- Wilson, J.M.G.; Jungner, G. Principles and Practice of Screening for Disease; Public health papers; World Health Organization: Geneva, Switzerland, 1968; Volume 34. [Google Scholar]
- Petersen, P.H.; Horder, M. 2.3 Clinical Test Evaluation. Unimodal and Bimodal Approaches. Scand. J. Clin. Lab. Investig. 1992, 52, 51–57. [Google Scholar] [CrossRef]
- Pires, A.M.; Amado, C. Interval Estimators for a Binomial Proportion: Comparison of Twenty Methods. Revstat Stat. J. 2008, 6, 165–197. [Google Scholar] [CrossRef]
- Schmoyeri, R.L.; Beauchamp, J.J.; Brandt, C.C.; Hoffman, F.O. Difficulties with the Lognormal Model in Mean Estimation and Testing. Environ. Ecol. Stat. 1996, 3, 81–97. [Google Scholar] [CrossRef]
- Bhaumik, D.K.; Kapur, K.; Gibbons, R.D. Testing Parameters of a Gamma Distribution for Small Samples. Technometrics 2009, 51, 326–334. [Google Scholar] [CrossRef]
- Williams, A. Calculation of the Expanded Uncertainty for Large Uncertainties Using the Lognormal Distribution. Accredit. Qual. Assur. 2020, 25, 335–338. [Google Scholar] [CrossRef]
- Stephens, M. The Bayesian Lens and Bayesian Blinkers. Philos. Trans. A Math. Phys. Eng. Sci. 2023, 381, 20220144. [Google Scholar] [CrossRef] [PubMed]
- Joint Committee for Guides in Metrology. Evaluation of Measurement Data—Supplement 1 to the “Guide to the Expression of Uncertainty in Measurement”—Propagation of Distributions Using a Monte Carlo Method Joint Committee for Guides in Metrology; BIPM: Sèvres, France, 2008. [Google Scholar]
- Joint Committee for Guides in Metrology. Guide to the Expression of Uncertainty in Measurement—Part 6: Developing and Using Measurement Models; BIPM: Sèvres, France, 2020. [Google Scholar]
- Meneilly, G.S.; Elliott, T. Metabolic Alterations in Middle-Aged and Elderly Obese Patients with Type 2 Diabetes. Diabetes Care 1999, 22, 112–118. [Google Scholar] [CrossRef]
- Geer, E.B.; Shen, W. Gender Differences in Insulin Resistance, Body Composition, and Energy Balance. Gend. Med. 2009, 6 (Suppl. S1), 60–75. [Google Scholar] [CrossRef] [PubMed]
- Van Cauter, E.; Polonsky, K.S.; Scheen, A.J. Roles of Circadian Rhythmicity and Sleep in Human Glucose Regulation. Endocr. Rev. 1997, 18, 716–738. [Google Scholar] [CrossRef]
- Colberg, S.R.; Sigal, R.J.; Fernhall, B.; Regensteiner, J.G.; Blissmer, B.J.; Rubin, R.R.; Chasan-Taber, L.; Albright, A.L.; Braun, B.; American College of Sports Medicine; et al. Exercise and Type 2 Diabetes: The American College of Sports Medicine and the American Diabetes Association: Joint Position Statement. Diabetes Care 2010, 33, e147–e167. [Google Scholar] [CrossRef]
- Salmerón, J.; Manson, J.E.; Stampfer, M.J.; Colditz, G.A.; Wing, A.L.; Willett, W.C. Dietary Fiber, Glycemic Load, and Risk of Non-Insulin-Dependent Diabetes Mellitus in Women. JAMA 1997, 277, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Surwit, R.S.; van Tilburg, M.A.L.; Zucker, N.; McCaskill, C.C.; Parekh, P.; Feinglos, M.N.; Edwards, C.L.; Williams, P.; Lane, J.D. Stress Management Improves Long-Term Glycemic Control in Type 2 Diabetes. Diabetes Care 2002, 25, 30–34. [Google Scholar] [CrossRef]
- Pandit, M.K.; Burke, J.; Gustafson, A.B.; Minocha, A.; Peiris, A.N. Drug-Induced Disorders of Glucose Tolerance. Ann. Intern. Med. 1993, 118, 529–539. [Google Scholar] [CrossRef]
- Dupuis, J.; Langenberg, C.; Prokopenko, I.; Saxena, R.; Soranzo, N.; Jackson, A.U.; Wheeler, E.; Glazer, N.L.; Bouatia-Naji, N.; Gloyn, A.L.; et al. New Genetic Loci Implicated in Fasting Glucose Homeostasis and Their Impact on Type 2 Diabetes Risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar] [CrossRef]
- O’Hagan, A.; Buck, C.E.; Daneshkhah, A.; Richard Eiser, J.; Garthwaite, P.H.; Jenkinson, D.J.; Oakley, J.E.; Rakow, T. Uncertain Judgements: Eliciting Experts’ Probabilities; John Wiley & Sons: Hoboken, NJ, USA, 2006; ISBN 9780470033302. [Google Scholar]
- Berger, J.O. Statistical Decision Theory and Bayesian Analysis; Springer: Berlin/Heidelberg, Germany, 1985; ISBN 9780387960982. [Google Scholar]
