1. Introduction
Nowadays, imaging plays a central role in medicine. Large amounts of imaging data are constantly generated in daily clinical practice, leading to continuously expanding archives, and ever progressive efforts are being made across the world to build large-scale medical imaging repositories [
1,
2]. This trend is in line with the increasing medical image consumption needs, which have been studied and categorized into four groups: patient care-related, research-related, education-related, and other [
3].
In the era of big data, however, navigating through large-scale medical imaging archives is becoming, correspondingly, increasingly troublesome. Browsing any available, large-scale medical imaging repository through a conventional text-based search engine is time-consuming, severely hampered if the repository lacks curated or expert-annotated metadata, the search results display options are limited. Conversely, the need for collecting curated or expert-annotated metadata may, in turn, be preventing the building of large, multi-center, international medical imaging repositories that meet the medical imaging needs of today. In this scenario, there is an enormous need for efficiently archiving, organizing, managing, and mining massive medical image datasets on the basis of their visual content (e.g., shape, morphology, structure), and it may be expected that this demand will only become more substantial in the foreseeable future.
Accordingly, attempts have been made over the last few decades to complement search strategies of conventional text-based engines by means of advancing image content-based repository indexation technology as this may lead to novel search engine possibilities [
4]. Several approaches have been used to develop content-based image retrieval (CBIR) systems that allow for automatic navigation through large-scale medical image repositories [
4]. Such promising capability fuels research efforts in the fields of computer vision and deep learning.
Formally, a CBIR system is a quadruple {D, Q, F, R (qi, dj)}, where: (i) D is a set composed of representations for the images in a given collection, (ii) Q is a set of representations for user information needs, operationally known as queries, (iii) F is a representational framework that allows images, queries, and their relationships to be jointly modeled, and, finally, (iv) R(qi, dj) is a ranking function which associates a real number with a query qi in Q and an image dj in D. The ranking defines an ordering among the images in a given collection regarding the query q.
Regarding F, this framework can be learned using supervised or unsupervised approaches. In supervised approaches, the representational framework has a grounded dataset made up of pairs of images and queries along with their respective ranking scores. The main limitation of these approaches is the difficulty of building a large volume of curated data that allows the system to generalize for new queries and images not considered during the training phase of the system. On the other hand, unsupervised approaches do not require a grounded dataset to train the representational framework. Instead, this type of system is typically based on distances between vector representations of images and queries. We take this second approach for this proposal.
CBIR systems have the potential to deliver relevant technology for clinical imaging through several use cases in medical education, research, and care, including clinical diagnostics [
5]. In radiology, for instance, it has long been shown that CBIR can facilitate diagnosis, especially for less experienced users such as radiology residents, but also for radiologists in cases of less frequent or rare diseases [
6]. Noteworthy, relatively less explored has been the possibility that image content-based repository indexation technology brings about to augment search results display options. This indexation approach empowers the development and application of intelligent similarity maps to display search results, which may further boost efficiency of navigation through large-scale medical imaging repositories. If integrated with a PACS (Picture Archiving and Communication System), this approach would make it possible to perform novel, automatic, independent, medical inter-consultations using medical image content with similar cases available in the local and other linked archives without requiring the user to enter a priori keywords to drive the search towards the best visual content match. Furthermore, such a CBIR system and intelligent and interactive visual browser, thoughtfully designed to be fully interoperable with healthcare systems according to international standards, may stimulate a burst of novel opportunities for medical education, research, and care.
The literature on this topic has identified use cases even when technical limitations would only allow CBIR based on shallow features such as shape and texture, and at a time when medical imaging fields have counted on a narrow set of digital images of limited resolution. Novel deep learning architectures have paved the pathway towards unleashing the potential for innovative applications to solve long-standing medical image search problems. Moreover, features from deep convolutional neural networks have yielded state-of-the-art performance in CBIR systems by efficiently overcoming long-standing computational shortcomings [
7,
8,
9]. With current computing capacity increasingly supplying powerful image analysis techniques, a new surge of capabilities and applications have been devised. Novel deep learning architectures have also empowered internal image representation learning, specifically, latent representations, which can be used to implement ground-breaking image content search engines [
10,
11,
12].
