Genomic Characterization of VIM and MCR Co-Producers: The First Two Clinical Cases, in Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Presentation, Antimicrobial Susceptibility Test and Molecular Investigations
2.2. Conjugation/Transformation Assay
2.3. Whole-Genome Sequencing (WGS)
2.4. Whole-Genome-Sequencing-Data Analysis
3. Results
3.1. Isolates Susceptibility Profiles
3.2. Plasmid Transferability
3.3. Whole-Genome Characterization
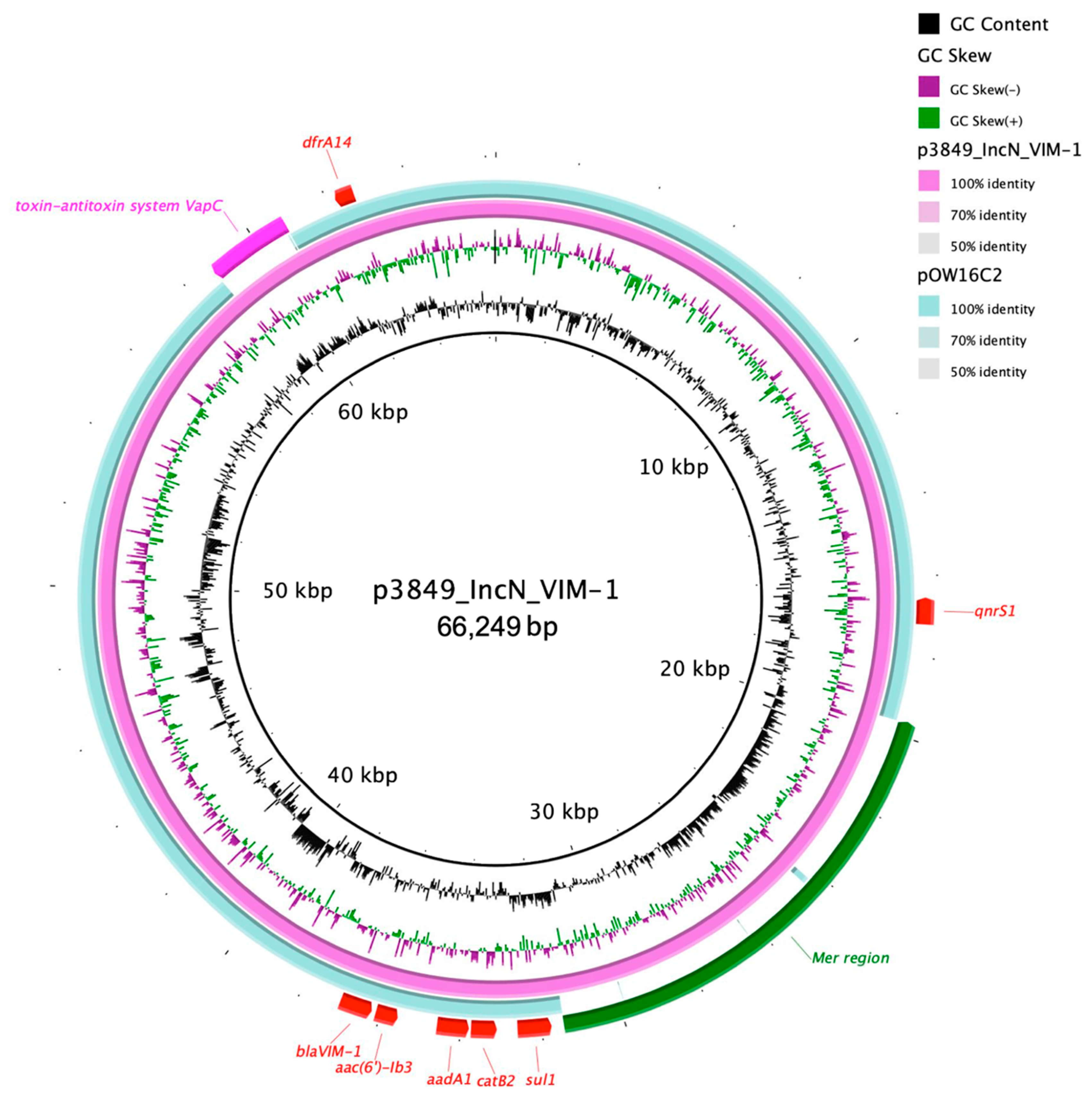
3.3.1. ENCL_3849
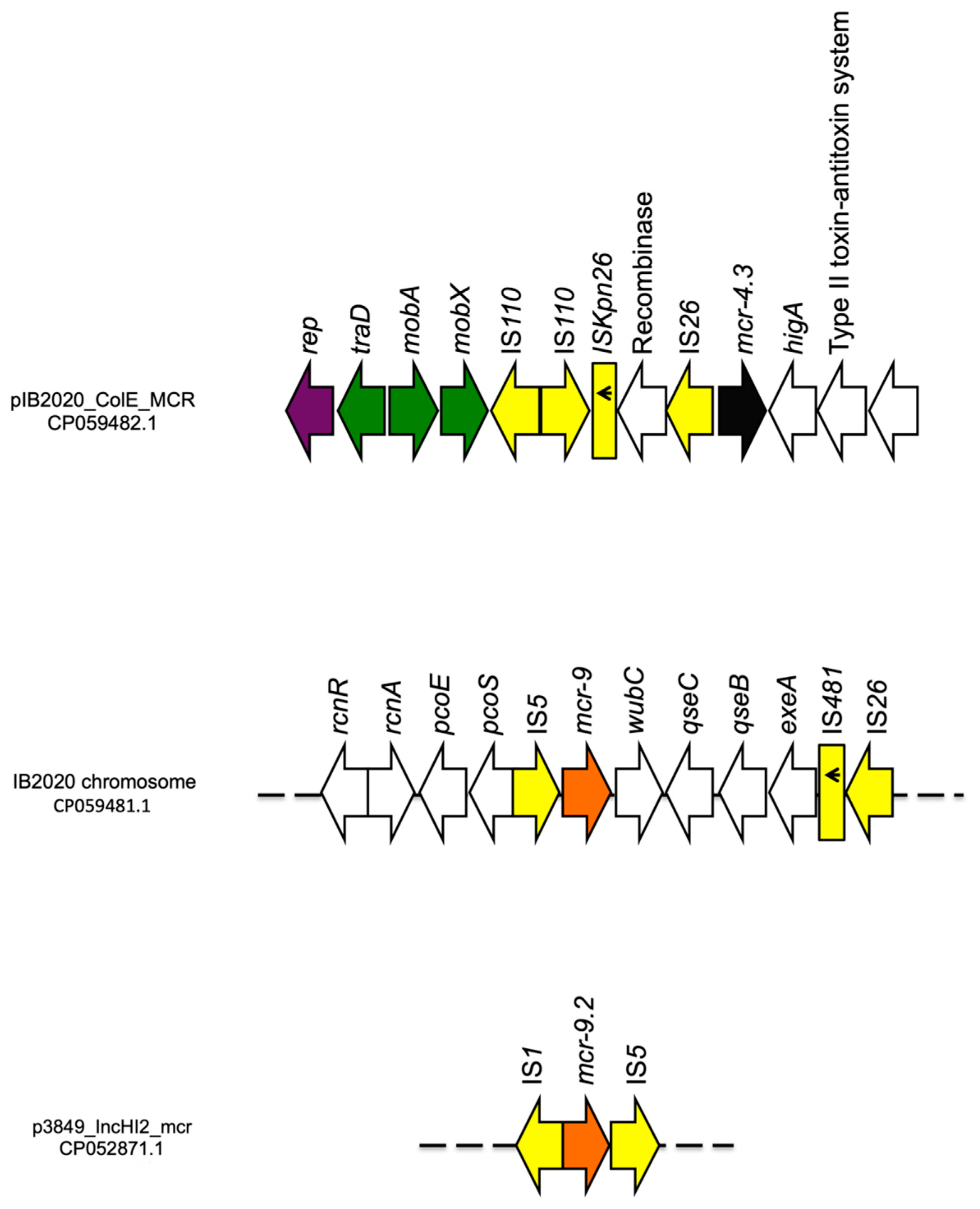
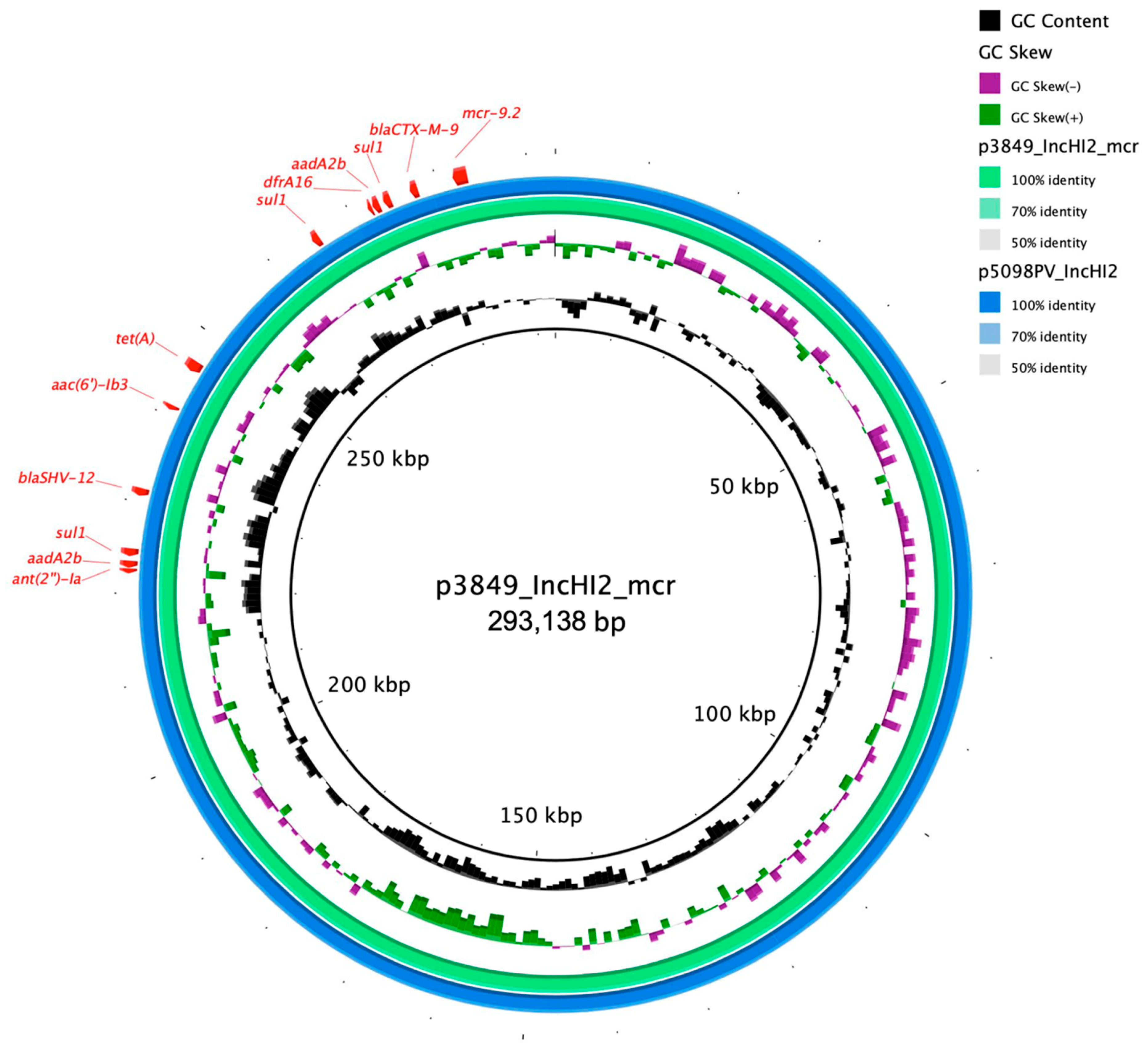
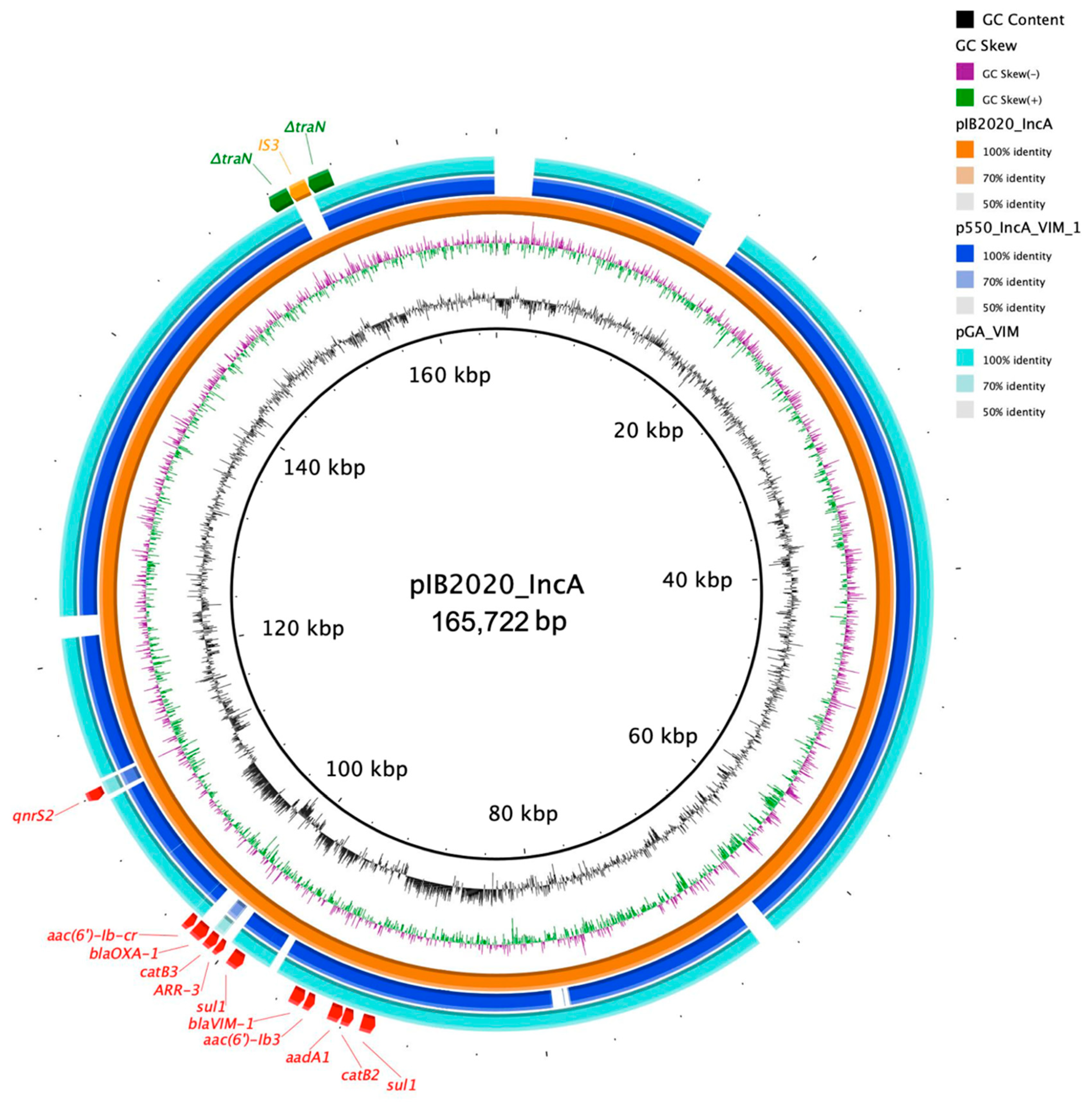
3.3.2. ENCB_IB2020
4. Discussion and Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mustafa, A.; Ibrahim, M.; Rasheed, M.A.; Kanwal, S.; Hussain, A.; Sami, A.; Ahmed, R.; Bo, Z. Genome-wide analysis of four enterobacter cloacae complex type strains: Insights into virulence and niche adaptation. Sci. Rep. 2020, 10, 8150. [Google Scholar] [CrossRef]
- Annavajhala, M.K.; Gomez-Simmonds, A.; Uhlemann, A.C. Multidrug-resistant enterobacter cloacae complex emerging as a global, diversifying threat. Front. Microbiol. 2019, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Petrosillo, N.; Taglietti, F.; Granata, G. Treatment options for colistin resistant Klebsiella pneumoniae: Present and future. J. Clin. Med. 2019, 8, 934. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Villa, L.; Feudi, C.; Curcio, L.; Orsini, S.; Luppi, A.; Pezzotti, G.; Magistrali, C.F. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017, 22, 30589. [Google Scholar] [CrossRef]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype typhimurium isolate. mBio 2019, 10, e00853-19. [Google Scholar] [CrossRef]
