Constructing the Schizophrenia Recognition Method Employing GLCM Features from Multiple Brain Regions and Machine Learning Techniques
Abstract
1. Introduction
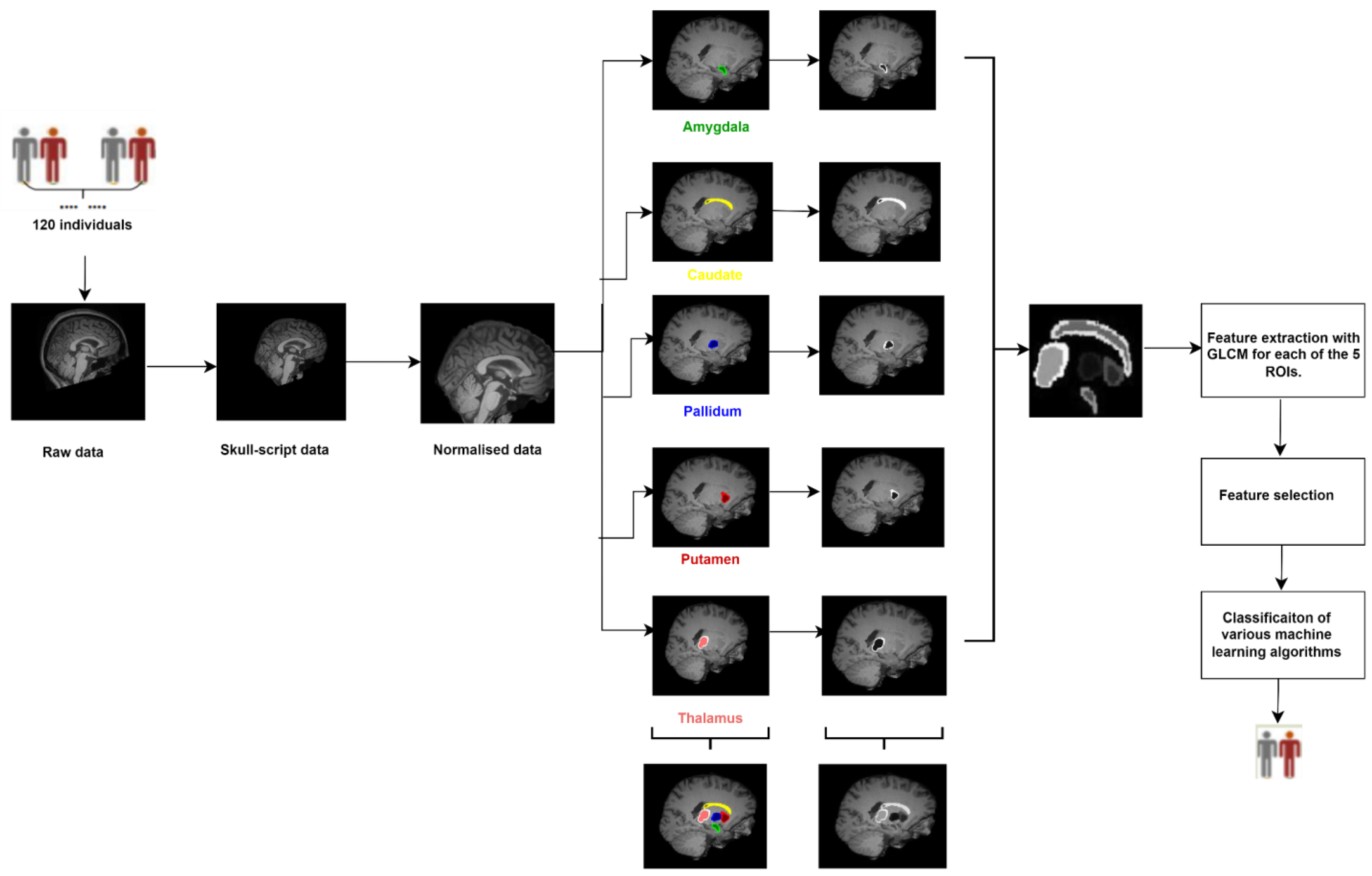
- This study focuses on subcortical structures such as the amygdala, caudate, pallidum, putamen, and thalamus. While these regions have been studied in relation to schizophrenia before, this study may offer new insights or findings specific to these regions.
