Accuracy of a Smartphone-Based Artificial Intelligence Application for Classification of Melanomas, Melanocytic Nevi, and Seborrheic Keratoses
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
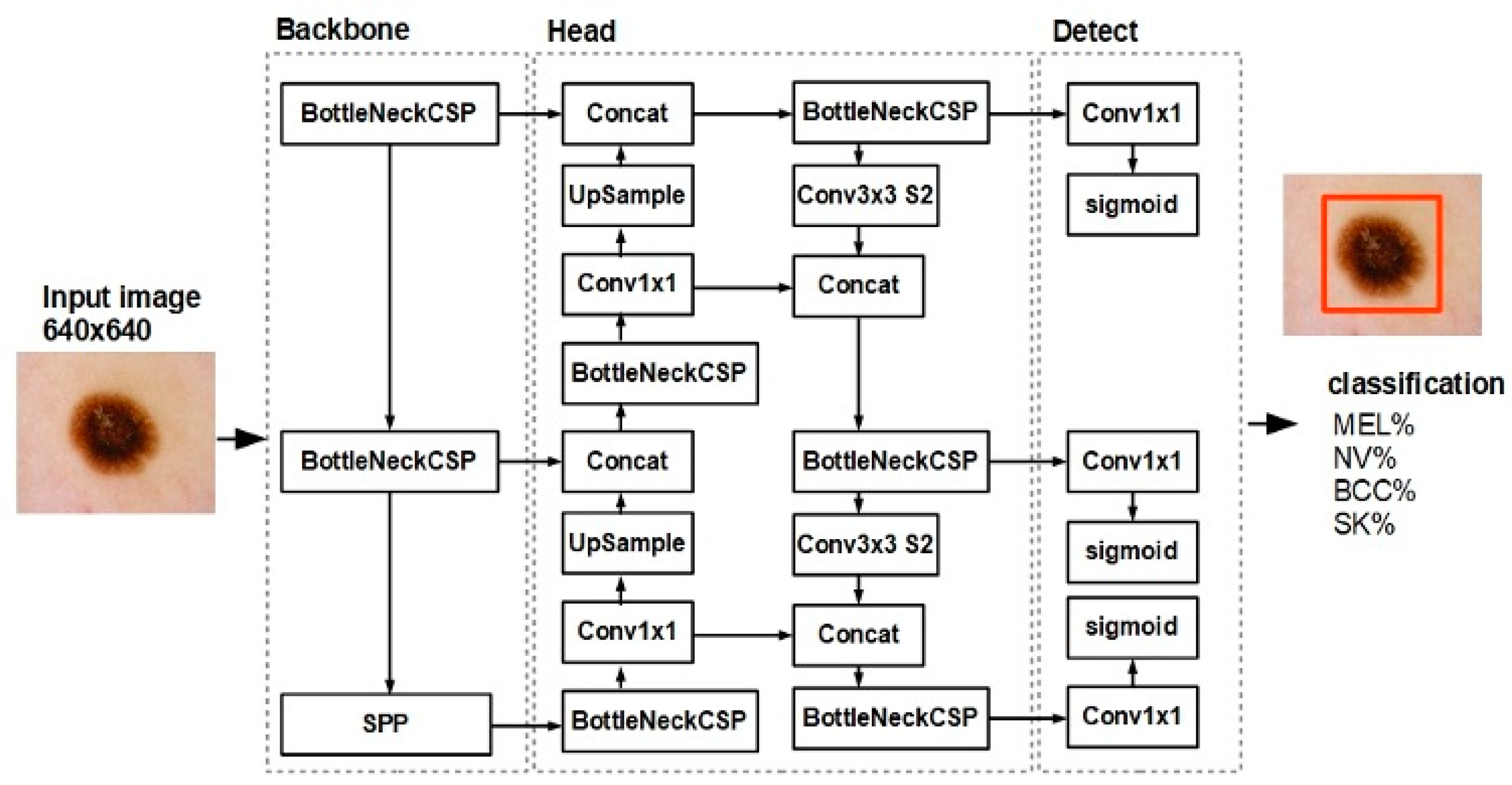
2.2. Skin Lesion Classification Model and Dataset
2.3. Test Dataset
2.4. Statistical Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arnold, M.; Singh, D.; Laversanne, M.; Vignat, J.; Vaccarella, S.; Meheus, F.; Cust, A.E.; de Vries, E.; Whiteman, D.C.; Bray, F. Global burden of cutaneous melanoma in 2020 and projections to 2040. JAMA Dermatol. 2022, 158, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Garbe, C.; Amaral, T.; Peris, K.; Hauschild, A.; Arenberger, P.; Basset-Seguin, N.; Bastholt, L.; Bataille, V.; del Marmol, V.; Dréno, B.; et al. European consensus-based interdisciplinary guideline for melanoma. Part 1: Diagnostics: Update 2022. Eur. J. Cancer 2022, 170, 236–255. [Google Scholar] [CrossRef]
- González-Cruz, C.; Descalzo, M.Á.; Arias-Santiago, S.; Molina-Leyva, A.; Gilaberte, Y.; Fernández-Crehuet, P.; Husein-ElAhmed, H.; Viera-Ramírez, A.; Fernández-Peñas, P.; Taberner, R.; et al. Proportion of Potentially Avoidable Referrals From Primary Care to Dermatologists for Cystic Lesions or Benign Neoplasms in Spain: Analysis of Data From the DIADERM Study. Actas Dermo Sifiliográficas 2019, 110, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.B.; Alfonso, S.V.; Hernandez, N.; Fernández, M.I. Examining the factors associated with past and present dermoscopy use among family physicians. Dermatol. Pract. Concept. 2017, 7, 63–70. [Google Scholar] [CrossRef]
- Westerhoff, K.; McCarthy, W.H.; Menzies, S.W. Increase in the sensitivity for melanoma diagnosis by primary care physicians using skin surface microscopy. Br. J. Dermatol. 2000, 143, 1016–1020. [Google Scholar] [CrossRef]
- Fee, J.A.; McGrady, F.P.; Hart, N.D. Dermoscopy use in primary care: A qualitative study with general practitioners. BMC Prim. Care 2022, 23, 47. [Google Scholar] [CrossRef] [PubMed]