- Lauretti, L.; Riccio, M.L.; Mazzariol, A.; Cornaglia, G.; Amicosante, G.; Fontana, R.; Rossolini, G.M. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 1999, 43, 1584–1590. [Google Scholar] [CrossRef]
- Piazza, A.; Comandatore, F.; Romeri, F.; Brilli, M.; Dichirico, B.; Ridolfo, A.; Antona, C.; Bandi, C.; Gismondo, M.R.; Rimoldi, S.G. Identification of blaVIM-1 gene in ST307 and ST661 Klebsiella pneumoniae clones in Italy: Old acquaintances for new combinations. Microb. Drug Resist. 2019, 25, 787–790. [Google Scholar] [CrossRef]
- Di Tella, D.; Tamburro, M.; Guerrizio, G.; Fanelli, I.; Sammarco, M.L.; Ripabelli, G. Molecular epidemiological insights into colistin-resistant and carbapenemases-producing clinical Klebsiella pneumoniae isolates. Infect. Drug Resist. 2019, 12, 3783–3795. [Google Scholar] [CrossRef]
- Simoni, S.; Caucci, S.; Brenciani, A.; Morroni, G.; Giovanetti, E.; Menzo, S.; Facinelli, B.; Mingoia, M. Increase and diversity of carbapenemase-producing Escherichia coli isolates, Italy. Future Microbiol. 2019, 14, 1035–1042. [Google Scholar] [CrossRef]
- Arcari, G.; Di Lella, F.M.; Bibbolino, G.; Mengoni, F.; Beccaccioli, M.; Antonelli, G.; Faino, L.; Carattoli, A. A multispecies cluster of VIM-1 carbapenemase-producing Enterobacterales linked by a novel, highly conjugative, and broad-host-range IncA plasmid forebodes the reemergence of VIM-1. Antimicrob. Agents Chemother. 2020, 64, e02435-19. [Google Scholar] [CrossRef] [PubMed]
