Diagnosing Burn Wounds Infection: The Practice Gap & Advances with MolecuLight Bacterial Imaging
Abstract
1. Introduction
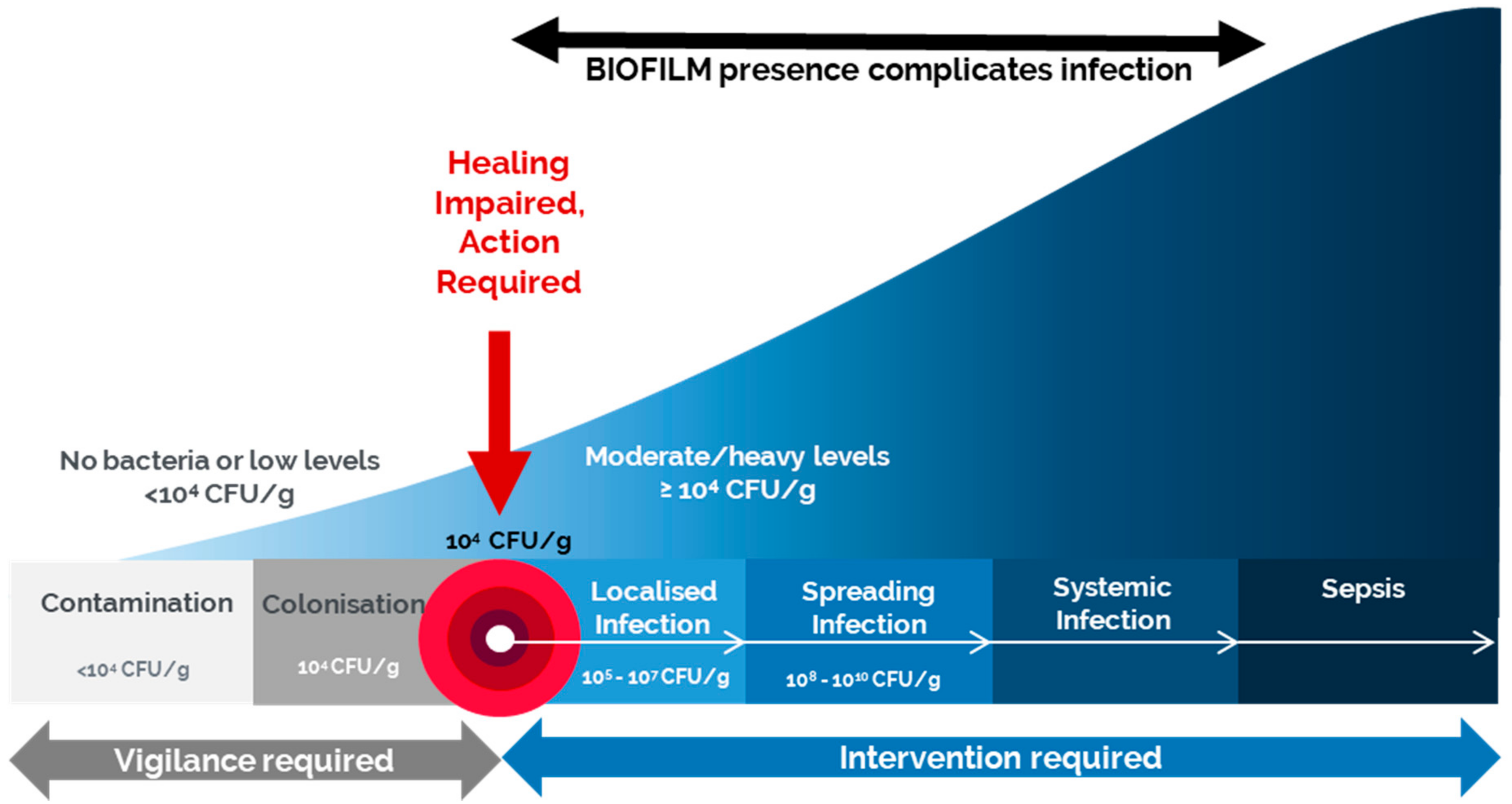
1.1. Pathogenesis of Burn Wounds Infection
- Contamination refers to the existence of non-proliferating microorganisms at a level that cannot trigger the immune response. Any open wound will contain some contamination with bacteria, typically natural flora, yet these bacteria are non-proliferating and at levels that do not evoke a host response or delay healing [14].
- Colonization is the presence of microorganisms with a limited proliferation rate without triggering the immune response or delaying healing. During these stages, vigilance is required but not necessarily antimicrobials [14].
- Spreading infection occurs as the bacteria increase in number and virulence and begin to invade the surrounding tissue and more overt signs of infection present like delayed wound healing, potentially erythema, wound breakdown, and dehiscence [14].
- Systemic infection is the most advanced stage which affects the whole body via vascular or lymphatic routes, leading to serious consequences such as sepsis and organ dysfunction [14].