Looking towards their implementation in the daily clinical workflow, however, there remain technical challenges. Large-scale repositories, reciprocally, are needed for CBIR systems to deliver appealing search results. For that purpose, in turn, local teams with experience in the use of healthcare integration standards are required. For multi-center collaborative efforts, there is also needed a data extractor inside each institution to anonymize, and transfer and convert data to a standardized semantic term.
The aim of the present project was to develop a CBIR system using learned latent image representation indexation, with a visual content, similarity-based, intelligent and interactive visual browser for efficient navigation. The system was developed using international standards to be fully interoperable to ease integration into routine clinical workflow and, thus, support current medical image demands throughout education, research and clinical care.
2. Materials and Methods
2.1. Building an Interoperable, Standardized and Anonymized Medical Image Repository
Integration with the hospital (Clinical Hospital University of Chile, Santiago, Chile) to have continuous feeding of medical images and metadata was composed of different microservices. First, a Mirth Connect integration system was used to receive data in different health standard types, DICOM channels, HL7 messages, and HL7-FHIR messages. Mirth Connect was integrated to an Anonymizer service, which eliminates all patients’ personal data and extracts healthcare data to feed an FHIR server using international standardized health terms (SNOMED-CT, ICD-9, ICD-10). This allows the development of an interoperable and standardized repository. The Institutional Review Board approved (METc 2020/035, on 8 July 2020) the storage and management of thorax computed tomography in this repository. All procedures were conducted in adherence to the declarations of Helsinki.
2.2. Deep Learning-Based Medical Image Indexation and Retrieval Strategy
Two image processing architectures based on deep learning were combined. The first, CE-Net (Context Encoder Network) [
13], was used to build a representation of the input image in a lower dimensional space, i.e., a latent representation. Using these image embeddings, a second neural architecture was trained, Xception [
14], which is capable of learning a new representation of the input image. This architecture is trained to solve a diagnosis classification task, which helps group images that coincide with their initial diagnosis.
2.2.1. CE-Net
The image segmentation architecture CE-Net [
13] was used first, which allows 2D medical images to be processed. The CE-Net architecture is an extension of the U-Net architecture [
15], which is an encoder-decoder architecture. Encoder-decoder architectures work with a tandem of neural layers, wherein the first architecture block, the encoder, is a sequence of layers responsible for building a representation of the input image in a lower dimensional space, known as latent space. The second architecture block, the decoder, is another sequence of layers responsible for transforming the encoding from latent space to original space, recovering the original dimensionality of the image. The layers of the U-Net architecture are convolutional, introduce max-pooling operators, and add residual connections to prevent the network from the loss of information between layers. This allows the construction of lower dimensional representations that retain the most important information from the encodings obtained in the previous layers. The architecture parameters are adjusted in such a way as to minimize the reconstruction error (defined as the difference between the original image and the reconstructed image). Thus, the encoder-decoder architectures are intended to encode the images in the latent space. The CE-Net architecture extends the U-Net architecture by incorporating two processing modules: the dense à-trous convolution (DAC) and the residual multi-kernel pooling (RMP) module. Both modules were designed to capture high-level characteristics, preserving spatial information throughout the encoder-decoder architecture. These representations were used to feed the second neural architecture used by the system, the Xception.
2.2.2. Xception
Xception [
14] is an architecture that extends the Inception [
16] architecture used in image classification. Classification models based on deep learning achieve good performance by incorporating representation learning mechanisms that enable adapting of these representations to the tasks for which they are trained. The Inception architecture uses modules based on convolutional operators that capture short-range dependencies in the input image; this allows learning of a new representation of the input image by identifying patterns between the pixels of the original image, which is useful for a better representation. The Xception architecture extends the Inception architecture by incorporating convolutional layers that allow capturing long-range dependencies, which has allowed this architecture to obtain good results in classification problems.
Yet, one of the disadvantages of architectures such as Xception, is that they have many parameters and, therefore, require a large volume of data to be trained. The Xception architecture was validated on an internal Google dataset, called JFT, which has more than 350 million high resolution images with labels on the order of 17,000 classes. The availability of labeled imaging data at this volume is unlikely to be reached in the medical field in the near future. Because the scale of the repositories with which this project works is much smaller, training an Xception architecture for medical images from scratch is not feasible.