- Giani, T.; Antonelli, A.; Sennati, S.; Di Pilato, V.; Chiarelli, A.; Cannatelli, A.; Gatsch, C.; Luzzaro, F.; Spanu, T.; Stefani, S.; et al. Results of the Italian infection-carbapenem resistance evaluation surveillance trial (iCREST-IT): Activity of ceftazidime/avibactam against Enterobacterales isolated from urine. J. Antimicrob. Chemother. 2020, 75, 979–983. [Google Scholar] [CrossRef] [PubMed]
- Calia, C.; Pazzani, C.; Oliva, M.; Scrascia, M.; Lovreglio, P.; Capolongo, C.; Dionisi, A.M.; Chiarelli, A.; Monno, R. Carbapenemases-producing Klebsiella pneumoniae in hospitals of two regions of Southern Italy. APMIS 2017, 125, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Principe, L.; Piazza, A.; Mauri, C.; Anesi, A.; Bracco, S.; Brigante, G.; Casari, E.; Agrappi, C.; Caltagirone, M.; Novazzi, F.; et al. Multicenter prospective study on the prevalence of colistin resistance in Escherichia coli: Relevance of mcr-1-positive clinical isolates in Lombardy, Northern Italy. Infect. Drug Resist. 2018, 11, 377–385. [Google Scholar] [CrossRef]
- Del Bianco, F.; Morotti, M.; Pedna, M.F.; Farabegoli, P.; Sambri, V. Microbiological surveillance of plasmid mediated colistin resistance in human Enterobacteriaceae isolates in Romagna (Northern Italy): August 2016–July 2017. Int. J. Infect. Dis. 2018, 69, 96–98. [Google Scholar] [CrossRef]
- Carretto, E.; Brovarone, F.; Nardini, P.; Russello, G.; Barbarini, D.; Pongolini, S.; Gagliotti, C.; Carattoli, A.; Sarti, M. Detection of mcr-4 positive Salmonella enterica serovar Typhimurium in clinical isolates of human origin, Italy, October to November 2016. Euro Surveill. 2018, 23, 17-00821. [Google Scholar] [CrossRef]