1.2. Standard of Care: Clinical Signs and Symptoms
1.3. Standard of Care: Microbiological Assessment
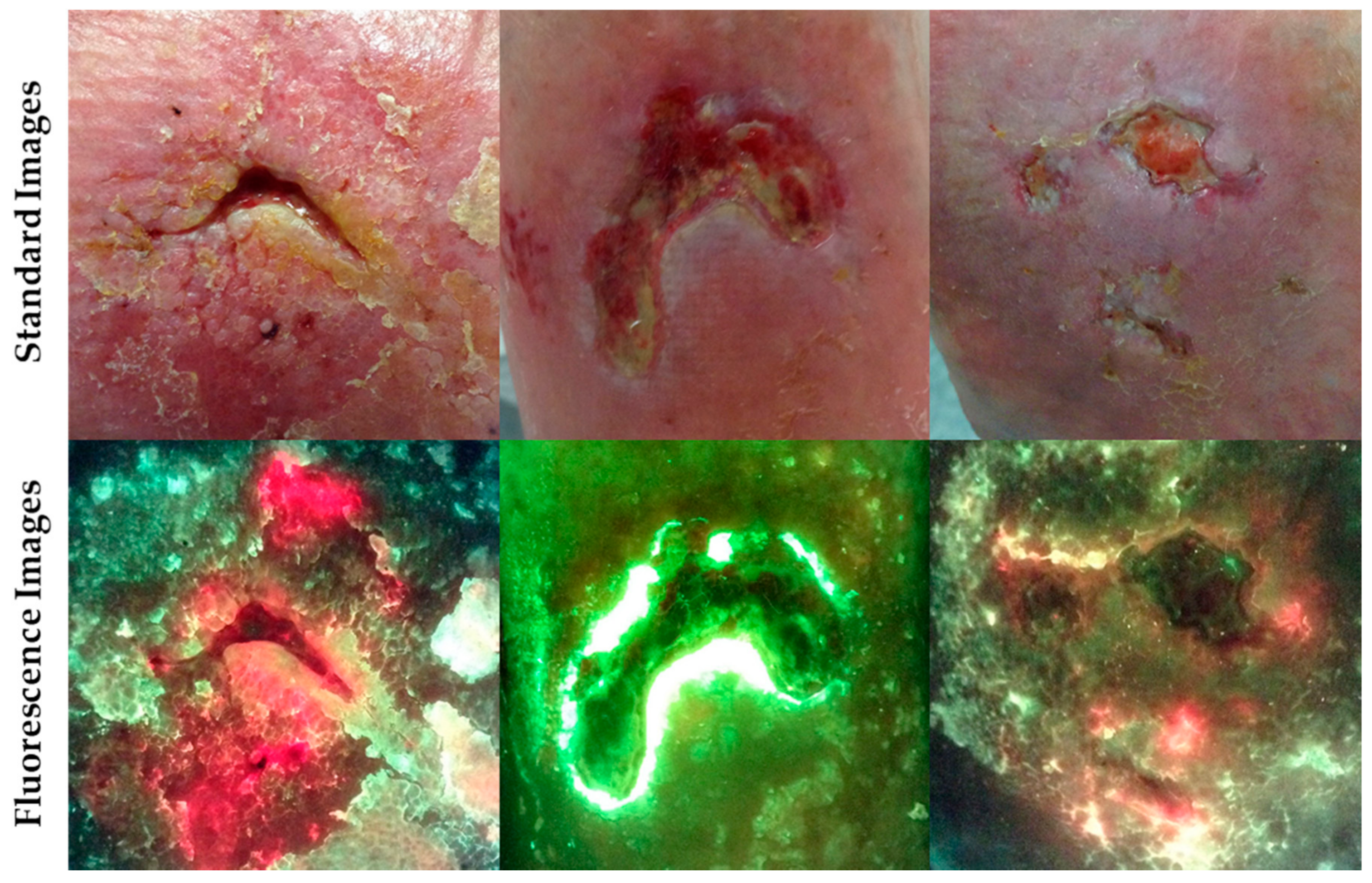
2. Fluorescence Imaging with MolecuLight i:X
Case Study 1
3. Diagnostic Accuracy of Fluorescence Imaging
3.1. Comparison of Fluorescence Imaging to Clinical Signs and Symptoms
3.2. The Role of Sampling Techniques in Fluorescence Imaging Diagnostic Accuracy
4. Fluorescence Imaging Shines a Light on Wound Microbiology
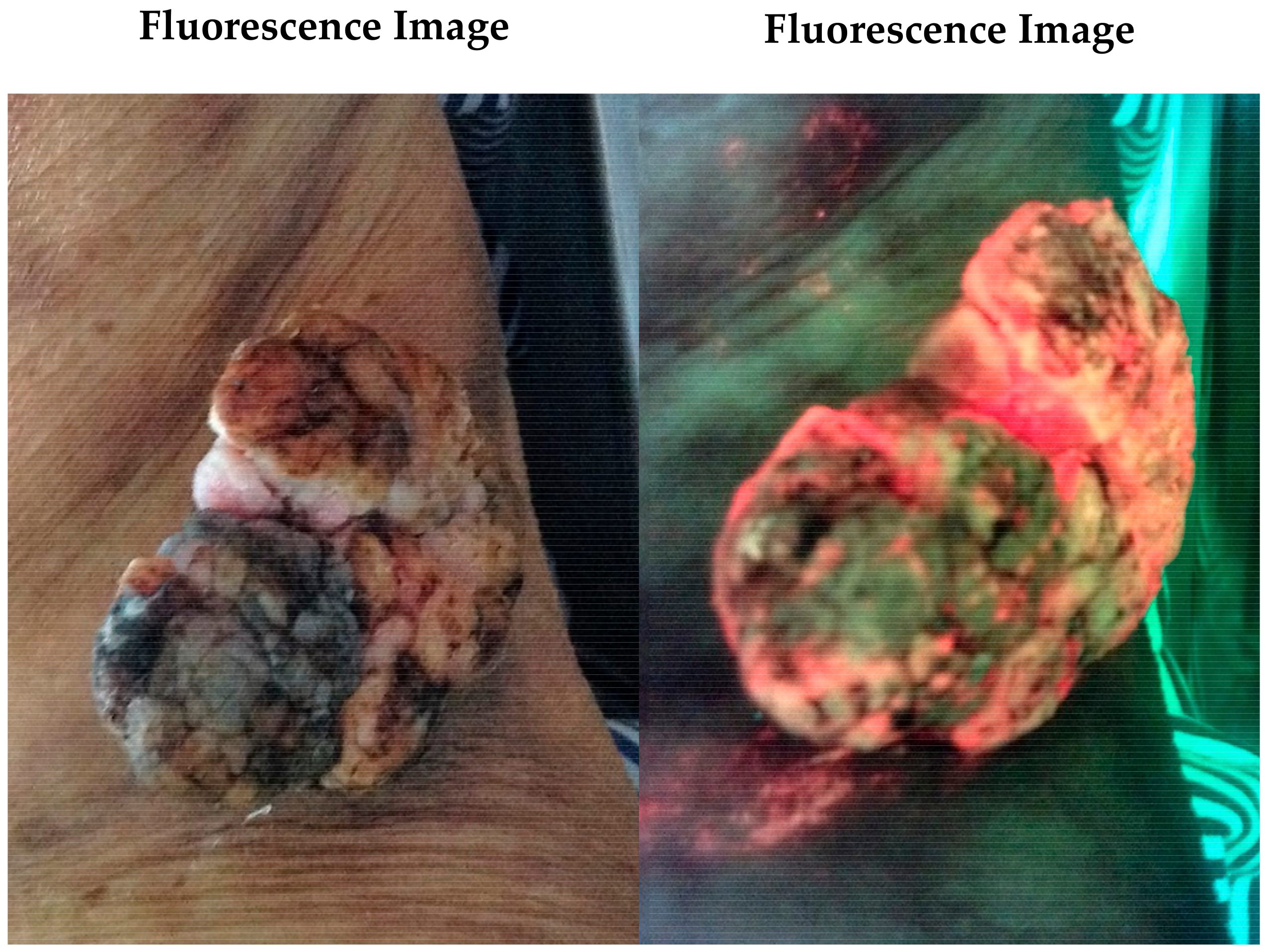
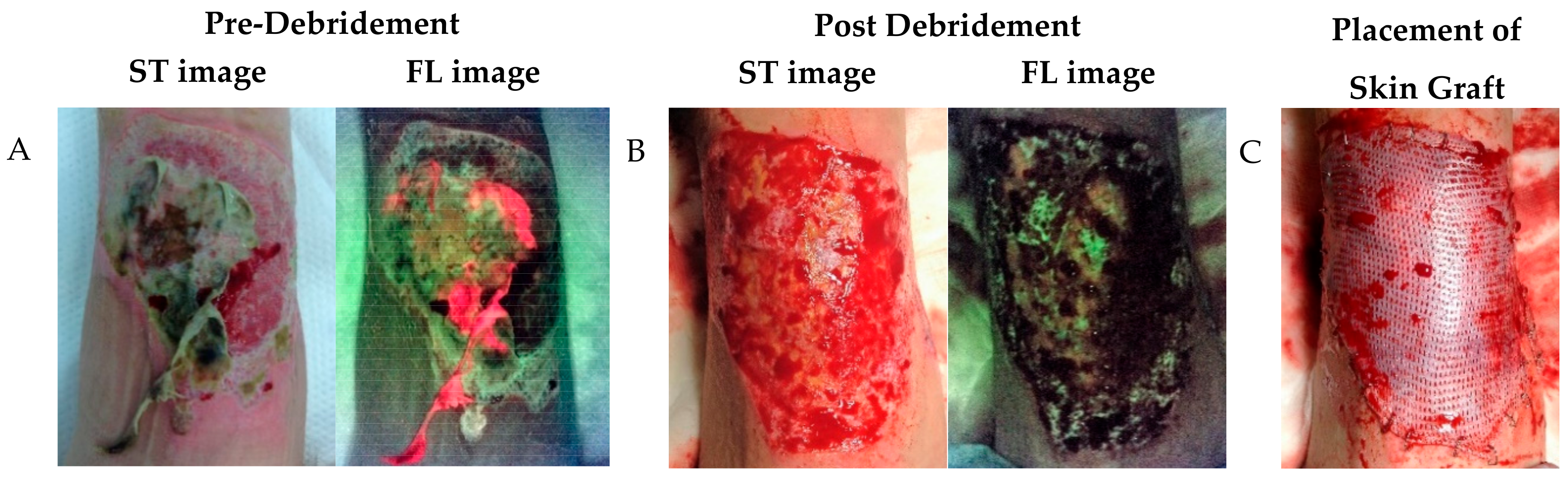
5. Impact of Fluorescence Imaging on Wound Care, Including Burns
Case Study 2
6. Limitations of Fluorescence Imaging: Another Tool in the Toolbox
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Guest, J.F.; Ayoub, N.; McIlwraith, T.; Uchegbu, I.; Gerrish, A.; Weidlich, D.; Vowden, K.; Vowden, P. Health economic burden that different wound types impose on the UK’s National Health Service. Int. Wound J. 2017, 14, 322–330. [Google Scholar] [CrossRef]
- Church, D.; Elsayed, S.; Reid, O.; Winston, B.; Lindsay, R. Burn wound infections. Clin. Microbiol. Rev. 2006, 19, 403–434. [Google Scholar] [CrossRef]
- Lachiewicz, A.M.; Hauck, C.G.; Weber, D.J.; Cairns, B.A.; van Duin, D. Bacterial Infections After Burn Injuries: Impact of Multidrug Resistance. Clin. Infect. Dis. 2017, 65, 2130–2136. [Google Scholar] [CrossRef]
- Rowan, M.P.; Cancio, L.C.; Elster, E.A.; Burmeister, D.M.; Rose, L.F.; Natesan, S.; Chan, R.K.; Christy, R.J.; Chung, K.K. Burn wound healing and treatment: Review and advancements. Crit. Care 2015, 19, 243. [Google Scholar] [CrossRef]
- Reddy, M.; Gill, S.S.; Wu, W.; Kalkar, S.R.; Rochon, P.A. Does this patient have an infection of a chronic wound? JAMA 2012, 307, 605–611. [Google Scholar] [CrossRef]
- Copeland-Halperin, L.R.; Kaminsky, A.J.; Bluefeld, N.; Miraliakbari, R. Sample procurement for cultures of infected wounds: A systematic review. J. Wound Care 2016, 25, S4–S6. [Google Scholar] [CrossRef] [PubMed]
- Le, L.; Baer, M.; Briggs, P.; Bullock, N.; Cole, W.; DiMarco, D.; Hamil, R.; Harrell, K.; Kasper, M.A.; Li, W.; et al. Diagnostic Accuracy of Point-of-Care Fluorescence Imaging for the Detection of Bacterial Burden in Wounds: Results from the 350-Patient FLAAG Trial. Adv. Wound Care 2020. [Google Scholar] [CrossRef] [PubMed]
- Rennie, M.Y.; Dunham, D.; Lindvere-Teene, L.; Raizman, R.; Hill, R.; Linden, R. Understanding Real-Time Fluorescence Signals from Bacteria and Wound Tissues Observed with the MolecuLight i:X(TM). Diagnostics 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.; Harding, K.G. Bacteria and wound healing. Curr. Opin. Infect. Dis. 2004, 17, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Bowler, P.G.; Duerden, B.I.; Armstrong, D.G. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 2001, 14, 244–269. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “house of biofilm cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef]
- Clinton, A.; Carter, T. Chronic Wound Biofilms: Pathogenesis and Potential Therapies. Lab. Med. 2015, 46, 277–284. [Google Scholar] [CrossRef]