Therefore, a different approach was introduced that allows reducing of the gap between the need for large volumes of data to use deep learning, and the actual availability of medium-scale datasets in the clinical setting. Two architectures trained in different tasks were combined. The CE-Net architecture was used to segment the images of the repository, and the latent representations were used as pre-trained vectors to adjust the Xception architecture according to diagnosis. By segmenting the images and working with their latent representations, their variability was reduced and placed in a common representation space, which provides better generalization abilities to the Xception because, instead of working with the original images, it works with images with reduced dimensionality. Thus, the Xception network can work with fewer parameters when solving the diagnostic classification task, making it possible to avoid the risk of overfitting attributable to limited volumes of data. Another advantage of this way of proceeding is that it enables working on images of different types. Since the CE-Net architecture builds a common representation space for images of different types, the Xception network can process these images indistinctly, solving the diagnostic classification problem on heterogeneous image sources.
Note that Xception is a supervised method. To solve the classification problem according to diagnosis, the model must encode the images to consistently activate the feature maps of the output layer in correspondence with the diagnosis. Accordingly, the encodings of the images that coincide in diagnosis decrease their relative distance, producing a clustering effect in the embedding space. Our representational framework takes advantage of these Xception features to improve the system’s search abilities in the target classes.
2.2.3. Ce-Net + Xception Assembling
We tested latent embeddings extracted from the DAC and RMP blocks of the Ce-Net, the latter delivering the best results in our system. The encoding extracted from the RMP block was fed into the Xception network. Accordingly, instead of working directly on the images, the Xception network works on the encodings of the images delivered by the Ce-Net RMP block. The RMP block gathers information from the DAC block using four different-sized pooling kernels. These features are fed into a 1 × 1 convolution to reduce the dimension of feature maps. Finally, these features are concatenated with the original features extracted from the DAC block, producing an encoding.
The Ce-Net processes images of 512 × 512 dimensions. The RMP block produces encodings with 384 × 344 dimensions. This encoding is ingested into the Xception, which outputs an activation map of the same dimensions. Both encodings, of the RMP block and for the output activation of the Xception, are flattened and concatenated, producing a one-dimensional embedding with 264,192 entries.
According to the formal definition of a CBIR system provided in
Section 1, each quadruple element corresponds to the following elements of our framework.
D is the set of images indexed by our system,
Q is an unbounded set of images used to query our system,
F is the representational framework defined by the Ce-Net + Xception ensemble, and
R(qi, di) is the distance computed by the nearest neighbor query engine. Note that the queries are unseen images, i.e., medical images not used during the representation learning training phase. In order to create the query encodings, we need to feed these images into the representational framework. Accordingly, for each query image, their embeddings are retrieved from the RMP block and the last feature activation map of the Xception network. Finally, these embeddings are used as query vector representations to feed the query engine.
One of the characteristics of our ensemble is that it combines two different capabilities in a single representational framework. On the one hand, the Ce-Net encodes images using convolutional filters of different sizes, which allows it to represent tumoral lesions of different sizes. This characteristic is mainly due to pooling kernels, which are used in the DAC and RMP blocks of the architecture. On the other hand, the Xception network allows images to be grouped according to diagnosis. The effect that this network produces in the latent space is to cluster the encodings, reducing the relative distances between the images that coincide in diagnosis. This capacity provides the system with better detection capabilities in the target classes. Due to the above, it is expected that images with lesions of different sizes or types of tissues, but that coincide in diagnosis, will cluster in the embedding space.
2.2.4. Training
The CE-Net can be trained for medical image segmentation tasks by showing original segmented image pairs to the network at the input and output of the network. This requires having a set of medical images together with their segmentation masks, generated by medical imaging specialists. To do this, we trained CE-Net with public datasets of different types of medical images.
The SARS-CoV-2 CT scan dataset was used to train the Xception network (SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification. Available online:
https://www.medrxiv.org/content/10.1101/2020.04.24.20078584v3, accessed on 27 July 2021). The dataset contains 1252 CT slices that are positive for SARS-CoV-2 infection (COVID-19) and 1230 CT slices for patients non-infected by SARS-CoV-2. Since each scan contains multiple slices, and some do not show ground-glass or other evidence of COVID-19, we used the Ce-Net to sample CT slices. The segmenter allowed CT slices to be sampled for COVID-19 patients who had evidence of ground-glass or pleural effusion. One axial slice was sampled per scan, maintaining the balance between healthy and sick patient slices.
