Combining Hepatic and Splenic CT Radiomic Features Improves Radiomic Analysis Performance for Liver Fibrosis Staging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Reference Standard
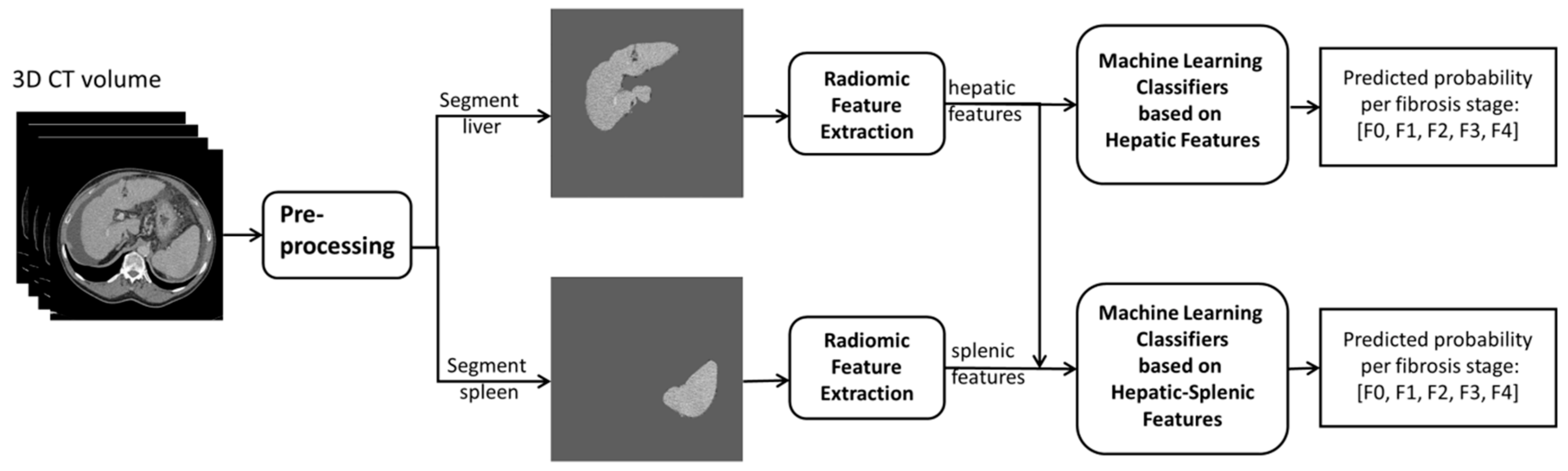
2.3. Image Processing and Feature Extraction for Radiomics Analysis
2.4. Radiomics Analysis for Liver Fibrosis Staging
2.5. Evaluation and Statistical Analysis
3. Results
3.1. Study Population
3.2. Radiomic Features
3.3. Performance of Liver Fibrosis Stage Classifiers
3.4. Top 5 Weighted Features of the Trained Classifiers
4. Discussion
5. Conclusions
Appendix A
Appendix A.1. Machine Learning Classifier for Liver Fibrosis Staging
Appendix A.2. Radiomic Features Used by Trained Classifiers
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cheemerla, S.; Balakrishnan, M. Global epidemiology of chronic liver disease. Clin. Liver Dis. 2021, 17, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.; Henderson, N.C. Antifibrotics in chronic liver disease: Tractable targets and translational challenges. Lancet Gastroenterol. Hepatol. 2016, 1, 328–340. [Google Scholar] [CrossRef]
- Moon, D.B.; Lee, S.G. Liver transplantation. Gut Liver 2009, 3, 145–165. [Google Scholar] [CrossRef] [PubMed]
- Tsochatzis, E.A.; Bosch, J.; Burroughs, A.K. Liver cirrhosis. Lancet 2014, 383, 1749–1761. [Google Scholar] [CrossRef]
- Thampanitchawong, P.; Piratvisuth, T. Liver biopsy: Complications and risk factors. World J. Gastroenterol. 1999, 5, 301–304. [Google Scholar] [CrossRef]
- Daginawala, N.; Li, B.; Buch, K.; Yu, H.; Tischler, B.; Qureshi, M.M.; Soto, J.A.; Anderson, S. Using texture analyses of contrast enhanced CT to assess hepatic fibrosis. Eur. J. Radiol. 2016, 85, 511–517. [Google Scholar] [CrossRef]
- Lubner, M.G.; Malecki, K.; Kloke, J.; Ganeshan, B.; Pickhardt, P.J. Texture analysis of the liver at MDCT for assessing hepatic fibrosis. Abdom. Radiol. 2017, 42, 2069–2078. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, S.S.; Park, B.; Yun, J.; Sung, Y.S.; Shim, W.H.; Shin, Y.M.; Kim, S.Y.; Lee, S.J.; Lee, M. Radiomics analysis of gadoxetic acid-enhanced MRI for staging liver fibrosis. Radiology 2019, 290, 380–387. [Google Scholar] [CrossRef]
- Choi, K.J.; Jang, J.K.; Lee, S.S.; Sung, Y.S.; Shim, W.H.; Kim, H.S.; Yun, J.; Choi, J.Y.; Lee, Y.; Kang, B.K.; et al. Development and validation of a deep learning system for staging liver fibrosis by using contrast agent-enhanced CT images in the liver. Radiology 2018, 289, 688–697. [Google Scholar] [CrossRef]
- Yasaka, K.; Akai, H.; Kunimatsu, A.; Abe, O.; Kiryu, S. Liver fibrosis: Deep convolutional neural network for staging by using gadoxetic acid-enhanced hepatobiliary phase MR images. Radiology 2018, 287, 146–155. [Google Scholar] [CrossRef]
