A Multivariant Surrogate Neutralization Assay Identifies Variant-Specific Neutralizing Antibody Profiles in Primary SARS-CoV-2 Omicron Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Multivariant Surrogate Virus Neutralization Test
2.3. Live-Virus Neutralization Test
2.4. Statistical Analyses
3. Results
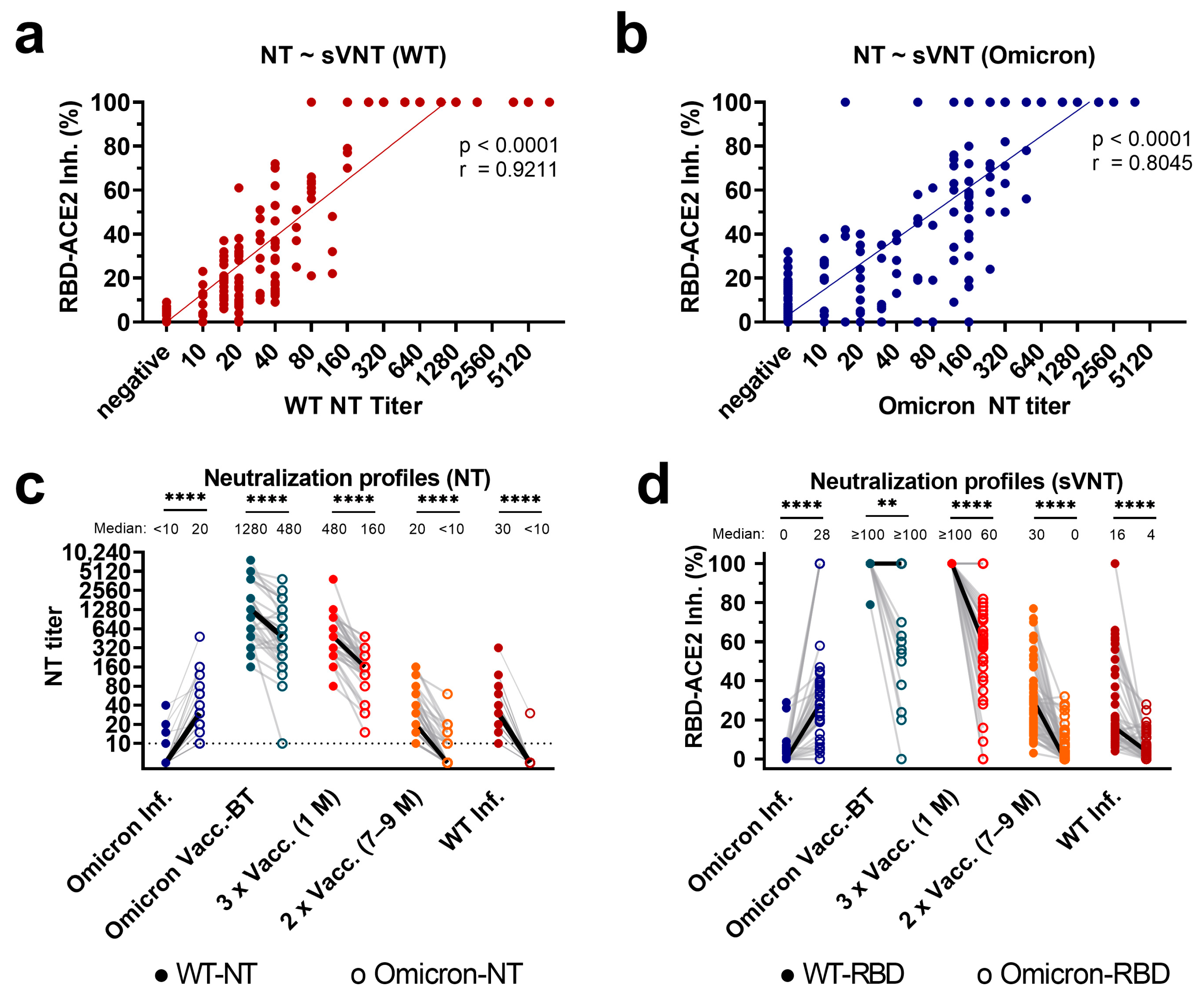
3.1. Correlation of Variant-Specific nAbs as Assessed with sVNT and NTs
3.2. Profiles of Variant-Specific nAbs as Assessed with sVNT and Live-Virus NTs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mykytyn, A.Z.; Rissmann, M.; Kok, A.; Rosu, M.E.; Schipper, D.; Breugem, T.I.; van den Doel, P.B.; Chandler, F.; Bestebroer, T.; de Wit, M.; et al. Antigenic cartography of SARS-CoV-2 reveals that Omicron BA.1 and BA.2 are antigenically distinct. Sci. Immunol. 2022, 7, eabq4450. [Google Scholar] [CrossRef] [PubMed]
- Medits, I.; Springer, D.N.; Graninger, M.; Camp, J.V.; Höltl, E.; Aberle, S.W.; Traugott, M.T.; Hoepler, W.; Deutsch, J.; Lammel, O.; et al. Different Neutralization Profiles After Primary SARS-CoV-2 Omicron BA.1 and BA.2 Infections. Front. Immunol. 2022, 13, 946318. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Huo, J.; Zhou, D.; Zahradník, J.; Supasa, P.; Liu, C.; Duyvesteyn, H.M.E.; Ginn, H.M.; Mentzer, A.J.; Tuekprakhon, A.; et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022, 185, 467–484.e415. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Henry, B.M.; Plebani, M. A Simple Epidemiologic Model for Predicting Impaired Neutralization of New SARS-CoV-2 Variants. Vaccines 2023, 11, 128. [Google Scholar] [CrossRef] [PubMed]
- Rössler, A.; Riepler, L.; Bante, D.; von Laer, D.; Kimpel, J. SARS-CoV-2 Omicron Variant Neutralization in Serum from Vaccinated and Convalescent Persons. N. Engl. J. Med. 2022, 386, 698–700. [Google Scholar] [CrossRef] [PubMed]
- Flemming, A. Omicron, the great escape artist. Nat. Rev. Immunol. 2022, 22, 75. [Google Scholar] [CrossRef]
- Callaway, E. Why does the Omicron sub-variant spread faster than the original? Nature 2022, 602, 556–557. [Google Scholar] [CrossRef]
- Schubert, M.; Bertoglio, F.; Steinke, S.; Heine, P.A.; Ynga-Durand, M.A.; Maass, H.; Sammartino, J.C.; Cassaniti, I.; Zuo, F.; Du, L.; et al. Human serum from SARS-CoV-2-vaccinated and COVID-19 patients shows reduced binding to the RBD of SARS-CoV-2 Omicron variant. BMC Med. 2022, 20, 102. [Google Scholar] [CrossRef]