As a validation set, the COVID-CT-dataset (COVID-CT. Available online:
https://github.com/UCSD-AI4H/COVID-CT, accessed on 27 July 2021) was used as a testing set. The dataset contains 349 CT slices from 216 COVID-19 patients and 463 non-COVID CT slices. The dataset contains images acquired with different media, for example, CT slices post-processed by cell phone cameras and some images with very low resolution. For these reasons, the dataset represents the real conditions of image acquisition for a system of this kind.
To generate a balanced set of queries, the Clinical Hospital of the University of Chile supported us with eight CT scans where half of them had suffered from COVID-19. From these CTs, 25 slices with COVID-19 and 25 slices without COVID-19 were extracted. Each of these queries was used to query our system.
As the Ce-Net is a network with many parameters, and to avoid overfitting, we initialized their weights using ImageNet samples [
17] (ImageNet. Available online:
https://image-net.org/, accessed on 27 July 2021).
2.2.5. Validation
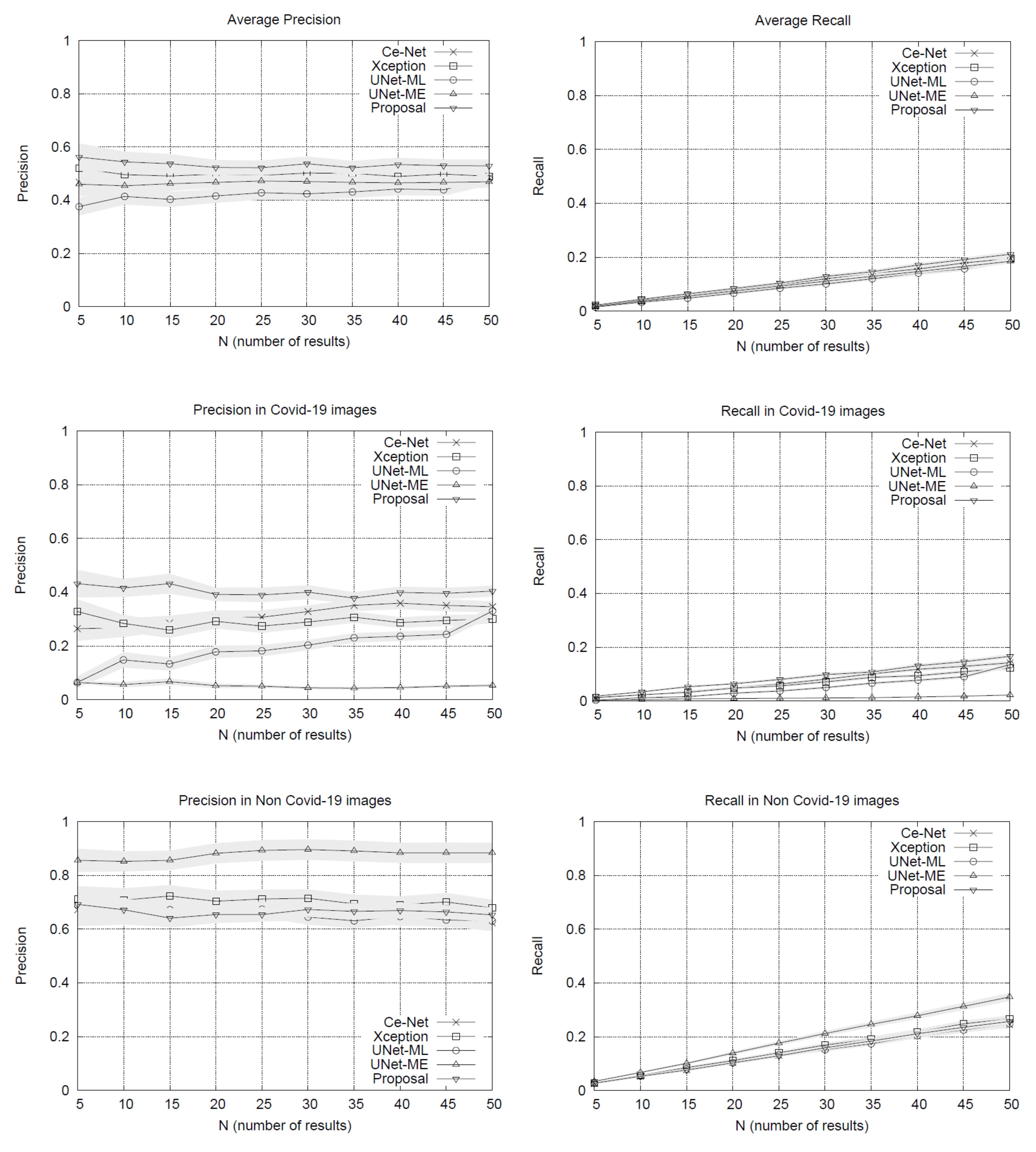
The performance of our search system was evaluated using precision and recall measures. To compute these metrics, the CT slices’ ground labels are considered according to SARS-CoV-2 diagnosis, counting matches between image examples labels and their list results. This proposal was evaluated considering four alternative methods:
- -
Ce-Net [
13]: Corresponds to a search system based on the encoding of the testing images obtained from the Ce-Net using the RMP block.