- Haggenmüller, S.; Maron, R.C.; Hekler, A.; Utikal, J.S.; Barata, C.; Barnhill, R.L.; Beltraminelli, H.; Berking, C.; Betz-Stablein, B.; Blum, A.; et al. Skin cancer classification via convolutional neural networks: Systematic review of studies involving human experts. Eur. J. Cancer 2021, 156, 202–216. [Google Scholar] [CrossRef]
- Tschandl, P.; Rinner, C.; Apalla, Z.; Argenziano, G.; Codella, N.; Halpern, A.; Janda, M.; Lallas, A.; Longo, C.; Malvehy, J.; et al. Human-computer collaboration for skin cancer recognition. Nat. Med. 2020, 26, 1229–1234. [Google Scholar] [CrossRef]
- Bechelli, S.; Delhommelle, J. Machine learning and deep learning algorithms for skin cancer classification from dermoscopic images. Bioengineering 2022, 9, 97. [Google Scholar] [CrossRef]
- Hekler, A.; Kather, J.N.; Krieghoff-Henning, E.; Utikal, J.S.; Meier, F.; Gellrich, F.F.; Upmeier Zu Belzen, J.; French, L.; Schlager, J.G.; Ghoreschi, K.; et al. Effects of label noise on deep learning-based skin cancer classification. Front. Med. 2020, 7, 177. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, real-time object detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar] [CrossRef]
- Guo, L.; Yang, Y.; Ding, H.; Zheng, H.; Yang, H.; Xie, J.; Li, Y.; Lin, T.; Ge, Y. A deep learning-based hybrid artificial intelligence model for the detection and severity assessment of vitiligo lesions. Ann. Transl. Med. 2022, 10, 590. [Google Scholar] [CrossRef]
- Ünver, H.M.; Ayan, E. Skin lesion segmentation in dermoscopic images with combination of YOLO and GrabCut algorithm. Diagnostics 2019, 9, 72. [Google Scholar] [CrossRef]
- Banerjee, S.; Singh, S.K.; Chakraborty, A.; Das, A.; Bag, R. Melanoma diagnosis using deep learning and fuzzy logic. Diagnostics 2020, 10, 577. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Holland-Letz, T.; et al. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur. J. Cancer 2019, 113, 47–54. [Google Scholar] [CrossRef]