- Unlike the datasets used in previous studies that targeted specific brain regions mentioned in the literature, the COBRE dataset was utilized providing an opportunity to examine the relationship between subcortical structures and schizophrenia using a specific dataset.
- For the first time, feature extraction was performed using the GLCM technique on the subcortical focused brain regions.
- High classification accuracies were achieved using various classification algorithms in three conditions: right hemisphere, left hemisphere, and bilateral hemispheres, based on the GLCM features of these subcortical regions.
- The detectability of hidden patterns in structural MR images belonging to five focused regions outside the whole brain using GLCM features was analyzed.
2. Materials and Methods
2.1. Dataset
2.2. Structural MRI Data Acquisition
2.3. Image Preprocessing and Segmentation
2.4. Feature Extraction
2.5. Feature Selection
2.6. Classification
3. Results
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jablensky, A. The diagnostic concept of schizophrenia: Its history, evolution, and future prospects. Dialogues Clin. Neurosci. 2010, 12, 271–287. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Chon, M.-W.; Kim, H.; Rathi, Y.; Bouix, S.; Shenton, M.E.; Kubicki, M. Diagnostic value of structural and diffusion imaging measures in schizophrenia. NeuroImage. Clin. 2018, 18, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Ettinger, U.; Meyhöfer, I.; Steffens, M.; Wagner, M.; Koutsouleris, N. Genetics, cognition, and neurobiology of schizotypal personality: A review of the overlap with schizophrenia. Front. Psychiatry 2014, 5, 18. [Google Scholar] [CrossRef]
- Lee, D.-K.; Lee, H.; Park, K.; Joh, E.; Kim, C.-E.; Ryu, S. Common gray and white matter abnormalities in schizophrenia and bipolar disorder. PLoS ONE 2020, 15, e0232826. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Sun, H.; Shi, S.; Jiang, D.; Tao, B.; Zhao, Y.; Zhang, W.; Gong, Q.; Sweeney, J.A.; Lui, S. White Matter Abnormalities in Never-Treated Patients with Long-Term Schizophrenia. Am. J. Psychiatry 2018, 175, 1129–1136. [Google Scholar] [CrossRef]
- Zhang, W.; Deng, W.; Yao, L.; Xiao, Y.; Li, F.; Liu, J.; Sweeney, J.A.; Lui, S.; Gong, Q. Brain Structural Abnormalities in a Group of Never-Medicated Patients with Long-Term Schizophrenia. Am. J. Psychiatry 2015, 172, 995–1003. [Google Scholar] [CrossRef]
- Svancer, P.; Spaniel, F. Brain ventricular volume changes in schizophrenia. A narrative review. Neurosci. Lett. 2021, 759, 136065. [Google Scholar] [CrossRef]
- Madre, M.; Canales-Rodríguez, E.J.; Fuentes-Claramonte, P.; Alonso-Lana, S.; Salgado-Pineda, P.; Guerrero-Pedraza, A.; Moro, N.; Bosque, C.; Gomar, J.J.; Ortíz-Gil, J.; et al. Structural abnormality in schizophrenia versus bipolar disorder: A whole brain cortical thickness, surface area, volume and gyrification analyses. NeuroImage. Clin. 2020, 25, 102131. [Google Scholar] [CrossRef]
- Raucher-Chéné, D.; Lavigne, K.M.; Makowski, C.; Lepage, M. Altered Surface Area Covariance in the Mentalizing Network in Schizophrenia: Insight Into Theory of Mind Processing. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2022, 7, 706–715. [Google Scholar] [CrossRef]