- -
Xception [
14]: Corresponds to a search system based on the encoding of the testing images obtained from the Xception using its last layer.
- -
U-Net-ME (manifold embeddings): Corresponds to a search system based on the encoding of the testing images obtained from the architecture of Baur et al. [
18], which extended U-Net and was trained for 5 epochs for segmentation and then 5 epochs with the manifold embedding loss. The embeddings were obtained using the last layer of the architecture.
- -
U-Net-ML (mixed learning): Corresponds to a model based on [
18], for searches over the encoding of the testing images obtained from a U-Net architecture. Trained for 5 epochs for segmentation, then 5 more epochs for classification. We conducted tests with several layers of the encoding but those that obtained the best results were the embeddings obtained using the last layer of the architecture.
The results of the experiments are shown in
Figure 1. The performance plots on the whole set of testing queries (at the top of
Figure 1) show that our proposal outperforms the other methods in precision. As we might expect, the precision drops slightly as the list of results grows. The variance around the mean precision also decreases gradually. The recall of all the methods is quite similar, reaching around 20% in lists with 50 image results.
By separating the testing set between COVID-19 and non COVID-19 queries, the results in
Figure 1 show that our method obtains advantages over other methods when using queries of patients with COVID-19, surpassing by a significant margin the most direct competitor, the Xception network. The other methods have lower performances. U-Net-ME performs well in the healthy patient class. However, this model exhibits overfitting to this class as its performance in the COVID-19 class is very low. Our proposal surpasses the other methods in COVID-19 images regarding recall rates, while U-Net-ME generates a better recall in images of healthy patients. The results confirm that our proposal is suitable for searching for images of COVID-19 patients, surpassing all its competitors in precision and without generating overfitting to any of the classes.
With the Ce-Net + Xception network validated, latent vectors were indexed in a nearest neighbors query engine, which is the basis of the intelligent visual browser.
2.3. Intelligent Interactive Visual Browser for Medical Images
The data ingestion pipeline (content + metadata) to the PACS has been systematically tested in its different stages, with different image sources. The software architecture design of the CBIR system was developed following a model of microservices grouped in the back-end and front-end of the system. These services are communicated via REST API (representational state transfer) in order to have a scalable system for a correct incorporation of new data and their respective descriptive metadata. This pipeline makes available the metadata information necessary for the results of the CBIR system. In the case of content (images), the procedures for their incorporation into the PACS go together with the metadata in the data ingestion pipeline, thus being available as potential search results for the CBIR system. The design of the system allows us to function in a decoupled way to the nature of the image for which the architecture of neural networks are designed to work. In the event that the image format is changed, it would only be necessary to update the network architecture.
2.3.1. Back-End
The combination CE-Net + Xception constitutes the back-end of the browser. The back-end provides a vector representation of all the images in the repository, i.e., the latent representations constructed using the CE-Net + Xception allow obtaining continuous and dense vectors of the same dimensionality for all the images in the repository. It is noteworthy that combining these architectures makes metadata availability requirements more flexible since none of the networks require metadata for ingested imaging datasets. This means that images of different types could be received without metadata, and all of them would have a latent representation in the same latent space that Xception uses for classification. Moreover, images of latent representations retrieved by Xception are expected to be separated by types of images (as a result of the CE-Net segmentation model) and by diagnosis (as a result of the Xception classification model). The clustering hypothesis according to type of image and diagnosis is supported by the combination of both architectures in tandem. This means that while the segmenter allows different types of medical images to be represented in the same representation space, the Xception network helps to separate them by diagnosis. It is worth mentioning that if diagnostic metadata is not available, the Xception network will obtain the representation of the unlabeled images using them as a testing partition.