- Balcázar, J.L.; Subirats, J.; Borrego, C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015, 6, 1216. [Google Scholar] [CrossRef]
- Wound infection in clinical practice (IWII). Wounds Int. 2016.
- Xu, L.; McLennan, S.V.; Lo, L.; Natfaji, A.; Bolton, T.; Liu, Y.; Twigg, S.M.; Yue, D.K. Bacterial load predicts healing rate in neuropathic diabetic foot ulcers. Diabetes Care 2007, 30, 378–380. [Google Scholar] [CrossRef] [PubMed]
- Lipsky, B.A.; Dryden, M.; Gottrup, F.; Nathwani, D.; Seaton, R.A.; Stryja, J. Antimicrobial stewardship in wound care: A Position Paper from the British Society for Antimicrobial Chemotherapy and European Wound Management Association. J. Antimicrob. Chemother. 2016, 71, 3026–3035. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, M.D. Bacteria and Antibiotics in Wound Healing. Surg. Clin. N. Am. 2020, 100, 757–776. [Google Scholar] [CrossRef] [PubMed]
- Breidenbach, W.C.; Trager, S. Quantitative culture technique and infection in complex wounds of the extremities closed with free flaps. Plast. Reconstr. Surg. 1995, 95, 860–865. [Google Scholar] [CrossRef]
- World Union of Wound Healing Societies. Principles of best practice: Diagnostics and wounds. A consensus document; London MEP Ltd.: London, UK, 2008. [Google Scholar]
- Gutowski, K.A. Grabb & Smith’s Plastic Surgery, 6th Edition. Plast. Reconstr. Surg. 2007, 120. [Google Scholar]
- Gardner, S.E.; Frantz, R.A.; Doebbeling, B.N. The validity of the clinical signs and symptoms used to identify localized chronic wound infection. Wound Repair Regen. 2001, 9, 178–186. [Google Scholar] [CrossRef]
- Serena, T.E.; Hanft, J.R.; Snyder, R. The lack of reliability of clinical examination in the diagnosis of wound infection: Preliminary communication. Int. J. Low. Extrem. Wounds 2008, 7, 32–35. [Google Scholar] [CrossRef]
- Gardner, S.E.; Hillis, S.L.; Frantz, R.A. Clinical signs of infection in diabetic foot ulcers with high microbial load. Biol. Res. Nurs. 2009, 11, 119–128. [Google Scholar] [CrossRef]
- Panuncialman, J.; Hammerman, S.; Carson, P.; Falanga, V. Wound edge biopsy sites in chronic wounds heal rapidly and do not result in delayed overall healing of the wounds. Wound Repair Regen. 2010, 18, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.E.; Robinowitz, N.; Chaulk, P.; Johnson, K. Comparison of chronic wound culture techniques: Swab versus curetted tissue for microbial recovery. Br. J. Community Nurs. 2014, 19, S22–S26. [Google Scholar] [CrossRef]
- Nelson, E.A.; Wright-Hughes, A.; Brown, S.; Lipsky, B.A.; Backhouse, M.; Bhogal, M.; Ndosi, M.; Reynolds, C.; Sykes, G.; Dowson, C.; et al. Concordance in diabetic foot ulceration: A cross-sectional study of agreement between wound swabbing and tissue sampling in infected ulcers. Health Technol. Assess. 2016, 20, 1–176. [Google Scholar] [CrossRef] [PubMed]
- Cross, H.H. Obtaining a wound swab culture specimen. Nursing2020 2014, 44, 68–69. [Google Scholar] [CrossRef]
- Mutluoglu, M.; Uzun, G.; Turhan, V.; Gorenek, L.; Ay, H.; Lipsky, B.A. How reliable are cultures of specimens from superficial swabs compared with those of deep tissue in patients with diabetic foot ulcers? J. Diabetes Complicat. 2012, 26, 225–229. [Google Scholar] [CrossRef]