- Marchetti, M.A.; Codella, N.C.F.; Dusza, S.W.; Gutman, D.A.; Helba, B.; Kalloo, A.; Mishra, N.; Carrera, C.; Celebi, M.E.; DeFazio, J.L.; et al. Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J. Am. Acad. Dermatol. 2018, 78, 270–277. [Google Scholar] [CrossRef]
- Marchetti, M.A.; Liopyris, K.; Dusza, S.W.; Codella, N.C.F.; Gutman, D.A.; Helba, B.; Kalloo, A.; Halpern, A.C. Computer algorithms show potential for improving dermatologists’ accuracy to diagnose cutaneous melanoma: Results of the International Skin Imaging Collaboration 2017. J. Am. Acad. Dermatol. 2020, 82, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.K.; Blum, A.; Kommoss, K.; Enk, A.; Toberer, F.; Rosenberger, A.; Haenssle, H.A. Assessment of diagnostic performance of dermatologists cooperating with a convolutional neural network in a prospective clinical study: Human with machine. JAMA Dermatol. 2023, e230905. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Combalia, M.; Codella, N.; Rotemberg, V.; Carrera, C.; Dusza, S.; Gutman, D.; Helba, B.; Kittler, H.; Kurtansky, N.R.; Liopyris, K.; et al. Validation of artificial intelligence prediction models for skin cancer diagnosis using dermoscopy images: The 2019 International Skin Imaging Collaboration Grand Challenge. Lancet Digit. Health 2022, 4, e330–e339. [Google Scholar] [CrossRef] [PubMed]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Haenssle, H.A.; Fink, C.; Toberer, F.; Winkler, J.; Stolz, W.; Deinlein, T.; Hofmann-Wellenhof, R.; Lallas, A.; Emmert, S.; Buhl, T.; et al. Man against machine reloaded: Performance of a market-approved convolutional neural network in classifying a broad spectrum of skin lesions in comparison with 96 dermatologists working under less artificial conditions. Ann. Oncol. 2020, 31, 137–143. [Google Scholar] [CrossRef]
- Haenssle, H.A.; Winkler, J.K.; Fink, C.; Toberer, F.; Enk, A.; Stolz, W.; Deinlein, T.; Hofmann-Wellenhof, R.; Kittler, H.; Tschandl, P.; et al. Skin lesions of face and scalp—Classification by a market-approved convolutional neural network in comparison with 64 dermatologists. Eur. J. Cancer 2021, 144, 192–199. [Google Scholar] [CrossRef]