- Lee, K.; Lim, Y.S.; Yong, D.; Yum, J.H.; Chong, Y. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 2003, 41, 4623–4629. [Google Scholar] [CrossRef]
- Doi, Y.; Potoski, B.A.; Adams-Haduch, J.M.; Sidjabat, H.E.; Pasculle, A.W.; Paterson, D.L. Simple disk-based method for detection of Klebsiella pneumoniae carbapenemase-type-lactamase by use of a boronic acid compound. J. Clin. Microbiol. 2008, 46, 4083–4086. [Google Scholar] [CrossRef]
- Glupczynski, Y.; Huang, T.-D.; Bouchahrouf, W.; De Castro, R.R.; Bauraing, C.; Gérard, M.; Verbruggen, A.-M.; Deplano, A.; Denis, O.; Bogaerts, P. Rapid emergence and spread of OXA-48-producing carbapenem-resistant Enterobacteriaceae isolates in Belgian hospitals. Int. J. Antimicrob. Agents 2012, 39, 168–172. [Google Scholar] [CrossRef]
- Ataei, B.; Shirani, K.; Roshandel, F. Antibiotic resistance pattern and evaluation of metallo-beta lactamase genes (VIM and IMP) in Pseudomonas aeruginosa strains producing MBL enzyme, isolated from patients with secondary immunodeficiency. Adv. Biomed. Res. 2016, 5, 124. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST. org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and Plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Moura, A.; Soares, M.; Pereira, C.; Leitao, N.; Henriques, I.; Correia, A. INTEGRALL: A database and search engine for integrons, integrases and gene cassettes. Bioinformatics 2009, 25, 1096–1098. [Google Scholar] [CrossRef]