- Loesche, M.; Gardner, S.E.; Kalan, L.; Horwinski, J.; Zheng, Q.; Hodkinson, B.P.; Tyldsley, A.S.; Franciscus, C.L.; Hillis, S.L.; Mehta, S.; et al. Temporal Stability in Chronic Wound Microbiota Is Associated With Poor Healing. J. Investig. Dermatol. 2017, 137, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, D.D.; Wolcott, R.D.; Sun, Y.; Dowd, S.E. Comparison of culture and molecular identification of bacteria in chronic wounds. Int. J. Mol. Sci. 2012, 13, 2535–2550. [Google Scholar] [CrossRef]
- Jones, L.M.; Rennie, M.Y.; Teene, L.; D’Souza, A.; Serena, T.E. Quantitative vs. Semi-Quantitative Measurements of Bacterial Load in Wounds: Assessment of 1053 Data Points from a 350-Patient Trial. In Proceedings of the 2020 Symposium on Advanced Wound Care (SAWC) Fall, Phoenix, AZ, USA, 4–6 November 2020. Virtual Symposium. [Google Scholar]
- Zekri, A.; King, W. Success of skin grafting on a contaminated recipient surface. Eur. J. Plast. Surg. 1995, 18, 40–42. [Google Scholar] [CrossRef]
- Høgsberg, T.; Bjarnsholt, T.; Thomsen, J.S.; Kirketerp-Møller, K. Success Rate of Split-Thickness Skin Grafting of Chronic Venous Leg Ulcers Depends on the Presence of Pseudomonas aeruginosa: A Retrospective Study. PLoS ONE 2011, 6, e20492. [Google Scholar] [CrossRef]
- Rennie, M.Y.; Lindvere-Teene, L.; Tapang, K.; Linden, R. Point-of-care fluorescence imaging predicts the presence of pathogenic bacteria in wounds: A clinical study. J. Wound Care 2017, 26, 452–460. [Google Scholar] [CrossRef]
- Hurley, C.M.; McClusky, P.; Sugrue, R.M.; Clover, J.A.; Kelly, J.E. Efficacy of a bacterial fluorescence imaging device in an outpatient wound care clinic: A pilot study. J. Wound Care 2019, 28, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Serena, T.E.; Harrell, K.; Serena, L.; Yaakov, R.A. Real-time bacterial fluorescence imaging accurately identifies wounds with moderate-to-heavy bacterial burden. J. Wound Care 2019, 28, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.; Woo, K.Y. A prospective multi-site observational study incorporating bacterial fluorescence information into the UPPER / LOWER wound infection checklists. Wounds 2020, 32, 299–308. [Google Scholar] [PubMed]
- Monici, M. Cell and tissue autofluorescence research and diagnostic applications. Biotechnol. Annu. Rev. 2005, 11, 227–256. [Google Scholar] [CrossRef] [PubMed]
- McGinley, K.J.; Webster, G.F.; Leyden, J.J. Facial follicular porphyrin fluorescence: Correlation with age and density of Propionibacterium acnes. Br. J. Dermatol. 1980, 102, 437–441. [Google Scholar] [CrossRef]
- Holt, P.; El-Dars, L.; Kenny, A.; Lake, A. Serial photography and Wood’s light examination as an aid to the clinical diagnosis of dermatitis artefacta. J. Vis. Commun. Med. 2013, 36, 31–34. [Google Scholar] [CrossRef]
- Kaliyadan, F.; Kuruvilla, J. Using a hand-held black-light source instead of a Wood’s lamp. J. Am. Acad. Dermatol. 2015, 72, e153–e154. [Google Scholar] [CrossRef]
- Ponka, D.; Baddar, F. Wood lamp examination. Can. Fam. Physician 2012, 58, 976. [Google Scholar] [PubMed]