- Habets, P.; Marcelis, M.; Gronenschild, E.; Drukker, M.; van Os, J. Reduced cortical thickness as an outcome of differential sensitivity to environmental risks in schizophrenia. Biol. Psychiatry 2011, 69, 487–494. [Google Scholar] [CrossRef]
- Goldman, A.L.; Pezawas, L.; Mattay, V.S.; Fischl, B.; Verchinski, B.A.; Chen, Q.; Weinberger, D.R.; Meyer-Lindenberg, A. Widespread reductions of cortical thickness in schizophrenia and spectrum disorders and evidence of heritability. Arch. Gen. Psychiatry 2009, 66, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.; Falkai, P.; Papiol, S. Neurodevelopmental disturbances in schizophrenia: Evidence from genetic and environmental factors. J. Neural Transm. 2023, 130, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Owen, M.J.; O’Donovan, M.C.; Thapar, A.; Craddock, N. Neurodevelopmental hypothesis of schizophrenia. Br. J. Psychiatry 2011, 198, 173–175. [Google Scholar] [CrossRef] [PubMed]
- Nour, M.M.; Howes, O.D. Interpreting the neurodevelopmental hypothesis of schizophrenia in the context of normal brain development and ageing. Proc. Natl. Acad. Sci. USA 2015, 112, E2745. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.H.; Antoniades, M.; Schnack, H.G.; Kahn, R.S.; Frangou, S. Brain age prediction in schizophrenia: Does the choice of machine learning algorithm matter? Psychiatry Res. Neuroimaging 2021, 310, 111270. [Google Scholar] [CrossRef]
- Avram, M.; Grothe, M.J.; Meinhold, L.; Leucht, C.; Leucht, S.; Borgwardt, S.; Brandl, F.; Sorg, C. Lower cholinergic basal forebrain volumes link with cognitive difficulties in schizophrenia. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2021, 46, 2320–2329. [Google Scholar] [CrossRef]
- Alkan, E.; Davies, G.; Evans, S.L. Cognitive impairment in schizophrenia: Relationships with cortical thickness in fronto-temporal regions, and dissociability from symptom severity. NPJ Schizophr. 2021, 7, 20. [Google Scholar] [CrossRef]
- Oh, J.; Oh, B.-L.; Lee, K.-U.; Chae, J.-H.; Yun, K. Identifying Schizophrenia Using Structural MRI with a Deep Learning Algorithm. Front. Psychiatry 2020, 11, 16. [Google Scholar] [CrossRef]
- Cabral, C.; Kambeitz-Ilankovic, L.; Kambeitz, J.; Calhoun, V.D.; Dwyer, D.B.; von Saldern, S.; Urquijo, M.F.; Falkai, P.; Koutsouleris, N. Classifying Schizophrenia Using Multimodal Multivariate Pattern Recognition Analysis: Evaluating the Impact of Individual Clinical Profiles on the Neurodiagnostic Performance. Schizophr. Bull. 2016, 42 (Suppl. 1), S110–S117. [Google Scholar] [CrossRef]
- Lei, D.; Pinaya, W.H.L.; Young, J.; van Amelsvoort, T.; Marcelis, M.; Donohoe, G.; Mothersill, D.O.; Corvin, A.; Vieira, S.; Huang, X.; et al. Integrating machining learning and multimodal neuroimaging to detect schizophrenia at the level of the individual. Hum. Brain Mapp. 2020, 41, 1119–1135. [Google Scholar] [CrossRef]