- Richardson, S.I.; Madzorera, V.S.; Spencer, H.; Manamela, N.P.; van der Mescht, M.A.; Lambson, B.E.; Oosthuysen, B.; Ayres, F.; Makhado, Z.; Moyo-Gwete, T.; et al. SARS-CoV-2 Omicron triggers cross-reactive neutralization and Fc effector functions in previously vaccinated, but not unvaccinated, individuals. Cell Host Microbe 2022, 30, 880–886.e4. [Google Scholar] [CrossRef]
- Rössler, A.; Netzl, A.; Knabl, L.; Schäfer, H.; Wilks, S.H.; Bante, D.; Falkensammer, B.; Borena, W.; von Laer, D.; Smith, D.J.; et al. BA.2 and BA.5 omicron differ immunologically from both BA.1 omicron and pre-omicron variants. Nat. Commun. 2022, 13, 7701. [Google Scholar] [CrossRef]
- Rössler, A.; Knabl, L.; von Laer, D.; Kimpel, J. Neutralization Profile after Recovery from SARS-CoV-2 Omicron Infection. N. Engl. J. Med. 2022, 386, 1764–1766. [Google Scholar] [CrossRef] [PubMed]
- Springer, D.N.; Perkmann, T.; Jani, C.M.; Mucher, P.; Prüger, K.; Marculescu, R.; Reuberger, E.; Camp, J.V.; Graninger, M.; Borsodi, C.; et al. Reduced Sensitivity of Commercial Spike-Specific Antibody Assays after Primary Infection with the SARS-CoV-2 Omicron Variant. Microbiol. Spectr. 2022, 10, e0212922. [Google Scholar] [CrossRef] [PubMed]
- Migueres, M.; Chapuy-Regaud, S.; Miédougé, M.; Jamme, T.; Lougarre, C.; Da Silva, I.; Pucelle, M.; Staes, L.; Porcheron, M.; Diméglio, C.; et al. Current immunoassays and detection of antibodies elicited by Omicron SARS-CoV-2 infection. J. Med. Virol. 2023, 95, e28200. [Google Scholar] [CrossRef]
- Fenwick, C.; Turelli, P.; Pellaton, C.; Farina, A.; Campos, J.; Raclot, C.; Pojer, F.; Cagno, V.; Nusslé, S.G.; D’Acremont, V.; et al. A high-throughput cell- and virus-free assay shows reduced neutralization of SARS-CoV-2 variants by COVID-19 convalescent plasma. Sci Transl. Med. 2021, 13, eabi8452. [Google Scholar] [CrossRef] [PubMed]
- Fenwick, C.; Croxatto, A.; Coste, A.T.; Pojer, F.; André, C.; Pellaton, C.; Farina, A.; Campos, J.; Hacker, D.; Lau, K.; et al. Changes in SARS-CoV-2 Spike versus Nucleoprotein Antibody Responses Impact the Estimates of Infections in Population-Based Seroprevalence Studies. J. Virol. 2021, 95, e01828-20. [Google Scholar] [CrossRef]
- Zaballa, M.E.; Perez-Saez, J.; de Mestral, C.; Pullen, N.; Lamour, J.; Turelli, P.; Raclot, C.; Baysson, H.; Pennacchio, F.; Villers, J.; et al. Seroprevalence of anti-SARS-CoV-2 antibodies and cross-variant neutralization capacity after the Omicron BA.2 wave in Geneva, Switzerland: A population-based study. Lancet Reg. Health–Eur. 2023, 24, 100547. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.N.; Chokkalingam, N.; Reuschel, E.L.; Purwar, M.; Xu, Z.; Gary, E.N.; Kim, K.Y.; Helble, M.; Schultheis, K.; Walters, J.; et al. SARS-CoV-2 Assays to Detect Functional Antibody Responses That Block ACE2 Recognition in Vaccinated Animals and Infected Patients. J. Clin. Microbiol. 2020, 58, 10–1128. [Google Scholar] [CrossRef]