- Breland, E.J.; Zhang, E.W.; Bermudez, T.; Martinez, C.R.; Hadjifrangiskou, M. The histidine residue of QseC is required for canonical signaling between QseB and PmrB in uropathogenic Escherichia coli. J. Bacteriol. 2017, 199, e00060-17. [Google Scholar] [CrossRef]
- Rahube, T.O.; Yost, C.K. Characterization of a mobile and multiple resistance plasmid isolated from swine manure and its detection in soil after manure application. J. Appl. Microbiol. 2012, 112, 1123–1133. [Google Scholar] [CrossRef]
- Matsumura, Y.; Peirano, G.; Devinney, R.; Bradford, P.A.; Motyl, M.R.; Adams, M.D.; Chen, L.; Kreiswirth, B.; Pitout, J.D.D. Genomic epidemiology of global VIM-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2017, 72, 2249–2258. [Google Scholar] [CrossRef]
- Mattioni Marchetti, V.; Bitar, I.; Piazza, A.; Mercato, A.; Fogato, E.; Hrabak, J.; Migliavacca, R. Genomic insight of VIM-harboring IncA plasmid from a clinical ST69 Escherichia coli strain in Italy. Microorganisms 2020, 8, 1232. [Google Scholar] [CrossRef] [PubMed]
- Chavda, K.D.; Westblade, L.F.; Satlin, M.J.; Hemmert, A.C.; Castanheira, M.; Jenkins, S.G.; Chen, L.; Kreiswirth, B.N. First report of blaVIM-4- and mcr-9-coharboring enterobacter species isolated from a pediatric patient. mSphere 2019, 4, e00629-19. [Google Scholar] [CrossRef] [PubMed]
- Soliman, A.M.; Maruyama, F.; Zarad, H.O.; Ota, A.; Nariya, H.; Shimamoto, T.; Shimamoto, T. Emergence of a multidrug-resistant enterobacter hormaechei clinical isolate from Egypt co-harboring mcr-9 and blaVIM-4. Microorganisms 2020, 8, 595. [Google Scholar] [CrossRef] [PubMed]