- Okada, N.; Fukunaga, M.; Yamashita, F.; Koshiyama, D.; Yamamori, H.; Ohi, K.; Yasuda, Y.; Fujimoto, M.; Watanabe, Y.; Yahata, N.; et al. Abnormal asymmetries in subcortical brain volume in schizophrenia. Mol. Psychiatry 2016, 21, 1460–1466. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Xiang, H.; Tan, S.; Yang, F.; Fan, H.; Guo, H.; Kochunov, P.; Wang, Z.; Hong, L.E.; Tan, Y. Subcortical structures and cognitive dysfunction in first episode schizophrenia. Psychiatry Res. Neuroimaging 2019, 286, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M. Fast Robust Automated Brain Extraction. Hum. Brain Mapp. 2002, 17, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Patenaude, B.; Smith, S.M.; Kennedy, D.N.; Jenkinson, M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage 2011, 56, 907–922. [Google Scholar] [CrossRef]
- Al-Awadi, J.; Aljobouri, H.; Hasan, A. MRI Brain Scans Classification Using Extreme Learning Machine on LBP and GLCM. Int. J. Online Biomed. Eng. 2023, 19, 134–149. [Google Scholar] [CrossRef]
- Gade, A.; Vyavahare, A. Feature Extraction using GLCM for Dietary Assessment Application. Int. J. Multimed. Image Process. 2018, 8, 409–413. [Google Scholar] [CrossRef]
- Mohanaiah, P.; Sathyanarayana, P.; GuruKumar, L. Image Texture Feature Extraction Using GLCM Approach. Int. J. Sci. Res. Publ. 2013, 3, 1–5. [Google Scholar]
- Brynolfsson, P.; Nilsson, D.; Torheim, T.; Asklund, T.; Karlsson, C.T.; Trygg, J.; Nyholm, T.; Garpebring, A. Haralick texture features from apparent diffusion coefficient (ADC) MRI images depend on imaging and pre-processing parameters. Sci. Rep. 2017, 7, 4041. [Google Scholar] [CrossRef]
- Sumaiya Thaseen, I.; Aswani Kumar, C. Intrusion detection model using fusion of chi-square feature selection and multi class SVM. J. King Saud Univ. Comput. Inf. Sci. 2017, 29, 462–472. [Google Scholar] [CrossRef]
- Schapire, R.E. Explaining AdaBoost. In Empirical Inference: Festschrift in Honor of Vladimir N. Vapnik; Schölkopf, B., Luo, Z., Vovk, V., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 37–52. [Google Scholar]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Leo, B. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar]
- Yang, J.; Yu, H.; Kunz, W. An Efficient LDA Algorithm for Face Recognition. In Proceedings of the Sixth International Conference on Control, Automation, Robotics and Vision, Singapore, 5–8 December 2000. [Google Scholar]
- Chen, C.-H.; Huang, W.-T.; Tan, T.-H.; Chang, C.-C.; Chang, Y.-J. Using K-Nearest Neighbor Classification to Diagnose Abnormal Lung Sounds. Sensors 2015, 15, 13132–13158. [Google Scholar] [CrossRef] [PubMed]
- John, G.H.; Langley, P. Estimating continuous distributions in Bayesian classifiers. In Proceedings of the Eleventh Conference and Uncertainty in Artificial İntelligence, Montréal, QC, Canada, 18–20 August 1995; pp. 338–345. [Google Scholar]
- Wang, Y.; Shi, J.; Xiao, H. Using Regularized Multi-Task Learning for Schizophrenia MRI Data Classification. J. Integr. Neurosci. 2022, 21, 119. [Google Scholar] [CrossRef] [PubMed]
- Manohar, L.; Ganesan, K. Diagnosis of Schizophrenia Disorder in MR Brain Images Using Multi-objective BPSO Based Feature Selection with Fuzzy SVM. J. Med. Biol. Eng. 2018, 38, 917–932. [Google Scholar] [CrossRef]