2.3.2. Front-End
The front-end is responsible for enabling the search engine on the repository. To do this efficiently, the images are indexed using an efficient data structure that allows searching for close neighbors. This data structure is Multiple Random Projection Trees (MRPT) [
19], which is considered to be state-of-the-art in the approximate search for close neighbors. MRPT allows the building of a search index with recall guarantees. This means that we can indicate a minimum recall rate and the structure is built in a way that satisfies this restriction. This element is important because if the repository grows in volume, an index that scales to a larger volume of data will be required. Once the index has been built, queries of close neighbors can be run. The user determines the number of nearest neighbors from the repository and returns the identifiers of the corresponding images. Queries are new images that do not need to be labeled, i.e., they do not need to be entered with any accompanying text. To enter it as a query, the image is first segmented using the CE-Net network pre-trained on the repository images. After retrieving its latent vector and ingesting it in the Xception network, the latent vector is retrieved from the Xception network, which is used as a representation of the image. This vector is in the same latent space as the images indexed in MRPT, so it can be used as a query vector to retrieve its nearest neighbors. As the representation is built on the same pipeline with which the repository images have been processed, the nearest neighbors will correspond to images that are similar in content, both in segmentation structure and diagnosis.
Figure 2 shows the pipeline of our proposed assistance system, including the query pipeline implemented on the front-end to enable the intelligent interactive visual browser for medical images.
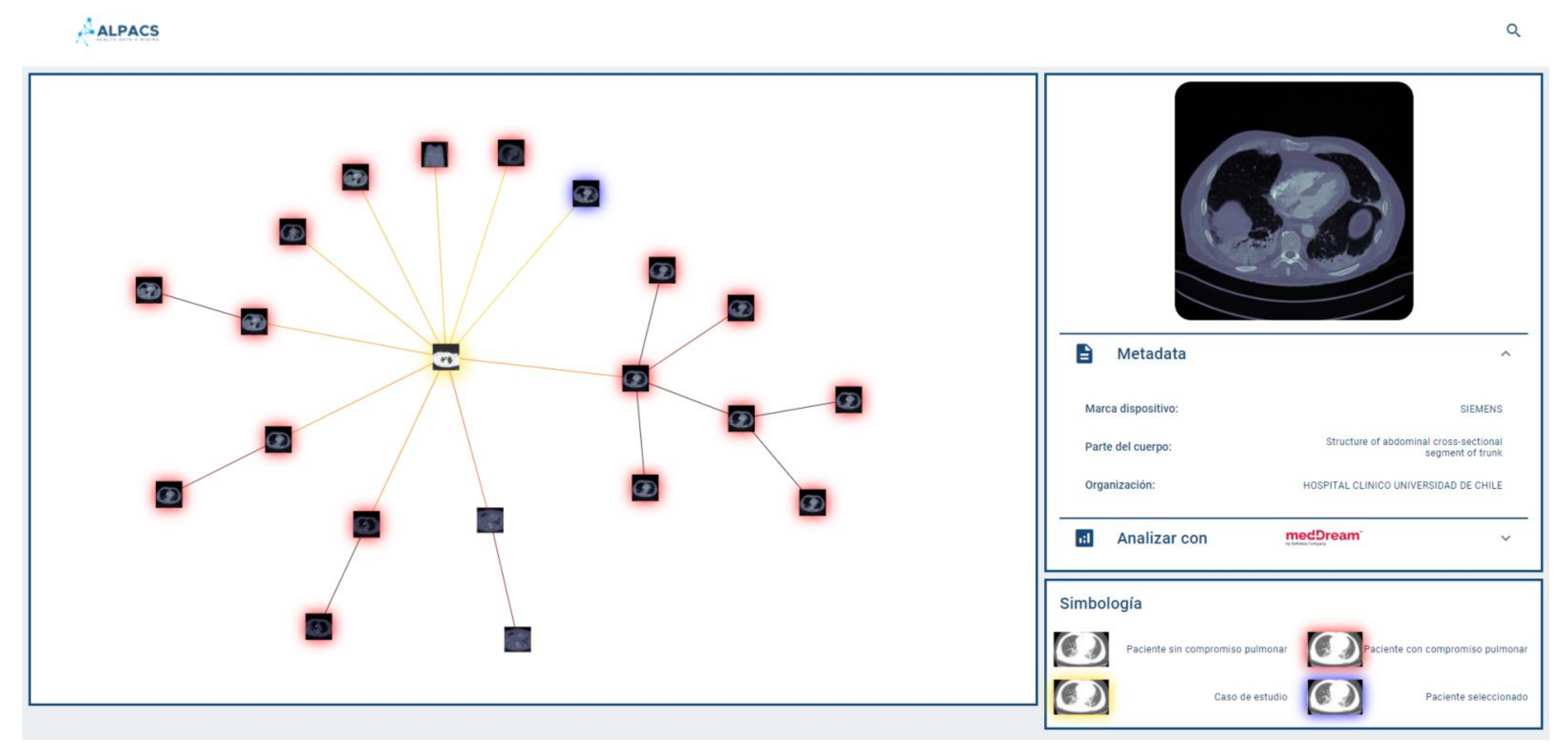
Since the browser is based on proximity searches, creating an ego network around the query image would provide valuable information to the user as to understand the different groups of images that make up the set of results. Because displaying the results according to similarities may be expected to be more informative than conventional displaying fashion in the form of a list, the visual browser was designed taking into consideration that once the nearest neighbors of the query image are retrieved, the results could be displayed by implementing a proximity graph.