- Byrnes, J.R.; Zhou, X.X.; Lui, I.; Elledge, S.K.; Glasgow, J.E.; Lim, S.A.; Loudermilk, R.P.; Chiu, C.Y.; Wang, T.T.; Wilson, M.R.; et al. Competitive SARS-CoV-2 Serology Reveals Most Antibodies Targeting the Spike Receptor-Binding Domain Compete for ACE2 Binding. mSphere 2020, 5, e00802-20. [Google Scholar] [CrossRef]
- AGES. Österreichische Gesellschaft für Ernährungssicherheit und Gesundheit GmBH—Report of Weekly Detected Variants of Concern in 2022 in Austria. Available online: https://www.ages.at/mensch/krankheit/krankheitserreger-von-a-bis-z/coronavirus#c12438 (accessed on 30 May 2023).
- Koblischke, M.; Traugott, M.T.; Medits, I.; Spitzer, F.S.; Zoufaly, A.; Weseslindtner, L.; Simonitsch, C.; Seitz, T.; Hoepler, W.; Puchhammer-Stöckl, E.; et al. Dynamics of CD4 T Cell and Antibody Responses in COVID-19 Patients With Different Disease Severity. Front. Med. 2020, 7, 592629. [Google Scholar] [CrossRef]
- Graninger, M.; Camp, J.V.; Aberle, S.W.; Traugott, M.T.; Hoepler, W.; Puchhammer-Stöckl, E.; Weseslindtner, L.; Zoufaly, A.; Aberle, J.H.; Stiasny, K. Heterogeneous SARS-CoV-2-Neutralizing Activities After Infection and Vaccination. Front. Immunol. 2022, 13, 888794. [Google Scholar] [CrossRef]
- Buchta, C.; Springer, D.; Jovanovic, J.; Borsodi, C.; Weidner, L.; Sareban, N.; Radler, U.; Müller, M.M.; Griesmacher, A.; Puchhammer-Stöckl, E.; et al. Three rounds of a national external quality assessment reveal a link between disharmonic anti-SARS-CoV-2 antibody quantifications and the infection stage. Clin. Chem. Lab. Med. 2023, 61, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Davis-Gardner, M.E.; Lai, L.; Wali, B.; Samaha, H.; Solis, D.; Lee, M.; Porter-Morrison, A.; Hentenaar, I.T.; Yamamoto, F.; Godbole, S.; et al. mRNA bivalent booster enhances neutralization against BA.2.75.2 and BQ.1.1. bioRxiv 2022. [Google Scholar] [CrossRef]
- Walker, M.R.; Podlekareva, D.; Johnsen, S.; Leerhøy, B.; Fougeroux, C.; Søgaard, M.; Salanti, A.; Ditlev, S.B.; Barfod, L. SARS-CoV-2 RBD-Specific Antibodies Induced Early in the Pandemic by Natural Infection and Vaccination Display Cross-Variant Binding and Inhibition. Viruses 2022, 14, 1861. [Google Scholar] [CrossRef]
- Gruell, H.; Vanshylla, K.; Tober-Lau, P.; Hillus, D.; Schommers, P.; Lehmann, C.; Kurth, F.; Sander, L.E.; Klein, F. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat. Med. 2022, 28, 477–480. [Google Scholar] [CrossRef] [PubMed]