- Philipp-Dormston, W.K.; Doss, M. Comparison of porphyrin and heme biosynthesis in various heterotrophic bacteria. Enzyme 1973, 16, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Nitzan, Y.; Salmon-Divon, M.; Shporen, E.; Malik, Z. ALA induced photodynamic effects on Gram positive and negative bacteria. Photochem. Photobiol. Sci. 2004, 3, 430–435. [Google Scholar] [CrossRef]
- Nitzan, Y.; Kauffman, M. Endogenous Porphyrin Production in Bacteria by $δ$-Aminolaevulinic Acid and Subsequent Bacterial Photoeradication. Lasers Med. Sci. 1999, 14, 269–277. [Google Scholar] [CrossRef]
- Meyer, J.M.; Abdallag, M.A. The Fluorescent Pigment of Pseudomonas fluorescens: Biosynthesis, Purification and Physicochemical Properties. Microbiology 1978, 107, 319–328. [Google Scholar] [CrossRef]
- Cody, Y.S.; Gross, D.C. Characterization of Pyoverdin(pss), the Fluorescent Siderophore Produced by Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 1987, 53, 928–934. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.M.; Lopez, A.; Rennie, M.Y.; Dunham, D.; Dacosta, R.S.; Smith, A.C. In Vivo and In Vitro Detection of Porphyrin-Producing Wound Pathogens, Planktonic and in Biofilm, with Real-Time Bacterial Fluorescence Imaging. Presented at Wounds Canada 2019, Niagara Falls, ON, Canada, 4–6 October 2019. [Google Scholar]
- Farhan, N.; Jeffery, S. Utility of MolecuLight i:X for Managing Bacterial Burden in Pediatric Burns. J. Burn Care Res. 2020, 41, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Ottolino-Perry, K.; Chamma, E.; Blackmore, K.M.; Lindvere-Teene, L.; Starr, D.; Tapang, K.; Rosen, C.F.; Pitcher, B.; Panzarella, T.; Linden, R.; et al. Improved detection of clinically relevant wound bacteria using autofluorescence image-guided sampling in diabetic foot ulcers. Int. Wound J. 2017, 14, 833–841. [Google Scholar] [CrossRef]
- Chew, B.J.W.; Griffin, M.; Butler, P.E.; Mosahebi, A. The use of MolecuLight i:X device in acute hand trauma. J. Plast. Reconstr. Aesthet. Surg. 2020, 73, 1357–1404. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.M.; Jacob, A.; Rennie, M.Y. A prospective, multi-site study evaluating the use of fluorescence imaging for the identification of bacteria-containing wounds in the context of long-term care facilities. In Proceedings of the 2020 Symposium on Advanced Wound Care (SAWC) Fall, Phoenix, AZ, USA, 4–6 November 2020. Virtual Symposium. [Google Scholar]
- Alawi, S.A.; Limbourg, A.; Strauss, S.; Vogt, P.M. Imaging of bacteria in burn wounds treated with split-thicknessgrafts in MEEK/MESH technique: A pilot study with first experiences in clinical wound evaluation with autofluorescence. Handchir. Mikrochir. Plast. Chir. 2019, 51, 130–138. [Google Scholar] [CrossRef]
- Blackshaw, E.L.; Jeffery, S.L.A. Efficacy of an imaging device at identifying the presence of bacteria in wounds at a plastic surgery outpatients clinic. J. Wound Care 2018, 27, 20–26. [Google Scholar] [CrossRef]
