A Deep Learning-Based Automated CT Segmentation of Prostate Cancer Anatomy for Radiation Therapy Planning-A Retrospective Multicenter Study
Abstract
1. Introduction
2. Materials and Methods
2.1. DL Delineation Model
2.2. Study Patients
2.3. Contouring
- An MC group contoured in the study into an empty dataset to meet clinical acceptance.
- An AC group that included the nonedited automatically created structure set.
- An AEC group that included the automatically created structures with expert modifications to meet clinical acceptance.
2.4. Contouring Analysis
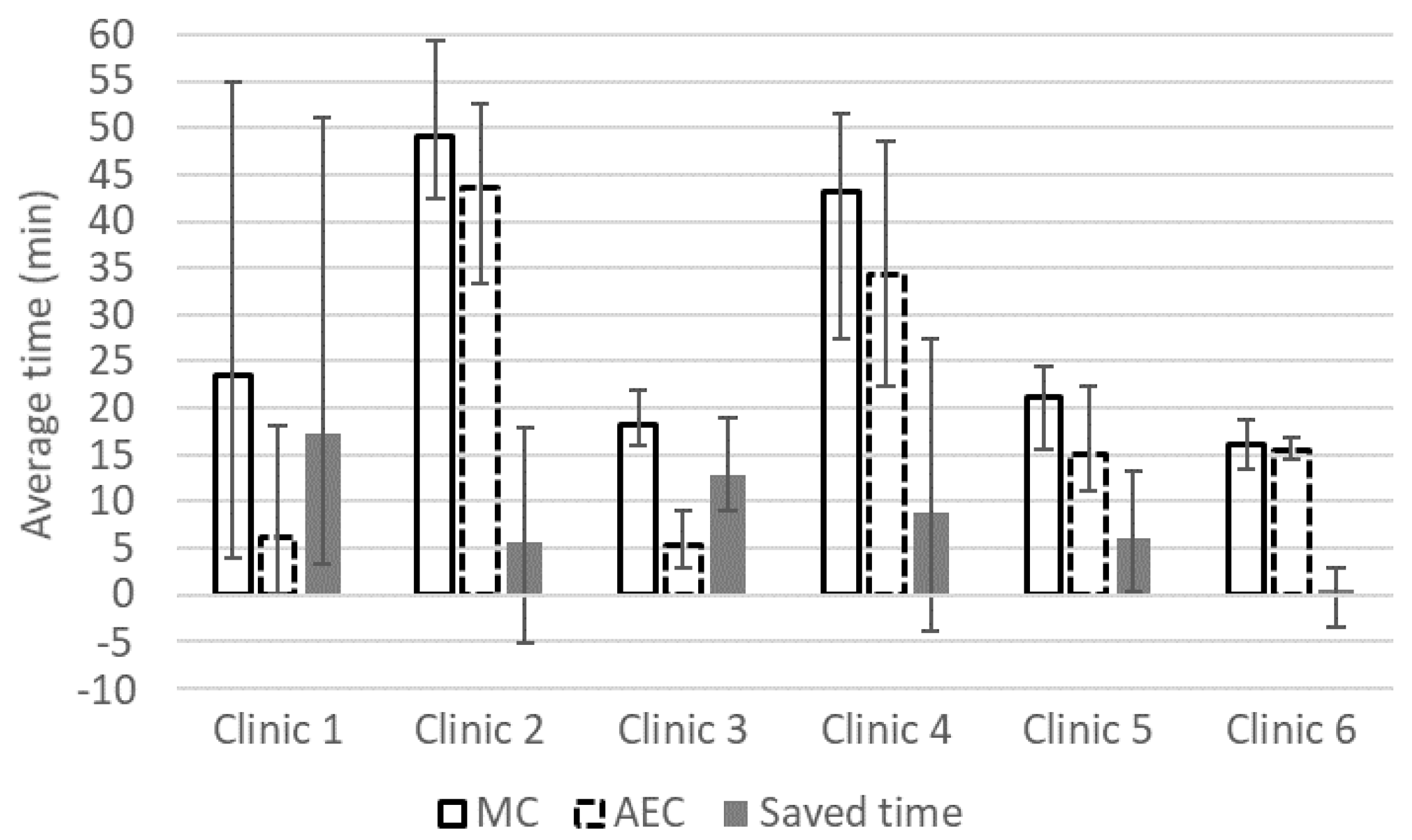
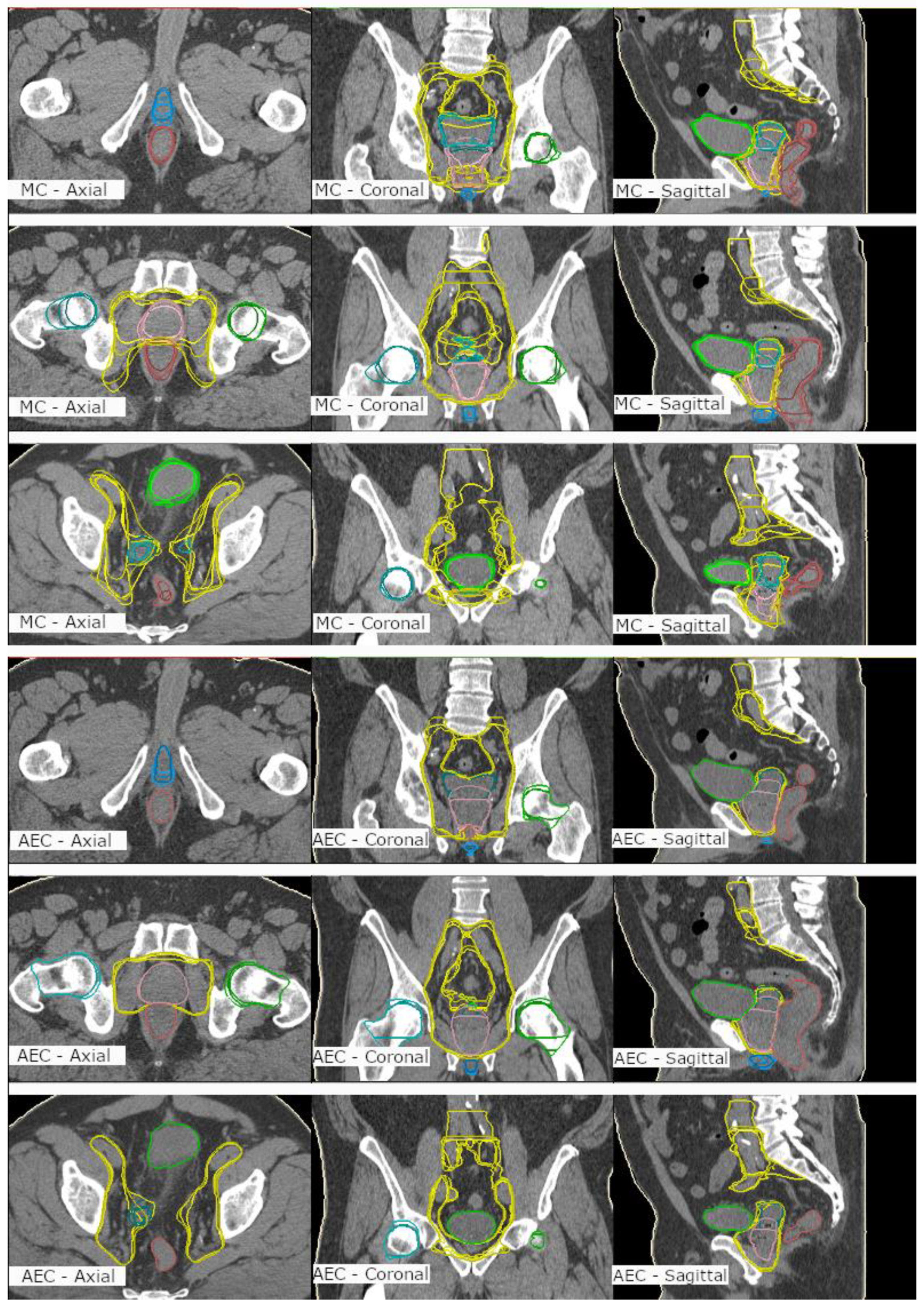
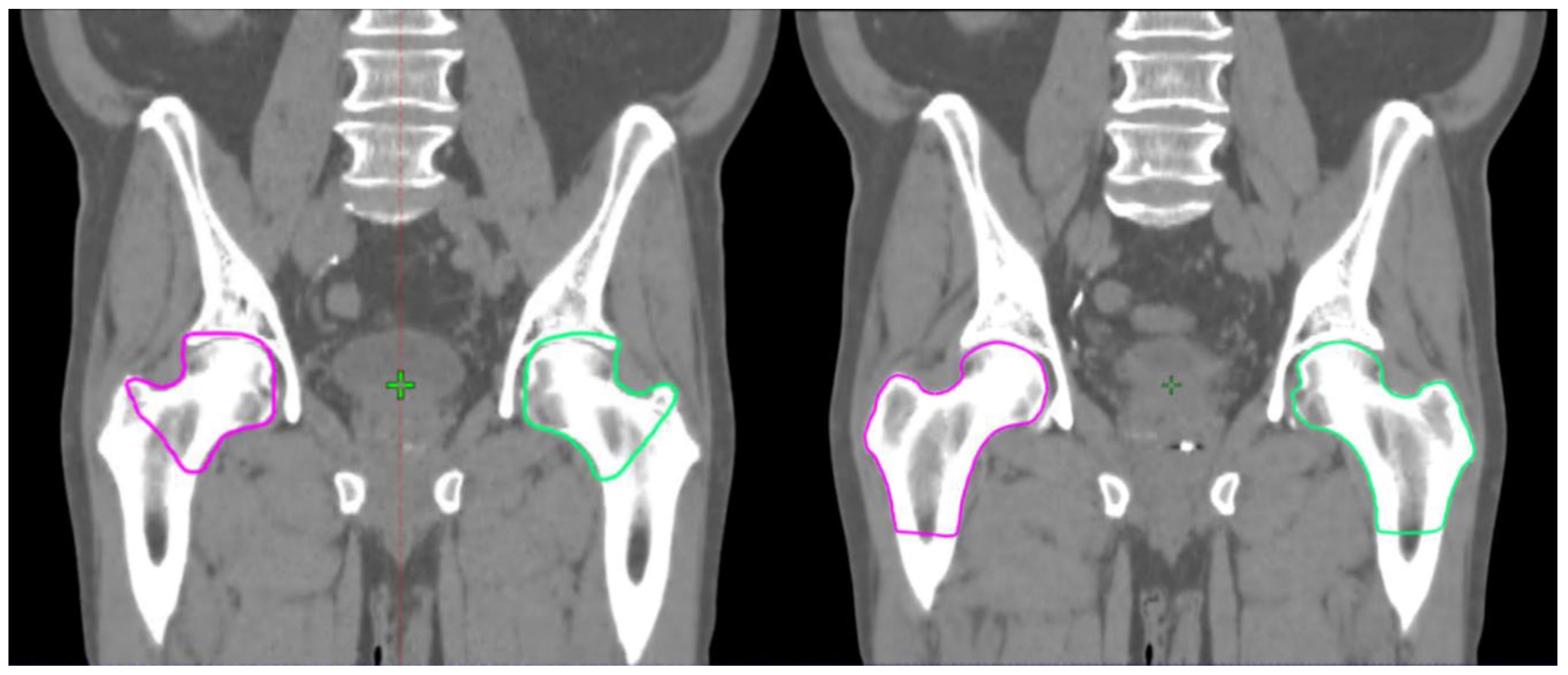
3. Results
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Mohler, J.L.; Antonarakis, E.S.; Armstrong, A.J.; D’Amico, A.V.; Davis, B.J.; Dorff, T.; Eastham, J.A.; Enke, C.A.; Farrington, T.A.; Higano, C.S.; et al. Prostate Cancer, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. JNCCN 2019, 17, 479–505. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.W.; Drinkwater, K.J. Geographical Variation in Radiotherapy Services across the UK in 2007 and the Effect of Deprivation. Clin. Oncol. 2009, 21, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Moon, D.H.; Efstathiou, J.A.; Chen, R.C. What is the best way to radiate the prostate in 2016? Urol. Oncol. 2017, 35, 59–68. [Google Scholar] [CrossRef]
- Chen, A.M.; Chin, R.; Beron, P.; Yoshizaki, T.; Mikaeilian, A.G.; Cao, M. Inadequate target volume delineation and local–Regional recurrence after intensity-modulated radiotherapy for human papillomavirus-positive oropharynx cancer. Radiother. Oncol. 2017, 123, 412–418. [Google Scholar] [CrossRef]