- Schnack, H.G.; Nieuwenhuis, M.; van Haren, N.E.M.; Abramovic, L.; Scheewe, T.W.; Brouwer, R.M.; Hulshoff Pol, H.E.; Kahn, R.S. Can structural MRI aid in clinical classification? A machine learning study in two independent samples of patients with schizophrenia, bipolar disorder and healthy subjects. Neuroimage 2014, 84, 299–306. [Google Scholar] [CrossRef]
- Janousova, E.; Schwarz, D.; Kasparek, T. Combining various types of classifiers and features extracted from magnetic resonance imaging data in schizophrenia recognition. Psychiatry Res. Neuroimaging 2015, 232, 237–249. [Google Scholar] [CrossRef]
- Lu, X.; Yang, Y.; Wu, F.; Gao, M.; Xu, Y.; Zhang, Y.; Yao, Y.; Du, X.; Li, C.; Wu, L.; et al. Discriminative analysis of schizophrenia using support vector machine and recursive feature elimination on structural MRI images. Medicine 2016, 95, e3973. [Google Scholar] [CrossRef]
- Liu, J.; Li, M.; Pan, Y.; Wu, F.-X.; Chen, X.; Wang, J. Classification of Schizophrenia Based on Individual Hierarchical Brain Networks Constructed From Structural MRI Images. IEEE Trans. Nanobioscience 2017, 16, 600–608. [Google Scholar] [CrossRef]
- Zhuang, H.; Liu, R.; Wu, C.; Meng, Z.; Wang, D.; Liu, D.; Liu, M.; Li, Y. Multimodal classification of drug-naïve first-episode schizophrenia combining anatomical, diffusion and resting state functional resonance imaging. Neurosci. Lett. 2019, 705, 87–93. [Google Scholar] [CrossRef]
- Chen, Z.; Yan, T.; Wang, E.; Jiang, H.; Tang, Y.; Yu, X.; Zhang, J.; Liu, C. Detecting Abnormal Brain Regions in Schizophrenia Using Structural MRI via Machine Learning. Comput. Intell. Neurosci. 2020, 2020, 6405930. [Google Scholar] [CrossRef]
- Yamamoto, M.; Bagarinao, E.; Kushima, I.; Takahashi, T.; Sasabayashi, D.; Inada, T.; Suzuki, M.; Iidaka, T.; Ozaki, N. Support vector machine-based classification of schizophrenia patients and healthy controls using structural magnetic resonance imaging from two independent sites. PLoS ONE 2020, 15, e0239615. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Qiu, J.; Lu, W. Support Vector Machine-Based Schizophrenia Classification Using Morphological Information from Amygdaloid and Hippocampal Subregions. Brain Sci. 2020, 10, 562. [Google Scholar] [CrossRef] [PubMed]
- Chilla, G.S.; Yeow, L.Y.; Chew, Q.H.; Sim, K.; Prakash, K.N.B. Machine learning classification of schizophrenia patients and healthy controls using diverse neuroanatomical markers and Ensemble methods. Sci. Rep. 2022, 12, 2755. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Liao, W.; Long, Z.; Tao, B.; Zhao, Q.; Luo, C.; Tamminga, C.A.; Keshavan, M.S.; Pearlson, G.D.; Clementz, B.A.; et al. Subtyping Schizophrenia Patients Based on Patterns of Structural Brain Alterations. Schizophr. Bull. 2022, 48, 241–250. [Google Scholar] [CrossRef]
- Hu, M.; Qian, X.; Liu, S.; Koh, A.J.; Sim, K.; Jiang, X.; Guan, C.; Zhou, J.H. Structural and diffusion MRI based schizophrenia classification using 2D pretrained and 3D naive Convolutional Neural Networks. Schizophr. Res. 2022, 243, 330–341. [Google Scholar] [CrossRef]
- Vyškovský, R.; Schwarz, D.; Churová, V.; Kašpárek, T. Structural MRI-Based Schizophrenia Classification Using Autoencoders and 3D Convolutional Neural Networks in Combination with Various Pre-Processing Techniques. Brain Sci. 2022, 12, 615. [Google Scholar] [CrossRef]