- Blumenthal, E.; Jeffery, S.L.A. The Use of the MolecuLight i:X in Managing Burns: A Pilot Study. J. Burn Care Res. 2018, 39, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.M.; Dunham, D.; Rennie, M.Y.; Kirman, J.; Lopez, A.J.; Keim, K.C.; Little, W.; Gomez, A.; Bourke, J.; Ng, H.; et al. In vitro detection of porphyrin-producing wound bacteria with real-time fluorescence imaging. Future Microbiol. 2020, 15, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Moelleken, M.; Jockenhofer, F.; Benson, S.; Dissemond, J. Prospective clinical study on the efficacy of bacterial removal with mechanical debridement in and around chronic leg ulcers assessed with fluorescence imaging. Int. Wound J. 2020, 17, 1011–1018. [Google Scholar] [CrossRef]
- Raizman, R.; Dunham, D.; Lindvere-Teene, L.; Jones, L.M.; Tapang, K.; Linden, R.; Rennie, M.Y. Use of a bacterial fluorescence imaging device: Wound measurement, bacterial detection and targeted debridement. J. Wound Care 2019, 28, 824–834. [Google Scholar] [CrossRef]
- Sapico, F.L.; Ginunas, V.J.; Thornhill-Joynes, M.; Canawati, H.N.; Capen, D.A.; Klein, N.E.; Khawam, S.; Montgomerie, J.Z. Quantitative microbiology of pressure sores in different stages of healing. Diagn. Microbiol. Infect. Dis. 1986, 5, 31–38. [Google Scholar] [CrossRef]
- Raizman, R. Fluorescence imaging guided dressing change frequency during negative pressure wound therapy: A case series. J. Wound Care 2019, 28, S28–S37. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.; Rennie, M.Y.; Douglas, J. Using Bacterial Fluorescence Imaging and Antimicrobial Stewardship to Guide Wound Management Practices: A Case Series. Ostomy Wound Manag. 2018, 64, 18–28. [Google Scholar] [CrossRef]
- Jeffery, S. The utility of MolecuLight bacterial sensing in the management of burns and traumatic wounds. In Proceedings of the Photonic Diagnosis and Treatment of Infections and Inflammatory Diseases II; International Society for Optics and Photonics: San Francisco, CA, USA, 2019; Volume 10863, p. 1086304. [Google Scholar]
- Kleintjes, W.G.; Plast, M.; Plast, F.C.; Sa, S.; Kotzee, E.P.; Chb, M.B. MolecuLight i:X: A new tool for wound infection diagnosis. South Afr. J. Plast. Reconstr. Aesthetic Surg. Burn. 2019, 2, 68–71. [Google Scholar] [CrossRef]
- Kim, P.J.; Attinger, C.E.; Bigham, T.; Hagerty, R.; Platt, S.; Anghel, E.; Steinberg, J.S.; Evans, K.K. Clinic-based Debridement of Chronic Ulcers Has Minimal Impact on Bacteria. Wounds Compend. Clin. Res. Pract. 2018, 30, 114–119. [Google Scholar]
- Schalk, I.J.; Guillon, L. Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: Implications for metal homeostasis. Environ. Microbiol. 2013, 15, 1661–1673. [Google Scholar] [CrossRef]
- Cavallaro, G.; Decaria, L.; Rosato, A. Genome-based analysis of heme biosynthesis and uptake in prokaryotic systems. J. Proteome Res. 2008, 7, 4946–4954. [Google Scholar] [CrossRef] [PubMed]
- Choby, J.E.; Skaar, E.P. Heme Synthesis and Acquisition in Bacterial Pathogens. J. Mol. Biol. 2016, 428, 3408–3428. [Google Scholar] [CrossRef] [PubMed]
- Wolcott, R.D.; Hanson, J.D.; Rees, E.J.; Koenig, L.D.; Phillips, C.D.; Wolcott, R.A.; Cox, S.B.; White, J.S. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2016, 24, 163–174. [Google Scholar] [CrossRef]
- Serena, T.; Serena, K.; Serena, L.; Sabo, M.; Patel, K.; Le, L.; Halperin, G.; Briggs, P.; Baer, M.; Thibodeaux, K. Bacterial fluorescence image guidance of antimicrobial decision making and stewardship. Presented at European Wound Management Association (EWMA) Conference, Gothenburg, Sweden, 5–7 June 2019. [Google Scholar]