- Sies, K.; Winkler, J.K.; Fink, C.; Bardehle, F.; Toberer, F.; Kommoss, F.K.F.; Buhl, T.; Enk, A.; Rosenberger, A.; Haenssle, H.A. Dark corner artefact and diagnostic performance of a market-approved neural network for skin cancer classification. J. Dtsch. Dermatol. Ges. 2021, 19, 842–850. [Google Scholar] [CrossRef]
- Winkler, J.K.; Sies, K.; Fink, C.; Toberer, F.; Enk, A.; Abassi, M.S.; Fuchs, T.; Blum, A.; Stolz, W.; Coras-Stepanek, B.; et al. Collective human intelligence outperforms artificial intelligence in a skin lesion classification task. J. Dtsch. Dermatol. Ges. 2021, 19, 1178–1184. [Google Scholar] [CrossRef]
- Veronese, F.; Branciforti, F.; Zavattaro, E.; Tarantino, V.; Romano, V.; Meiburger, K.M.; Salvi, M.; Seoni, S.; Savoia, P. The role in teledermoscopy of an inexpensive and easy-to-use smartphone device for the classification of three types of skin lesions using convolutional neural networks. Diagnostics 2021, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Udrea, A.; Mitra, G.D.; Costea, D.; Noels, E.C.; Wakkee, M.; Siegel, D.M.; de Carvalho, T.M.; Nijsten, T.E.C. Accuracy of a smartphone application for triage of skin lesions based on machine learning algorithms. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Sangers, T.; Reeder, S.; van der Vet, S.; Jhingoer, S.; Mooyaart, A.; Siegel, D.M.; Nijsten, T.; Wakkee, M. Validation of a market-approved artificial intelligence mobile health app for skin cancer screening: A prospective multicenter diagnostic accuracy study. Dermatology 2022, 238, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Rezatofighi, H.; Tsoi, N.; Gwak, J.; Sadeghian, A.; Reid, I.; Savarese, S. Generalized intersection over union: A metric and a loss for bounding box regression. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 658–666. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Combalia, M.; Codella, N.C.; Rotemberg, V.; Helba, B.; Vilaplana, V.; Reiter, O.; Carrera, C.; Barreiro, A.; Halpern, A.C.; Puig, S.; et al. BCN20000: Dermoscopic lesions in the wild. arXiv 2019, preprint. arXiv:1908.02288. [Google Scholar]