- Cazzaniga, L.F.; Marinoni, M.A.; Bossi, A.; Bianchi, E.; Cagna, E.; Cosentino, D.; Scandolaro, L.; Valli, M.; Frigerio, M. Interphysician variability in defining the planning target volume in the irradiation of prostate and seminal vesicles. Radiother. Oncol. 1998, 47, 293–296. [Google Scholar] [CrossRef]
- Fiorino, C.; Reni, M.; Bolognesi, A.; Cattaneo, G.M.; Calandrino, R. Intra- and inter-observer variability in contouring prostate and seminal vesicles, implications for conformal treatment planning. Radiother. Oncol. 1998, 47, 285–292. [Google Scholar] [CrossRef]
- Valicenti, R.K.; Sweet, J.W.; Hauck, W.W.; Hudes, R.S.; Lee, T.; Dicker, A.P.; Waterman, F.M.; Anne, P.R.; Corn, B.W.; Galvin, J.M. Variation of clinical target volume in three-dimensional conformal radiation therapy for prostate cancer. Int. J Radiot. Oncol. Biol. Phys. 1999, 44, 931–935. [Google Scholar] [CrossRef]
- Njeh, C.F. Tumor delineation, the weakest link in the search for accuracy in radiotherapy. J. Med. Phys. 2008, 33, 136–140. [Google Scholar] [CrossRef]
- Sharp, G.; Fritscher, K.D.; Pekar, V.; Peroni, M.; Shusharina, N.; Veeraraghavan, H.; Yang, J. Vision 20/20: Perspectives on automated image segmentation for radiotherapy. Med. Phys. 2014, 41, 050902. [Google Scholar] [CrossRef]
- Geraghty, J.P.; Grogan, G.; Ebert, M.A. Automatic segmentation of male pelvic anatomy on computed tomography images: A comparison with multiple observers in the context of a multicentre clinical trial. Radiat. Oncol. 2013, 8, 106. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Meyer, P.; Noblet, V.; Mazzara, C.; Lallement, A. Survey on deep learning for radiotherapy. Biol. Med. 2018, 98, 126–146. [Google Scholar] [CrossRef]