- Soh, L.-K.; Tsatsoulis, C. Texture Analysis of Sar Sea Ice Imagery Using Gray Level Co-Occurrence Matrices. IEEE Trans. Geosci. Remote Sens. 1999, 37, 780–795. [Google Scholar]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar]
- Clausi, D.A. An Analysis of Co-Occurrence Texture Statistics as a Function of Grey Level Quantization. Can. J. Remote Sens. 2002, 28, 45–62. [Google Scholar]
- Balakumar, B.; Raviraj, P.; Divya, D.E. Brain Tumor Classification Using Machine Learning Algorithms. Elysium J. Eng. Res. Manag. 2016, 4, 30–41. [Google Scholar]
| Individuals with Schizophrenia (n = 60) | Healthy Controls (n = 60) | |
|---|---|---|
| Age | 34.03 | 31.87 |
| Gender (Male/Female) | 48/12 | 40/20 |
| Handedness (Right/Left/Both) | (50/9/1) | 52/7/1 |
| Age of first episode | 20.6 | - |
| Age of onset (Years) | 21.1 | - |
| Positive (PANSS) | 15.0 | - |
| Negative (PANSS) | 14.8 | - |
| General (PANSS) | 29.6 | - |
| Right Hemisphere | AUC | Accuracy | Sensitivity | Specificity | F1 |
|---|---|---|---|---|---|
| Adaboost | 82.31 | 80.43 | 84.65 | 80.0 | 72.70 |
| GBoost | 88.29 | 83.13 | 71.43 | 90.19 | 86.96 |
| XGBoost | 79.01 | 83.34 | 88.90 | 76.78 | 82.35 |
| Random forest | 86.25 | 87.88 | 75.00 | 100 | 90.89 |
| LDA | 95.38 | 88.90 | 92.31 | 80.0 | 80.02 |
| kNN | 84.42 | 83.34 | 100 | 72.7 | 84.2 |
| NB(G) | 85.0 | 77.78 | 80.0 | 75.0 | 75.0 |
| Left Hemisphere | AUC | Accuracy | Sensitivity | Specificity | F1 |
|---|---|---|---|---|---|
| Adaboost | 96.15 | 91.44 | 92.31 | 100 | 90.3 |
| GBoost | 100 | 94.35 | 85.71 | 100 | 95.65 |
| XGBoost | 86.63 | 83.34 | 66.57 | 100 | 95.65 |
| Random forest | 100 | 89.01 | 100 | 80.0 | 88.78 |
| LDA | 100 | 94.4 | 92.31 | 100 | 91.90 |
| kNN | 90.0 | 88.89 | 100 | 80.0 | 89.04 |
| NB(G) | 98.75 | 83.92 | 70.0 | 100 | 84.21 |
| Right + Left Hemispheres | AUC | Accuracy | Sensitivity | Specificity | F1 |
|---|---|---|---|---|---|
| Adaboost | 96.25 | 88.98 | 87.5 | 90.0 | 90.0 |
| GBoost | 97.14 | 91.67 | 93.33 | 90.47 | 92.68 |
| XGBoost | 97.19 | 91.67 | 90.0 | 93.75 | 90.91 |
| Random forest | 88.57 | 85.47 | 86.67 | 80.95 | 85.0 |
| LDA | 97.84 | 86.11 | 94.44 | 77.78 | 84.85 |
| kNN | 90.0 | 83.33 | 93.75 | 75.0 | 83.34 |
| NB(G) | 90.26 | 83.74 | 90.91 | 71.42 | 76.92 |
| Refs. | Dataset | Schizophrenia /Healthy Control (Numbers) | Brain Regions | Extracted Features | Machine Learning Techniques | Accuracy (%) |
|---|---|---|---|---|---|---|
| [38] 2014 | Collected data | 66/66 | Whole brain | Gray matter densities | SVM | 90 |
| [39] 2015 | Collected data | 49/49 | Whole brain | Imaging features: deformations MR intensities, Gray matter densities | The modified maximum uncertainty linear discriminant analysis | 81.6 |
| [40] 2016 | Collected data | 41/42 | Gray matter and white matter | Gray matter and white matter volumes | SVM | 88.4 |
| [41] 2017 | Collected data | 38/38 | Described cortical regions | Cortical thickness features | SVM | 88.72 |
| [37] 2018 | NAMIC database | 20/20 | Whole brain |
Hu moments, GLCM, Zernika moments, Structure tensor | SVM and Fuzzy SVM | 90 |