- Rotemberg, V.; Kurtansky, N.; Betz-Stablein, B.; Caffery, L.; Chousakos, E.; Codella, N.; Combalia, M.; Dusza, S.; Guitera, P.; Gutman, D.; et al. A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci. Data 2021, 8, 34. [Google Scholar] [CrossRef]
- Codella, N.; Rotemberg, V.; Tschandl, P.; Celebi, M.E.; Dusza, S.; Gutman, D.; Helba, B.; Kalloo, A.; Liopyris, K.; Marchetti, M.; et al. Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2019, arXiv:1902.03368. [Google Scholar]
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 168–172. [Google Scholar] [CrossRef]
- Gutman, D.; Codella, N.C.; Celebi, E.; Helba, B.; Marchetti, M.; Mishra, N.; Halpern, A. Skin lesion analysis toward melanoma detection: A challenge at the International Symposium on Biomedical Imaging (ISBI) 2016, hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2016, arXiv:1605.01397. [Google Scholar]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Gülseren, D.; Hofmann-Wellenhof, R. Evaluation of dermoscopic criteria for seborrheic keratosis on non-polarized versus polarized dermoscopy. Skin Res. Technol. 2019, 25, 801–804. [Google Scholar] [CrossRef]
- Dika, E.; Lambertini, M.; Patrizi, A.; Misciali, C.; Scarfì, F.; Pellacani, G.; Mandel, V.D.; Di Tullio, F.; Stanganelli, I.; Chester, J.; et al. Folliculotropism in head and neck lentigo maligna and lentigo maligna melanoma. J. Dtsch. Dermatol. Ges. 2021, 19, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Bartenstein, D.W.; Kelleher, C.M.; Friedmann, A.M.; Duncan, L.M.; Tsao, H.; Sober, A.J.; Hawryluk, E.B. Contrasting features of childhood and adolescent melanomas. Pediatr. Dermatol. 2018, 35, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Marghoob, N.G.; Liopyris, K.; Jaimes, N. Dermoscopy: A review of the structures that facilitate melanoma detection. J. Osteopath. Med. 2019, 119, 380–390. [Google Scholar] [CrossRef]