2.4. Results Display
A key aspect of our visual browser concerns its fashion for displaying search results. A graphic library called VivaGraph (VivaGraph. Available online
https://github.com/anvaka/VivaGraphJS, accessed on 27 July 2021) was selected, in JavaScript, which allows rendering results’ graphs in near real-time. The VivaGraph library allows a graph display with thousands of nodes in fractions of seconds, making it ideal for the purposes of this search engine. The rendering of the graph consists of generating a node for each result, and joining the results with edges whose length is inversely proportional to their proximity. In this way, the closest results are grouped into clusters and the most distant are more distantly displayed. The rendering is defined by a layout algorithm. A Force Atlas layout is used that produces more compact graphs than other layouts, which is useful for the tool. The graph is responsive, i.e., it allows the user to select an image and display a metadata box that complements its description (if available). Whenever metadata is available, the viewer will show it in a responsive selection box, which delivers a tab of an image selected by the user on the proximity graph. The system allows the user to provide text (e.g., diagnosis of the query image) as part of the repository consultation, in which case it will be used to filter out the results returned by the query engine. This case is called hybrid search (content + metadata). If the user does not provide text, the query is performed by using a content-only search (no metadata).
Figure 3 shows a query graph displayed using our system.
2.5. Survey among Professionals of the Healthcare Sector
To explore the potential applications that professionals of the healthcare care sector would foresee for a CBIR system with an intelligent interactive visual browser, a survey was performed among nurses, medical technologists, dentists, general practitioners, radiology residents, radiologists, and other medical specialists such as ophthalmologists, surgeons, gynecologists, urologists, pediatricians, and otolaryngologist, among others, at the Clinical Hospital University of Chile. The composition of the survey sample was: 56.7% medical doctors, 26.9% medical technologists, 7.4% nurses, 3% dentists, 3% medical informatics, 1.5% biochemists, and 1.5% students. On the other hand, the sample was distributed as 31.4% male, 34.3% female, and 34.3% unknown gender.