- Yasaka, K.; Akai, H.; Kunimatsu, A.; Abe, O.; Kiryu, S. Deep learning for staging liver fibrosis on CT: A pilot study. Eur. Radiol. 2018, 28, 4578–4585. [Google Scholar] [CrossRef] [PubMed]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Yakar, D.; Dierckx, R.A.J.O.; Mouridsen, K.B.; Kwee, T.C.; de Haas, R.J. Liver fibrosis staging by deep learning: A visual-based explanation of diagnostic decisions of the model. Eur. Radiol. 2021, 31, 9620–9627. [Google Scholar] [CrossRef] [PubMed]
- Bedossa, P.; Poynard, T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology 1996, 24, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Milletari, F.; Navab, N.; Ahmadi, S. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016. [Google Scholar] [CrossRef] [Green Version]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J.W.L. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Freund, Y.; Schapire, R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Statist. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar] [CrossRef] [Green Version]
- Strobl, C.; Boulesteix, A.L.; Kneib, T.; Augustin, T.; Zeileis, A. Conditional variable importance for random forests. BMC Bioinform. 2008, 9, 307. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Efron, B.; Tibshirani, R. An Introduction to the Bootstrap; Chapman & Hall: New York, NY, USA, 1994. [Google Scholar] [CrossRef] [Green Version]
- Nitsch, J.; Sack, J.; Halle, M.W.; Moltz, J.H.; Wall, A.; Rutherford, A.E.; Kikinis, R.; Meine, H. MRI-based radiomic feature analysis of end-stage liver disease for severity stratification. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 457–466. [Google Scholar] [CrossRef]
- Shafiq-Ul-Hassan, M.; Zhang, G.G.; Latifi, K.; Ullah, G.; Hunt, D.C.; Balagurunathan, Y.; Abdalah, M.A.; Schabath, M.B.; Goldgof, D.G.; Mackin, D.; et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med. Phys. 2017, 44, 1050–1062. [Google Scholar] [CrossRef] [PubMed]
- Balagurunathan, Y.; Gu, Y.; Wang, H.; Kumar, V.; Grove, O.; Hawkins, S.; Kim, J.; Goldgof, D.B.; Hall, L.O.; Gatenby, R.A.; et al. Reproducibility and Prognosis of Quantitative Features Extracted from CT Images. Transl. Oncol. 2014, 7, 72–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Variable | Total Cohort | Liver Fibrosis Stage | |||||
|---|---|---|---|---|---|---|---|
| F0 | F1 | F2 | F3 | F4 | |||
| Total number of patients | 252 | 134 | 8 | 10 | 18 | 82 | |
| Median age (interquartile range) | 59 (48−65) | 63 (50-−74) | 64 (38−71) | 57 (43−64) | 48 (43−62) | 60 (54−65) | |
| Gender | Male | 140 (55.6%) | 68 (50.7%) | 3 (37.5%) | 7 (70.0%) | 11 (61.1%) | 51 (62.2%) |
| Female | 112 (44.4%) | 66 (49.3%) | 5 (62.5%) | 3 (30.0%) | 7 (38.9%) | 31 (37.8%) | |
| Etiology of liver fibrosis | Alcoholic | 26 (22.0%) | - | 0 | 0 | 1 | 25 |