- Janowska, A.; Oranges, T.; Iannone, M.; Romanelli, M.; Dini, V. Seborrheic keratosis-like melanoma: A diagnostic challenge. Melanoma Res. 2021, 31, 407–412. [Google Scholar] [CrossRef]
- Zakaria, A.; Maurer, T.; Su, G.; Amerson, E. Impact of teledermatology on the accessibility and efficiency of dermatology care in an urban safety-net hospital: A pre-post analysis. J. Am. Acad. Dermatol. 2019, 81, 1446–1452. [Google Scholar] [CrossRef]
- Zakaria, A.; Miclau, T.A.; Maurer, T.; Leslie, K.S.; Amerson, E. Cost minimization analysis of a teledermatology triage system in a managed care setting. JAMA Dermatol. 2021, 157, 52–58. [Google Scholar] [CrossRef]


| Characteristics | Number | Percent |
|---|---|---|
| Patients | 100 | |
| Mean age (SD), years | 55.4 (±15.8) | |
| Sex | ||
| Male | 54 | 54 |
| Female | 46 | 46 |
| Assessed lesions | 100 | |
| Image datasets * | ||
| HAM 10000 | 38 | 38 |
| MSK-1 | 23 | 23 |
| MSK-2 | 14 | 14 |
| MSK-3 | 3 | 3 |
| MSK-4 | 16 | 16 |
| MSK-5 | 5 | 5 |
| UDA2 | 1 | 1 |
| Lesion classes | ||
| Melanoma | 32 | 32 |
| Melanocytic nevus | 35 | 35 |
| Seborrheic keratosis | 33 | 33 |
| Localization | ||
| Head and neck | 4 | 4 |
| Upper extremities | 19 | 19 |
| Lower extremities | 20 | 20 |
| Anterior torso | 16 | 16 |
| Lateral torso | 2 | 2 |
| Posterior torso | 26 | 26 |
| Not specified | 6 | 6 |
| Rater Level | Melanoma | Melanocytic Nevus | Seborrheic Keratosis | |||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
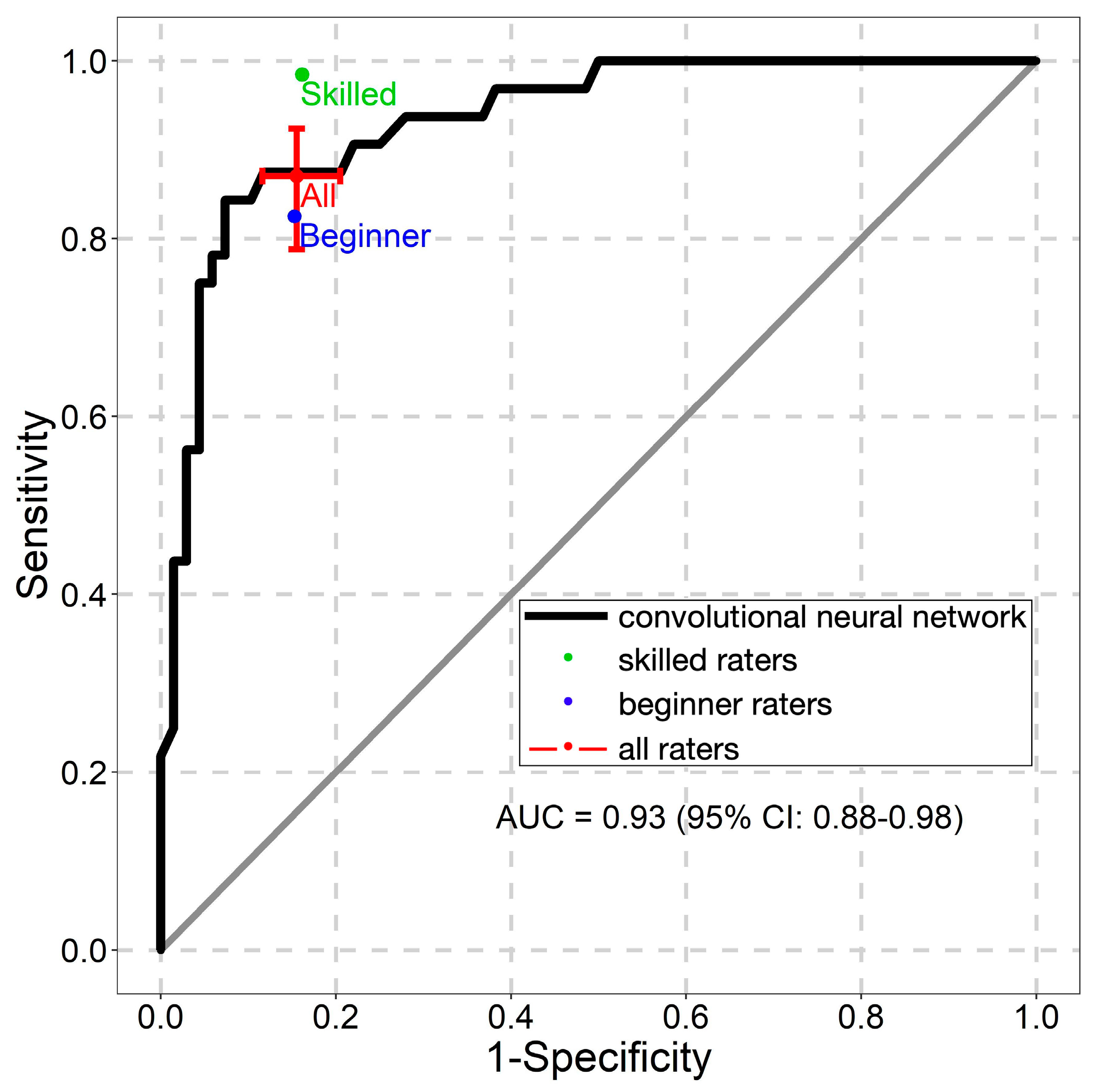
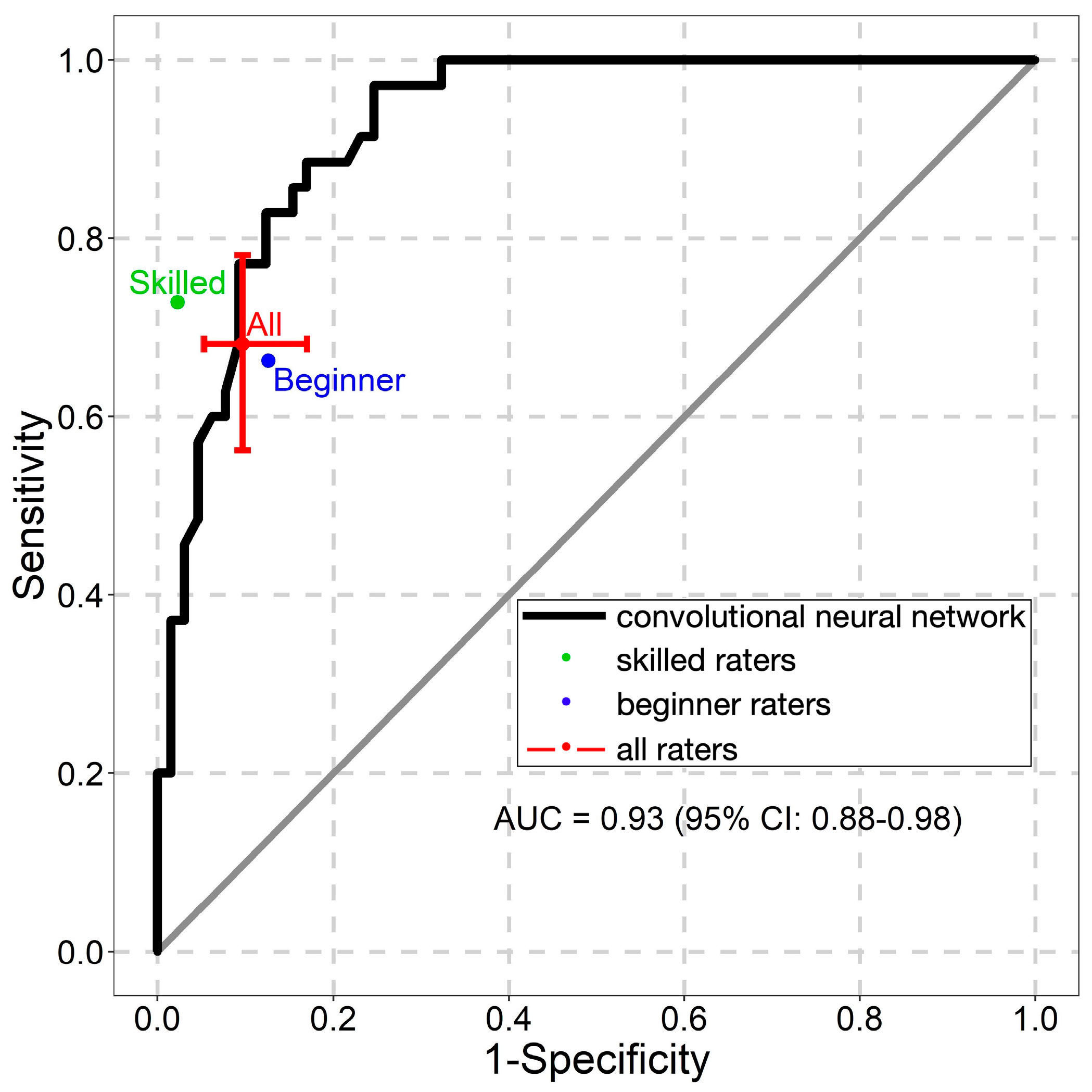
| Skilled | 0.98 (0.92–1.00) | 0.84 (0.51–0.96) | 0.73 (0.33–0.94) | 0.98 (0.88–1.00) | 0.89 (0.58–0.98) | 0.99 (0.92–1.00) |
| Beginners | 0.83 (0.77–0.87) | 0.85 (0.77–0.90) | 0.66 (0.57–0.74) | 0.87 (0.80–0.92) | 0.73 (0.52–0.87) | 0.89 (0.83–0.93) |
| All raters | 0.87 (0.79–0.92) | 0.84 (0.76–0.90) | 0.68 (0.56–0.78) | 0.90 (0.83–0.95) | 0.78 (0.60–0.89) | 0.91 (0.85–0.95) |