- Quandt, J.; Muik, A.; Salisch, N.; Lui, B.G.; Lutz, S.; Krüger, K.; Wallisch, A.K.; Adams-Quack, P.; Bacher, M.; Finlayson, A.; et al. Omicron BA.1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes. Sci. Immunol. 2022, 7, eabq2427. [Google Scholar] [CrossRef]
- Kaku, C.I.; Bergeron, A.J.; Ahlm, C.; Normark, J.; Sakharkar, M.; Forsell, M.N.E.; Walker, L.M. Recall of preexisting cross-reactive B cell memory after Omicron BA.1 breakthrough infection. Sci. Immunol. 2022, 7, eabq3511. [Google Scholar] [CrossRef] [PubMed]
- Springer, D.N.; Bauer, M.; Medits, I.; Camp, J.V.; Aberle, S.W.; Burtscher, C.; Höltl, E.; Weseslindtner, L.; Stiasny, K.; Aberle, J.H. Bivalent COVID-19 mRNA booster vaccination (BA.1 or BA.4/BA.5) increases neutralization of matched Omicron variants. medRxiv 2023. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Springer, D.N.; Traugott, M.; Reuberger, E.; Kothbauer, K.B.; Borsodi, C.; Nägeli, M.; Oelschlägel, T.; Kelani, H.; Lammel, O.; Deutsch, J.; et al. A Multivariant Surrogate Neutralization Assay Identifies Variant-Specific Neutralizing Antibody Profiles in Primary SARS-CoV-2 Omicron Infection. Diagnostics 2023, 13, 2278. https://doi.org/10.3390/diagnostics13132278
Springer DN, Traugott M, Reuberger E, Kothbauer KB, Borsodi C, Nägeli M, Oelschlägel T, Kelani H, Lammel O, Deutsch J, et al. A Multivariant Surrogate Neutralization Assay Identifies Variant-Specific Neutralizing Antibody Profiles in Primary SARS-CoV-2 Omicron Infection. Diagnostics. 2023; 13(13):2278. https://doi.org/10.3390/diagnostics13132278
Chicago/Turabian StyleSpringer, David Niklas, Marianna Traugott, Elisabeth Reuberger, Klaus Benjamin Kothbauer, Christian Borsodi, Michelle Nägeli, Theresa Oelschlägel, Hasan Kelani, Oliver Lammel, Josef Deutsch, and et al. 2023. "A Multivariant Surrogate Neutralization Assay Identifies Variant-Specific Neutralizing Antibody Profiles in Primary SARS-CoV-2 Omicron Infection" Diagnostics 13, no. 13: 2278. https://doi.org/10.3390/diagnostics13132278
APA StyleSpringer, D. N., Traugott, M., Reuberger, E., Kothbauer, K. B., Borsodi, C., Nägeli, M., Oelschlägel, T., Kelani, H., Lammel, O., Deutsch, J., Puchhammer-Stöckl, E., Höltl, E., Aberle, J. H., Stiasny, K., & Weseslindtner, L. (2023). A Multivariant Surrogate Neutralization Assay Identifies Variant-Specific Neutralizing Antibody Profiles in Primary SARS-CoV-2 Omicron Infection. Diagnostics, 13(13), 2278. https://doi.org/10.3390/diagnostics13132278