| Autoimmune hepatitis | 5 (4.2%) | - | 0 | 1 | 1 | 3 | |
| HBV | 3 (2.5%) | - | 0 | 0 | 0 | 3 | |
| HCV | 10 (8.5%) | - | 0 | 0 | 1 | 9 | |
| PSC | 3 (2.5%) | - | 0 | 2 | 1 | 0 | |
| Steato- hepatitis | 8 (6.8%) | - | 0 | 0 | 0 | 8 | |
| Wilson disease | 1 (0.8%) | - | 0 | 0 | 0 | 1 | |
| Other | 17 (14.4%) | - | 1 | 0 | 7 | 9 | |
| Unknown | 45 (38.1) | - | 7 | 7 | 7 | 24 | |
| Machine Learning Classifier Type | Training Features | Accuracy (%; 95% CI) | Microaveraged AUC (95% CI) | AUC (95% CI) | Accuracy (%; 95% CI) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Significant Fibrosis | Advanced Fibrosis | Cirrhosis | Significant Fibrosis | Advanced Fibrosis | Cirrhosis | ||||
| Logistic regression classifier with L1 penalty | Selected liver features | 76 (70,83) | 0.94 (0.93,0.97) | 0.95 (0.93,0.98) | 0.92 (0.88,0.95) | 0.91 (0.86,0.95) | 86 (78,90) | 80 (75,88) | 82 (78,100) |
| Selected liver & spleen features | 84 (80,90) | 0.95 * (0.93,0.98) | 0.93 (0.90,0.98) | 0.88 (0.84,0.94) | 0.94 (0.90,0.99) | 92 (83,95) | 86 (83,95) | 90 (88,100) | |
| Logistic regression classifier with L2 penalty | Selected liver features | 78 (73,85) | 0.95 (0.93,0.97) | 0.96 (0.95,0.99) | 0.91 (0.88,0.95) | 0.93 (0.90,0.97) | 88 (78,93) | 82 (78,88) | 84 (80,100) |
| Selected liver & spleen features | 80 (75,85) | 0.95 ** (0.93,0.97) | 0.95 (0.93,0.98) | 0.90 (0.86,0.95) | 0.94 (0.91,0.97) | 88 (78,93) | 81 (78,90) | 86 (83,100) | |
| AdaBoosting | Selected liver features | 74 (68,80) | 0.82 (0.79,0.86) | 0.74 (0.67,0.80) | 0.72 (0.63,0.79) | 0.82 (0.73,0.88) | 76 (70,84) | 80 (75,100) | 74 (67,80) |
| Selected liver & spleen features | 76 (70,83) | 0.84 *** (0.82,0.88) | 0.57 (0.48,0.66) | 0.61 (0.52,0.69) | 0.84 (0.75,0.90) | 78 (73,85) | 82 (78,100) | 57 (48,66) | |
| Gradient Boosting | Selected liver features | 76 (70,83) | 0.88 (0.85,0.93) | 0.85 (0.81,0.92) | 0.83 (0.78,0.89) | 0.88 (0.78,0.93) | 80 (75,85) | 80 (75,100) | 85 (81,92) |
| Selected liver & spleen features | 78 (73,85) | 0.91 **** (0.88,0.95) | 0.84 (0.80,0.90) | 0.88 (0.85,0.94) | 0.86 (0.78,0.90) | 80 (75,88) | 84 (80,100) | 84 (80,90) | |
| XGBoosting | Selected liver features | 80 (75,88) | 0.86 (0.82,0.91) | 0.78 (0.70,0.87) | 0.79 (0.71,0.89) | 0.90 (0.83,0.95) | 84 (80,90) | 86 (83,100) | 78 (70,87) |
| Selected liver & spleen features | 82 (78,88) | 0.93 ***** (0.90,0.96) | 0.86 (0.82,0.92) | 0.90 (0.86,0.94) | 0.90 (0.80,0.95) | 84 (80,93) | 88 (85,100) | 86 (82,92) | |
| Weight Ranking | Radiomic Feature Names | |||||||
|---|---|---|---|---|---|---|---|---|
| L1 Penalty and F0 | L1 Penalty and F4 | L2 Penalty and F0 | L2 Penalty and F4 | |||||
| Feature Category | Feature Name | Feature Category | Feature Name | Feature Category | Feature Name | Feature Category | Feature Name | |