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallett, S.; QUADAS-2 Steering Group. A Systematic Review Classifies Sources of Bias and Variation in Diagnostic Test Accuracy Studies. J. Clin. Epidemiol. 2013, 66, 1093–1104. [Google Scholar] [CrossRef] [PubMed]
- Salameh, J.-P.; Bossuyt, P.M.; McGrath, T.A.; Thombs, B.D.; Hyde, C.J.; Macaskill, P.; Deeks, J.J.; Leeflang, M.; Korevaar, D.A.; Whiting, P.; et al. Preferred Reporting Items for Systematic Review and Meta-Analysis of Diagnostic Test Accuracy Studies (PRISMA-DTA): Explanation, Elaboration, and Checklist. BMJ 2020, 370, m2632. [Google Scholar] [CrossRef] [PubMed]
- Schlattmann, P. Tutorial: Statistical Methods for the Meta-Analysis of Diagnostic Test Accuracy Studies. Clin. Chem. Lab. Med. 2023, 61, 777–794. [Google Scholar] [CrossRef] [PubMed]












| Diabetic Participants | Nondiabetic Participants | |
|---|---|---|
| n | 154 | 822 |
| Mean | 120.7 | 102.6 |
| Median | 117.0 | 102.0 |
| Standard Deviation | 19.1 | 10.9 |
| Skewness | 1.448 | 0.523 |
| Kurtosis | 6.354 | 3.445 |
| Diabetic Participants | Nondiabetic Participants | |||
|---|---|---|---|---|
| Estimated Distribution | ||||
| Mean Uncertainty | 1.586 | 0 | 1.028 | 0 |
| Mean | 120.7 | 120.7 | 102.6 | 102.6 |
| Median | 119.4 | 119.4 | 102.1 | 102.1 |
| Standard Deviation | 17.8 | 17.7 | 10.9 | 10.7 |
| Skewness | 0.446 | 0.444 | 0.315 | 0.312 |
| Kurtosis | 3.355 | 3.352 | 3.177 | 3.174 |
| p-value (Cramér–von Mises test) | 0.294 | 0.295 | 0.281 | 0.299 |
| Settings | Figure 5 and Figure 6 | Figure 7 | Figure 8 and Figure 9 | Figure 10 | Figure 11 and Figure 12 | Figure 13 | Figure 14 and Figure 15 | Figure 16 | Figure 17 and Figure 18 | Figure 19 |
|---|---|---|---|---|---|---|---|---|---|---|
| p | - | 0.95 | - | 0.95 | - | 0.95 | - | 0.95 | - | 0.95 |
| x | 31.0–192.0 | 31.0–192.0 | 126.0 | 126.0 | 126.0 | 126.0 | 126.0 | 126.0 | 126.0 | 126.0 |
| 120.7 | 120.7 | 120.7 | 120.7 | 120.7 | 120.7 | 120.7 | 120.7 | 120.7 | 120.7 | |
| 17.7 | 17.7 | 17.7 | 17.7 | 17.7 | 17.7 | 17.7 | 17.7 | 17.7 | 17.7 | |
| 154 | 154 | 154 | 154 | 154 | 154 | - | - | 154 | 154 | |
| 102.7 | 102.7 | 102.7 | 102.7 | 102.7 | 102.7 | 102.7 | 102.7 | 102.7 | 102.7 | |
| 10.7 | 10.7 | 10.7 | 10.7 | 10.7 | 10.7 | 10.7 | 10.7 | 10.7 | 10.7 | |
| 822 | 822 | 822 | 822 | 822 | 822 | - | - | 822 | 822 | |
| n | - | - | - | - | - | - | 65–5000 | 65–5000 | - | - |
| r | - | - | - | - | - | - | 0.158 | 0.158 | - | - |
| 0.866 | 0.866 | 0.0–0.161 | 0.0–0.161 | 0.866 | 0.866 | 0.866 | 0.866 | 0.866 | 0.866 | |
| 0.0109 | 0.0109 | 0.0109 | 0.0109 | 0.0–0.1 | 0.0–0.1 | 0.0109 | 0.0109 | 0.0109 | 0.0109 | |
| - | 1350 | - | 1350 | - | 1350 | - | 1350 | - | 1350 | |
| lognormal | lognormal | lognormal | lognormal | lognormal | lognormal | lognormal | lognormal | normal lognormal gamma | normal lognormal gamma | |
| lognormal | lognormal | lognormal | lognormal | lognormal | lognormal | lognormal | lognormal | normal lognormal gamma | normal lognormal gamma |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatzimichail, T.; Hatjimihail, A.T. A Software Tool for Estimating Uncertainty of Bayesian Posterior Probability for Disease. Diagnostics 2024, 14, 402. https://doi.org/10.3390/diagnostics14040402
Chatzimichail T, Hatjimihail AT. A Software Tool for Estimating Uncertainty of Bayesian Posterior Probability for Disease. Diagnostics. 2024; 14(4):402. https://doi.org/10.3390/diagnostics14040402
Chicago/Turabian StyleChatzimichail, Theodora, and Aristides T. Hatjimihail. 2024. "A Software Tool for Estimating Uncertainty of Bayesian Posterior Probability for Disease" Diagnostics 14, no. 4: 402. https://doi.org/10.3390/diagnostics14040402
APA StyleChatzimichail, T., & Hatjimihail, A. T. (2024). A Software Tool for Estimating Uncertainty of Bayesian Posterior Probability for Disease. Diagnostics, 14(4), 402. https://doi.org/10.3390/diagnostics14040402