- Sadek, M.; Nariya, H.; Shimamoto, T.; Kayama, S.; Yu, L.; Hisatsune, J.; Sugai, M.; Nordmann, P.; Poirel, L.; Shimamoto, T. First genomic characterization of blaVIM-1 and mcr-9-coharbouring Enterobacter hormaechei isolated from food of animal origin. Pathogens 2020, 9, 687. [Google Scholar] [CrossRef] [PubMed]
- Bitar, I.; Papagiannitsis, C.C.; Kraftova, L.; Chudejova, K.; Marchetti, V.M.; Hrabak, J. Detection of five mcr-9-carrying Enterobacterales isolates in four Czech hospitals. mSphere 2020, 5, e01008-20. [Google Scholar] [CrossRef] [PubMed]
- Karlowsky, J.A.; Kazmierczak, K.M.; de Jonge, B.L.M.; Hackel, M.A.; Sahm, D.F.; Bradford, P.A. In vitro activity of aztreonam-avibactam against Enterobacteriaceae and Pseudomonas aeruginosa isolated by clinical laboratories in 40 countries from 2012 to 2015. Antimicrob. Agents Chemother. 2017, 61, e00472-17. [Google Scholar] [CrossRef] [PubMed]
- Zhanel, G.G.; Golden, A.R.; Zelenitsky, S.; Wiebe, K.; Lawrence, C.K.; Adam, H.J.; Idowu, T.; Domalaon, R.; Schweizer, F.; Zhanel, M.A.; et al. Cefiderocol: A siderophore cephalosporin with activity against carbapenem-resistant and multidrug-resistant gram-negative bacilli. Drugs 2019, 79, 271–289. [Google Scholar] [CrossRef]
- García-Fernández, A.; Villa, L.; Moodley, A.; Hasman, H.; Miriagou, V.; Guardabassi, L.; Carattoli, A. Multilocus sequence typing of IncN plasmids. J. Antimicrob. Chemother. 2011, 66, 1987–1991. [Google Scholar] [CrossRef]