- Aung, B. Can Fluorescence Imaging Predict the Success of CTPs for Wound Closure and Save Costs? Today’s Wound Clin. 2019, 13, 22–25. [Google Scholar]
- Cole, W.; Coe, S. Use of a bacterial fluorescence imaging system to target wound debridement and accelerate healing: A pilot study. J. Wound Care 2020, 29, S44–S52. [Google Scholar] [CrossRef]
- DaCosta, R.S.; Kulbatski, I.; Lindvere-Teene, L.; Starr, D.; Blackmore, K.; Silver, J.I.; Opoku, J.; Wu, Y.C.; Medeiros, P.J.; Xu, W.; et al. Point-of-care autofluorescence imaging for real-time sampling and treatment guidance of bioburden in chronic wounds: First-in-human results. PLoS ONE 2015, 10, e0116623. [Google Scholar] [CrossRef]
- Rahma, S.; Woods, J.; Nixon, J.; Brown, S.; Russell, D. The use of Point-of-Care Bacterial Autofluorescence Imaging in the Management of Diabetic Foot Ulcers: A Pilot Randomised Controlled Trial. In Proceedings of the 2020 Symposium on Advanced Wound Care (SAWC) Fall, Phoenix, AZ, USA, 4–6 November 2020. Virtual Symposium. [Google Scholar]
- Clement, M.; Daniel, G.; Trelles, M. Optimising the design of a broad-band light source for the treatment of skin. J. Cosmet. Laser Ther. Off. Publ. Eur. Soc. Laser Dermatol. 2005, 7, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Ash, C.; Dubec, M.; Donne, K.; Bashford, T. Effect of wavelength and beam width on penetration in light-tissue interaction using computational methods. Lasers Med. Sci. 2017, 32, 1909–1918. [Google Scholar] [CrossRef] [PubMed]
| Local Infection | Spreading Infection | ||||
|---|---|---|---|---|---|
| Covert (Subtle Signs) | Overt (Classic) Signs | ||||
| Hypergranulation (excessive “vascular” tissue) Epithelial bridging and pocketing in granulation tissue Wound breakdown and enlargement Delayed wound healing beyond expectations New or increased pain Increasing malodor | Erythema Local warmth Swelling Purulent discharge Delayed wound healing beyond expectations New or increasing pain Increased malodor | Extending induration Lymphangitis Crepitus Wound breakdown/dehiscence with or without satellite lesions Malaise/lethargy or non-specific general deterioration Loss of appetite Inflammation, swelling, or lymph glands | |||
| The number of covert signs present: | /7 | The number of overt signs present: | /7 | The number of spreading signs present: | /7 |
| Author | n | Study Design | Wound Types | Care Setting | Sampling Method | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Le et al. [7] | 350 | multi-center controlled, observational clinical trial | DFU, PU, VLU, SS, other | outpatient wound care centres | biopsy | 59 | 89 | 96 | 32 | 64 |
| Chew et al. [51] | 35 | observational study | hand trauma wounds | outpatient | swab | 100 | 97 | 67 | 100 | 97 |
| Jones et al. [56] | 36 | multi-site observational study | DFU, PU, VLU | long-term care | swab | 100 | 94 | 94 | ||
| Hill et al. [37] | 43 | multi-center prospective observational | DFU, PU, VLU, SS, other | inpatient, outpatient | swabs | 100 | 100 | 100 | 100 | 100 |