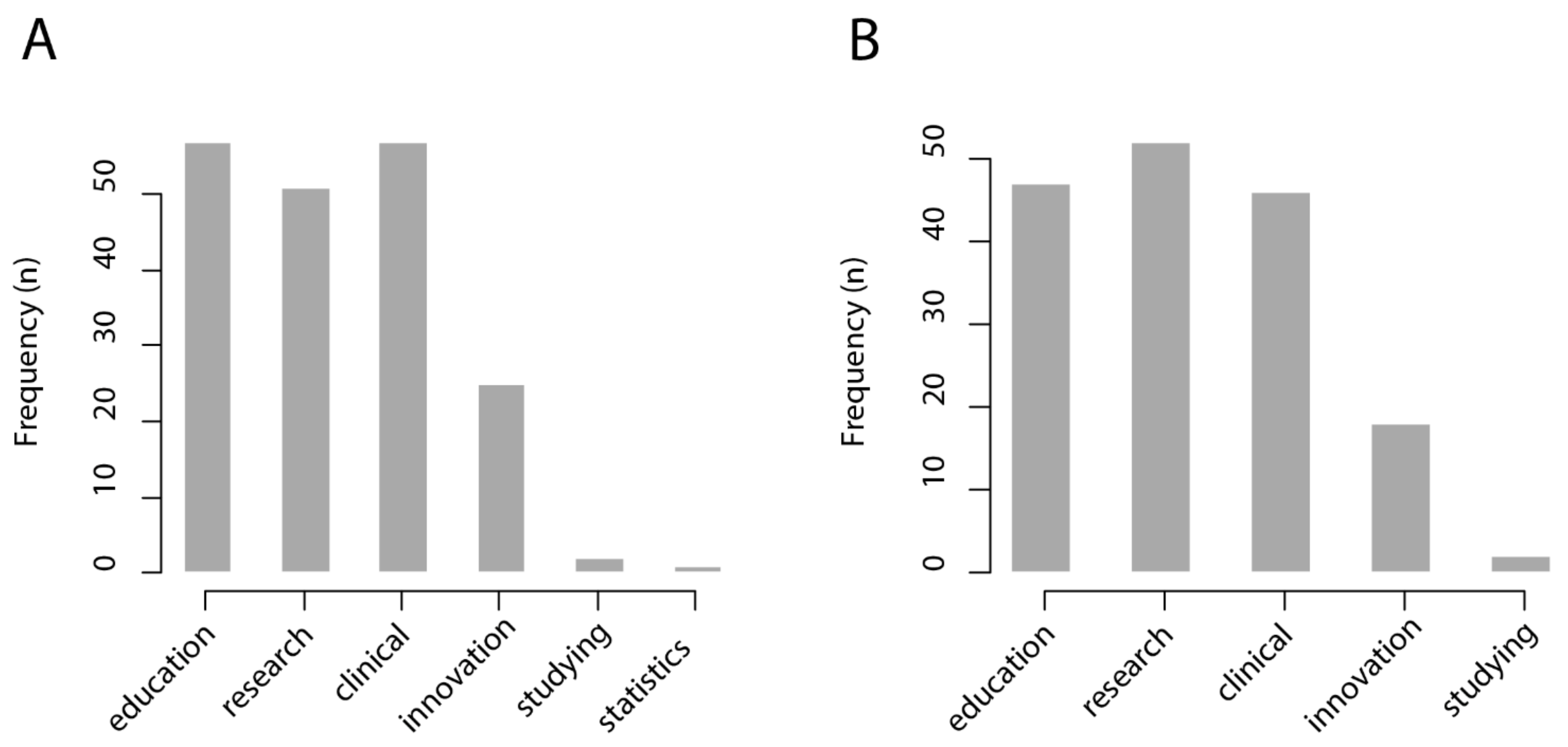
The invitation to participate was shared through digital media (electronic mailing and social networks). All participants were invited to, in turn, share the link to the survey with healthcare professionals of their working and social network. The survey was applied between November and December 2020. In total, 67 subjects completed the survey. For the questions “in which cases would a CBIR system be useful?” and “in which cases would an intelligent interactive visual browser be useful?”, respondents were asked to choose from six possible answers: medical education; research; clinical care; innovation/technological development; personal study; management statistics.
We found that most respondents from the healthcare sector foresee that a CBIR system would be useful for medical education (57/67 = 85%), research (51/67 = 76%), and clinical care (57/67 = 85%), while it would be less useful for innovation and technological development (25/67 = 37%), personal study (2/67 = 0.3%), and management statistics (1/67 = 0.1%). Similarly, most respondents from the healthcare sector foresee that an intelligent interactive visual browser would be useful for medical education (47/67 = 70%), research (52/67 = 78%), and clinical care (47/67 = 70%), while being less useful for innovation and technological development (18/67 = 27%) and personal study (2/67 = 0.3%). The results of this survey are shown in
Figure 4.
To further evaluate the system’s usability, the System Usability Scale will be applied in upcoming evaluations of the system once it is implemented and running in a corresponding environment for medical education, research, and care purposes.