- Hancock, S.J.; Phan, M.D.; Peters, K.M.; Forde, B.M.; Chong, T.M.; Yin, W.F.; Chan, K.G.; Paterson, D.L.; Walsh, T.R.; Beatson, S.A.; et al. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob. Agents Chemother. 2016, 61, e01740-16. [Google Scholar] [CrossRef]
- Marchetti, V.M.; Bitar, I.; Mercato, A.; Nucleo, E.; Bonomini, A.; Pedroni, P.; Hrabak, J.; Migliavacca, R. Complete nucleotide sequence of plasmids of two Escherichia coli strains carrying blaNDM-5 and blaNDM-5 and blaOXA-181 from the same patient. Front. Microbiol. 2020, 10, 3095. [Google Scholar] [CrossRef]
| Isolate | AMP | AMS | ATM | CTX | TET | CAZ | COL | TZP | ETP | MEM | GEN | TOB | TGC | SXT | PIP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. cloacae 3849 | >128 | >128 | >16 | >8 | >32 | >16 | 0.25 | 64 | >2 | 0.5 | 8 | >8 | 0.5 | >4 | >128 |
| E. coli A15*3849 | >128 | >128 | >16 | >8 | >32 | >16 | 0.25 | 64 | >2 | 0.5 | 8 | >8 | 0.25 | >4 | >128 |
| E. kobei IB2020 | >128 | >128 | 1 | >8 | 2 | >16 | 8 | 64 | >2 | 16 | 0.5 | 4 | 0.25 | 0.5 | >128 |
| ID | Species | MLST | Genetic Element | Replicon | pMLST | Antibiotic Resistance Genes |
|---|---|---|---|---|---|---|
| ENCB_IB2020 (2020/8240) | E. kobei | ST 54 | Chromosome | NA | NA | fosA, mcr-9, blaACT-9 |
| pIB2020_IncA | IncA | ST 12 | aac(6′)-lb-cr, aac(6′)-lb3, qnrS2, aadA1, sul1 *, arr-3, catB2, catB3, blaOXA-1, blaVIM-1 | |||
| pIB2020_ColE_MCR | ColE | - | mcr-4.3 | |||
| pIB2020_IncFIB | IncFIB | - | - | |||
| pIB2020_L | NT | - | - | |||
| pIB2020_N | NT | - | - | |||
| pIB2020_S | NT | - | - | |||
| ENCL_3849 | E. cloacae | ST 382 | Chromosome | NA | NA | fosA, blaACT-5 |