- El Naqa, I.; Ruan, D.; Valdes, G.; Dekker, A.; McNutt, T.; Ge, Y.; Wu, Q.J.; Hun Oh, J.; Thor, M.; Smith, W.; et al. Machine learning and modeling: Data, validation, communication challenges. Med. Phys. 2018, 45, e834–e840. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, A.W.M.J.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Balagopal, A.; Kazemifar, S.; Nguyen, D.; Lin, M.H.; Hannan, R.; Owrangi, A.; Jiang, S. Fully automated organ segmentation in male pelvic CT images. Phys. Med. Biol. 2018, 63, 245015. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Cao, X.; Shi, Y.; Nie, D.; Gao, Y.; Shen, D. Pelvic Organ Segmentation Using Distinctive Curve Guided Fully Convolutional Networks. IEEE Trans. Med. Imaging 2019, 38, 585–595. [Google Scholar] [CrossRef]
- Kearney, V.; Chan, J.W.; Wang, T. Attention-enabled 3D boosted convolutional neural networks for semantic CT segmentation using deep supervision. Phys. Med. Biol. 2019, 64, 135001. [Google Scholar] [CrossRef]
- Liu, C.; Gardner, S.J.; Wen, N.; Elshaikh, M.A.; Siddiqui, F.; Movsas, B.; Chetty, I.J. Automatic Segmentation of the Prostate on CT Images Using Deep Neural Networks (DNN). Int. J. Radiat. Oncol. Biol. Phys. 2019, 104, 924–932. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. arXiv 2016, arXiv:1606.06650. [Google Scholar]
- Chen, A.M.; Chin, R.; Beron, P.; Yoshizaki, T.; Mikaeilian, A.G.; Cao, M. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2016, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Nikolov, S.; Blackwell, S.; Mendes, R.; De Fauw, J.; Meyer, C.; Hughes, C.; Patel, Y.; Meyer, C.; Askham, H.; Romera-Paredes, B.; et al. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy. arXiv 2018, arXiv:1809.04430. [Google Scholar]