| 1st | GLRLM * | Low Gray Level Run Emphasis * | First order * | Robust Mean Absolute Deviation * | Shape * | Flatness * | NGTDM * | Coarseness * |
| 2nd | Shape * | Flatness * | Shape # | Maximum 2D Diameter Slice # | Shape * | Least Axis Length * | Shape # | Maximum 2D Diameter Slice # |
| 3rd | Shape * | Maximum 2D Diameter Row * | GLRLM * | Run Length Nonuniformity Normalized * | Shape * | Maximum 2D Diameter Row * | First order * | Robust Mean Absolute Deviation * |
| 4th | GLSZ * | Zone Entropy * | NGTDM * | Coarseness * | GLRLM * | Low Gray Level Run Emphasis * | Shape # | Surface Area # |
| 5th | Shape * | Least Axis Length * | GLCM # | Cluster Shade # | GLRLM * | Short Run Low Gray Level Emphasis * | Shape # | Minor Axis Length # |
| Weight Ranking | Radiomic Feature Names | |||||
|---|---|---|---|---|---|---|
| AdaBoosting | Gradient Boosting | XGBoosting | ||||
| Feature Category | Feature Name | Feature Category | Feature Name | Feature Category | Feature Name | |
| 1st | First order * | Total Energy * | NGTDM * | Strength * | NGTDM * | Strength * |
| 2nd | First order # | Median # | Shape * | Maximum 2D Diameter Row * | GLSZM * | Size Zone Nonuniformity * |
| 3rd | GLRLM # | Low Gray Level Run Emphasis # | GLCM # | Imc2 # | GLRLM # | Gray Level Variance # |
| 4th | GLSZM * | Size Zone Nonuniformity * | Shape * | Mesh Volume * | GLRLM * | Run Length Nonuniformity * |
| 5th | Shape * | Flatness * | GLSZM * | Zone Variance * | GLSZM # | Large Area High Gray Level Emphasis # |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, Y.; Yakar, D.; Dierckx, R.A.J.O.; Mouridsen, K.B.; Kwee, T.C.; de Haas, R.J. Combining Hepatic and Splenic CT Radiomic Features Improves Radiomic Analysis Performance for Liver Fibrosis Staging. Diagnostics 2022, 12, 550. https://doi.org/10.3390/diagnostics12020550
Yin Y, Yakar D, Dierckx RAJO, Mouridsen KB, Kwee TC, de Haas RJ. Combining Hepatic and Splenic CT Radiomic Features Improves Radiomic Analysis Performance for Liver Fibrosis Staging. Diagnostics. 2022; 12(2):550. https://doi.org/10.3390/diagnostics12020550
Chicago/Turabian StyleYin, Yunchao, Derya Yakar, Rudi A. J. O. Dierckx, Kim B. Mouridsen, Thomas C. Kwee, and Robbert J. de Haas. 2022. "Combining Hepatic and Splenic CT Radiomic Features Improves Radiomic Analysis Performance for Liver Fibrosis Staging" Diagnostics 12, no. 2: 550. https://doi.org/10.3390/diagnostics12020550
APA StyleYin, Y., Yakar, D., Dierckx, R. A. J. O., Mouridsen, K. B., Kwee, T. C., & de Haas, R. J. (2022). Combining Hepatic and Splenic CT Radiomic Features Improves Radiomic Analysis Performance for Liver Fibrosis Staging. Diagnostics, 12(2), 550. https://doi.org/10.3390/diagnostics12020550