| p3846_IncHI2_mcr | IncHI2 | ST 1 | qnrA1, aac(6′)-lb3, ant(2′’)-la, mcr-9.2, aadA2b *, sul1 *, dfrA16, tet(A), blaCTX-M-9, blaSHV-12 | |||
| p3846_IncN_VIM-1 | IncN | ST 7 | qnrS1, aac(6′)-lb3, aadA1, sul1, dfrA14, catB2, blaVIM-1 | |||
| p3846III | NT | - | - | |||
| p3849I | NT | - | - | |||
| p3849II | NT | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchetti, V.M.; Bitar, I.; Sarti, M.; Fogato, E.; Scaltriti, E.; Bracchi, C.; Hrabak, J.; Pongolini, S.; Migliavacca, R. Genomic Characterization of VIM and MCR Co-Producers: The First Two Clinical Cases, in Italy. Diagnostics 2021, 11, 79. https://doi.org/10.3390/diagnostics11010079
Marchetti VM, Bitar I, Sarti M, Fogato E, Scaltriti E, Bracchi C, Hrabak J, Pongolini S, Migliavacca R. Genomic Characterization of VIM and MCR Co-Producers: The First Two Clinical Cases, in Italy. Diagnostics. 2021; 11(1):79. https://doi.org/10.3390/diagnostics11010079
Chicago/Turabian StyleMarchetti, Vittoria Mattioni, Ibrahim Bitar, Mario Sarti, Elena Fogato, Erika Scaltriti, Chiara Bracchi, Jaroslav Hrabak, Stefano Pongolini, and Roberta Migliavacca. 2021. "Genomic Characterization of VIM and MCR Co-Producers: The First Two Clinical Cases, in Italy" Diagnostics 11, no. 1: 79. https://doi.org/10.3390/diagnostics11010079
APA StyleMarchetti, V. M., Bitar, I., Sarti, M., Fogato, E., Scaltriti, E., Bracchi, C., Hrabak, J., Pongolini, S., & Migliavacca, R. (2021). Genomic Characterization of VIM and MCR Co-Producers: The First Two Clinical Cases, in Italy. Diagnostics, 11(1), 79. https://doi.org/10.3390/diagnostics11010079