| Hurley et al. [35] | 33 | single-center prospective observational | lower-limb wounds | outpatient | swabs (43) | 100 | 78 | 95 | 100 | 96 |
| Serena et al. [36] | 19 | single-center prospective observational clinical trial | VLU, DFU | advanced-wound care centres | biopsy | 73 | 100 | 100 | 17 | 74 |
| Farhan & Jeffrey [49] | 16 | observational study | burn | pediatric burns outpatient centre | Levine swabs | 100 | 72 | 63 | 100 | 82 |
| Alawi et al. [53] | 14 | pilot observational study | burn | not reported | swabs | 87 | 88 | 82 | 90 | 87 |
| Blackshaw & Jeffrey [54] | 14 | observational study | burn, trauma | burns outpatient department | swabs | 100 | 89 | 89 | 100 | 94 |
| Blumenthal & Jeffrey [55] | 20 | observational study | burn | burns outpatient department | swabs | 81 | 75 | 93 | 50 | 80 |
| Ottolino-Perry [50] | 33 | non-randomised clinical trial | DFU | wound care centre | swabs | 78 | 78 | 64 | 88 | 78 |
| Average | 89 | 87 | 87 | 78 | 86 | |||||
| Weighted Average | 74 | 88 | 91 | 53 | 75 | |||||
| Detected in Vitro (Based on Red Porphyrin Fluorescence) | Detected in Clinical Studies (Based on Sampling in Areas of Red or Cyan Fluorescence) | |
|---|---|---|
| Genus | Species | |
| Staphylococcus | aureus | [7,8,34,35,36,37,49,50,54,55,58,60,61,62,63,64] |
| epidermidis | [7,50,58] | |
| capitis | [7] | |
| lugdunensis | [7,34,35,37] | |
| Pseudomonas | aeruginosa | [7,8,35,36,37,49,54,55,58,62,63,64] |
| putida | ||
| Escherichia | coli | [7,37,50,55,58,61,62] |
| Corynebacterium | striatum | [7,36] |
| Proteus | mirabilis | [7,8,34,35,52,58,62,63] |
| vulgaris | [7,60] | |
| Enterobacter | cloacae | [7,8,34,50,55,62] |
| Serratia | marcescens | [7,8,55,58] |
| Acinetobacter | baumannii | [7,52,61] |
| Klebsiella | pneumoniae | [7,34,36,37,55,58,64] |
| oxytoca | [7] | |
| Morganella | morganii | [7,60,61] |
| Propionibacterium | acnes | [7,36,60] |
| Stenotrophomonas | maltophilia | [7,55,62] |
| Bacteroides | fragilis | [7,60,62] |
| Aeromonas | hydrophila | |
| Alcaligenes | faecalis | [7] |
| Bacillus | cereus | |
| Citrobacter | koseri | [7,34] |
| freundii | [7,55] | |
| Clostridium | perfringens | [7,36] |
| Listeria | monocytogenes | |
| inocua | ||
| Peptostreptococcus | anaerobius | [7] |
| Veillonella | parvula | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farhan, N.; Jeffery, S. Diagnosing Burn Wounds Infection: The Practice Gap & Advances with MolecuLight Bacterial Imaging. Diagnostics 2021, 11, 268. https://doi.org/10.3390/diagnostics11020268
Farhan N, Jeffery S. Diagnosing Burn Wounds Infection: The Practice Gap & Advances with MolecuLight Bacterial Imaging. Diagnostics. 2021; 11(2):268. https://doi.org/10.3390/diagnostics11020268
Chicago/Turabian StyleFarhan, Nawras, and Steven Jeffery. 2021. "Diagnosing Burn Wounds Infection: The Practice Gap & Advances with MolecuLight Bacterial Imaging" Diagnostics 11, no. 2: 268. https://doi.org/10.3390/diagnostics11020268
APA StyleFarhan, N., & Jeffery, S. (2021). Diagnosing Burn Wounds Infection: The Practice Gap & Advances with MolecuLight Bacterial Imaging. Diagnostics, 11(2), 268. https://doi.org/10.3390/diagnostics11020268