- Pejavar, S.; Yom, S.S.; Hwang, A.; Speight, J.; Gottschalk, A.; Hsu, I.C.; Roach, M., III. Computer-Assisted, Atlas-Based Segmentation for Target Volume Delineation in Whole Pelvic IMRT for Prostate Cancer. Technol. Cancer Res. Treat. 2013, 3, 199–206. [Google Scholar] [CrossRef]
- La Macchia, M.; Fellin, F.; Amichetti, M.; Cianchetti, M.; Gianolini, S.; Paola, V.; Lomax, A.J.; Widesott, L. Systematic evaluation of three different commercial software solutions for automatic segmentation for adaptive therapy in head-and-neck, prostate and pleural cancer. Radiat. Oncol. 2012, 7, 160. [Google Scholar] [CrossRef]
- Simmat, I.; Georg, P.; Georg, D.; Birkfellner, W.; Goldner, G.; Stock, M. Assessment of accuracy and efficiency of atlas-based autosegmentation for prostate radiotherapy in a variety of clinical conditions. Strahlenther. Onkol. 2012, 188, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Hwee, J.; Louie, A.V.; Gaede, S.; Bauman, G.; D’Souza, D.; Sexton, T.; Lock, M.; Ahmad, B.; Rodrigues, G. Technology Assessment of Automated Atlas Based Segmentation in Prostate Bed Contouring. Radiat. Oncol. 2011, 6, 110. [Google Scholar] [CrossRef] [PubMed]
- Langmack, K.A.; Perry, C.; Sinstead, C.; Mills, J.; Saunders, D. The utility of atlas-based segmentation in the male pelvis is dependent on the interobserver agreement of the structures segmented. Br. J. Radiol. 2014, 87, 20140299. [Google Scholar] [CrossRef]
- Sjöberg, C.; Lundmark, M.; Granberg, C.; Johansson, S.; Ahnesjö, A.; Montelius, A. Clinical evaluation of multi-atlas based segmentation of lymph node regions in head and neck and prostate cancer patients. Radiat. Oncol. 2013, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Kuisma, A.; Ranta, I.; Keyriläinen, J.; Suilamo, S.; Wright, P.; Pesola, M.; Warner, L.; Löyttyniemi, E.; Minn, H. Validation of automated magnetic resonance image segmentation for radiation therapy planning in prostate cancer. PHIRO 2020, 13, 14–20. [Google Scholar] [CrossRef]