| NNM | 0.88 (0.71–0.96) | 0.87 (0.76–0.94) | 0.77 (0.60–0.90) | 0.91 (0.81–0.97) | 0.52 (0.34–0.69) | 0.93 (0.83–0.98) |
| Rater Level | Melanoma | Melanocytic Nevus | Seborrheic Keratosis | |||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| Skilled | −0.11 (−0.27, 0.00) | 0.03 (−0.13, 0.37) | 0.04 (−0.23, 0.46) | −0.07 (−0.17, 0.04) | −0.38 (−0.58, −0.02) | −0.06 (−0.15, 0.02) |
| Beginners | 0.05 (−0.12, 0.15) | 0.02 (−0.10, 0.13) | 0.11 (−0.08, 0.26) | 0.03 (−0.08, 0.13) | −0.22 (−0.45, 0.06) | 0.04 (−0.06, 0.12) |
| All raters | 0.00 (−0.17, 0.12) | 0.02 (−0.10, 0.13) | 0.09 (−0.11, 0.26) | 0.00 (−0.10, 0.10) | −0.26 (−0.48, −0.01) | 0.01 (−0.09, 0.09) |
| Rater Level | Melanoma | Melanocytic Nevus | Seborrheic Keratosis | |||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| Beginner 1 | 0.91 (0.75–0.98) | 0.93 (0.84–0.98) | 0.57 (0.39–0.74) | 0.97 (0.89–1.00) | 0.97 (0.84–1.00) | 0.82 (0.71–0.90) |
| Beginner 2 | 0.84 (0.67–0.95) | 0.84 (0.73–0.92) | 0.80 (0.63–0.92) | 0.82 (0.70–0.90) | 0.58 (0.39–0.75) | 0.96 (0.87–0.99) |
| Beginner 3 | 0.78 (0.60–0.91) | 0.90 (0.80–0.96) | 0.60 (0.42–0.76) | 0.92 (0.83–0.97) | 0.97 (0.84–1.00) | 0.85 (0.74–0.93) |
| Beginner 4 | 0.78 (0.60–0.91) | 0.85 (0.75–0.93) | 0.74 (0.57–0.88) | 0.86 (0.75–0.93) | 0.64 (0.45–0.80) | 0.87 (0.76–0.94) |
| Beginner 5 | 0.81 (0.64–0.93) | 0.72 (0.60–0.82) | 0.60 (0.42–0.76) | 0.80 (0.68–0.89) | 0.52 (0.34–0.69) | 0.94 (0.85–0.98) |
| Skilled 1 | 0.97 (0.84–1.00) | 0.71 (0.58–0.81) | 0.51 (0.34–0.69) | 0.95 (0.87–0.99) | 0.79 (0.61–0.91) | 0.97 (0.90–1.00) |
| Skilled 2 | 1.00 (0.89–0.96) | 0.97 (0.90–1.00) | 0.94 (0.81–0.99) | 1.00 (0.94–1.00) | 1.00 (0.89–1.00) | 1.00 (0.95–1.00) |
| Rater Level | Fleiss Kappa | |||
|---|---|---|---|---|
| Melanoma | Melanocytic Nevus | Seborrheic Keratosis | All Lesion Classes | |
| Skilled | 0.57 (0.37–0.77) | 0.49 (0.30–0.69) | 0.79 (0.59–0.98) | 0.62 (0.48–0.76) |
| Beginners | 0.56 (0.50–0.62) | 0.43 (0.37–0.50) | 0.50 (0.44–0.56) | 0.50 (0.46–0.54) |