| [42] 2019 | Collected data | 40/29 | Whole brain | Cortical thickness, gray matter volume, surface area, mean curvature, curvature index and folding index | Multi kernel SVM | 71.19 |
| [43] 2020 | COBRE dataset | 34/34 | Gray matter, white matter | Selected gray matter and white matter features | SVM | 85.27 |
| [44] 2020 | Collected data | 50/51 49/48 | Whole brain | Voxels of mean gray matter image | SVM |
72.2 72.3 |
| [45] 2020 |
COBRE dataset | 57/69 | Amygdaloid and hippocampal subregions | Morphological features | SVM | 81.75 |
| [46] 2022 | Collected data | 158/76 | Cortical and subcortical brain areas | Cortical and subcortical volume, cortical surface area, cortical mean curvature and cortical thickness | kNN, Logistic regression, SVM, Decision trees, Random forests | Range of 83–87 |
| [36] 2022 | Three different data centers | A: 137/132 B: 62/94 C: 144/181 | Gray matter, white matter, cerebrospinal fluid, and lateral ventricles | Texture features obtained by GLCM | SVM |
A: 66.67 B: 75.00 C: 70.83 |
| [47] 2022 | Collected data and B-SNIP dataset | 163/173 133/250 | Selected some brain regions | Subcortical volumes from seven regions (thalamus, caudate, putamen, pallidum, hippocampus, amygdala, and nucleus accumbens), cortical thickness and cortical surface area measures were extracted for 34 gray matter regions | Significant group difference at p < 0.05 | Significant differences were obtained between the groups |
| [48] 2022 | NUSDAST dataset, IMH dataset | 141/134 148/76 | Whole brain |
Probability maps of gray matter, white matter, and cerebrospinal fluid | Naive 3D CNN models |
79.27 70.98 |
| [49] 2022 | Collected data | 52/52 | Whole brain | (i) Image registration with skull stripping and two automated morphometry methods, (ii) voxel-based morphometry, and (iii) deformation-based morphometry | Autoencoders and 3D-CNN |
69.62 62.31 |
| Proposed study | COBRE dataset | 60/60 | Thalamus, caudate, putamen, pallidum, and amygdala | GLCM features | Adaboost, Gboost, XGboost, Random forest, LDA, kNN, NB |
94.4 with LDA from left hemisphere features |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gengeç Benli, Ş.; Andaç, M. Constructing the Schizophrenia Recognition Method Employing GLCM Features from Multiple Brain Regions and Machine Learning Techniques. Diagnostics 2023, 13, 2140. https://doi.org/10.3390/diagnostics13132140
Gengeç Benli Ş, Andaç M. Constructing the Schizophrenia Recognition Method Employing GLCM Features from Multiple Brain Regions and Machine Learning Techniques. Diagnostics. 2023; 13(13):2140. https://doi.org/10.3390/diagnostics13132140
Chicago/Turabian StyleGengeç Benli, Şerife, and Merve Andaç. 2023. "Constructing the Schizophrenia Recognition Method Employing GLCM Features from Multiple Brain Regions and Machine Learning Techniques" Diagnostics 13, no. 13: 2140. https://doi.org/10.3390/diagnostics13132140
APA StyleGengeç Benli, Ş., & Andaç, M. (2023). Constructing the Schizophrenia Recognition Method Employing GLCM Features from Multiple Brain Regions and Machine Learning Techniques. Diagnostics, 13(13), 2140. https://doi.org/10.3390/diagnostics13132140