| All raters | 0.56 (0.51–0.60) | 0.46 (0.42–0.50) | 0.56 (0.52–0.61) | 0.53 (0.50–0.56) |
| Study | Melanoma | Melanocytic Nevus | Seborrheic Keratosis | |||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| NNM | 0.88 (0.71–0.96) | 0.87 (0.76–0.94) | 0.77 (0.60–0.90) | 0.91 (0.81–0.97) | 0.52 (0.34–0.69) | 0.93 (0.83–0.98) |
| Veronese et al. [18] | 0.84 | 0.82 | N/A | N/A | N/A | N/A |
| Udrea et al. [19] * | 0.93 (0.88–0.96) | N/A | N/A | 0.78 (0.77–0.79) | N/A | 0.78 (0.77–0.79) |
| Sangers et al. [20] * | 0.82 (0.59–0.95) | 0.73 (0.66, 0.80) | N/A | 0.80 (0.76–0.84) | N/A | 0.80 (0.76–0.84) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liutkus, J.; Kriukas, A.; Stragyte, D.; Mazeika, E.; Raudonis, V.; Galetzka, W.; Stang, A.; Valiukeviciene, S. Accuracy of a Smartphone-Based Artificial Intelligence Application for Classification of Melanomas, Melanocytic Nevi, and Seborrheic Keratoses. Diagnostics 2023, 13, 2139. https://doi.org/10.3390/diagnostics13132139
Liutkus J, Kriukas A, Stragyte D, Mazeika E, Raudonis V, Galetzka W, Stang A, Valiukeviciene S. Accuracy of a Smartphone-Based Artificial Intelligence Application for Classification of Melanomas, Melanocytic Nevi, and Seborrheic Keratoses. Diagnostics. 2023; 13(13):2139. https://doi.org/10.3390/diagnostics13132139
Chicago/Turabian StyleLiutkus, Jokubas, Arturas Kriukas, Dominyka Stragyte, Erikas Mazeika, Vidas Raudonis, Wolfgang Galetzka, Andreas Stang, and Skaidra Valiukeviciene. 2023. "Accuracy of a Smartphone-Based Artificial Intelligence Application for Classification of Melanomas, Melanocytic Nevi, and Seborrheic Keratoses" Diagnostics 13, no. 13: 2139. https://doi.org/10.3390/diagnostics13132139
APA StyleLiutkus, J., Kriukas, A., Stragyte, D., Mazeika, E., Raudonis, V., Galetzka, W., Stang, A., & Valiukeviciene, S. (2023). Accuracy of a Smartphone-Based Artificial Intelligence Application for Classification of Melanomas, Melanocytic Nevi, and Seborrheic Keratoses. Diagnostics, 13(13), 2139. https://doi.org/10.3390/diagnostics13132139