- Lawton, C.A.; Michalski, J.; El-Naqa, I.; Buyyounouski, M.K.; Lee, W.R.; Menard, C.; O’Meara, M.D.E.; Rosenthal, S.A.; Ritter, M.; Seider, M.; et al. RTOG GU Radiation oncology specialists reach consensus on pelvic lymph node volumes for high-risk prostate cancer. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 383–387. [Google Scholar] [CrossRef]
- Salembier, C.; Villeirs, G.; De Bari, B.; Hoskin, P.; Pieters, B.R.; Van Vulpen, M.; Khoo, V.; Henry, A.; Bossi, A.; De Meerleer, G.; et al. ESTRO ACROP Consensus Guideline on CT- And MRI-based Target Volume Delineation for Primary Radiation Therapy of Localized Prostate Cancer. Radiother Oncol. 2018, 127, 49–61. [Google Scholar] [CrossRef]
| ROI | DSC | S-DSC (2.5 mm) | HD95 (mm) | Diff_cm3 (cm3) | Diff_% (%) |
|---|---|---|---|---|---|
| Bladder | 0.93 | 0.68 | 3.3 | −1.2 ± 12.0 | −1.7 ± 8.3 |
| Prostate | 0.82 | 0.38 | 6.1 | −13.5 ± 10.1 | −31.6 ± 26.1 |
| Femoral head L | 0.68 | 0.22 | 25.0 | −4.0 ± 32.4 | −37.9 ± 60.7 |
| Femoral head R | 0.69 | 0.22 | 24.7 | −3.0 ± 31.2 | −30.5 ± 38.7 |
| Lymph nodes * | 0.80 | 0.39 | 14.7 | 40.9 ± 86.3 | 5.2 ± 12.4 |
| Penile bulb | 0.51 | 0.33 | 7.7 | 1.2 ± 2.2 | −6.8 ± 67.0 |
| Rectum | 0.84 | 0.58 | 11.4 | −4.5 ± 11.4 | −9.6 ± 18.7 |
| Seminal vesicles | 0.72 | 0.52 | 7.1 | 0.6 ± 6.9 | −11.1 ± 31.2 |
| Oncologist 2 | Oncologist 3 | Oncologist 4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ROI | AEC | MC | Diff | AEC | MC | Diff | AEC | MC | Diff | |
| Oncologist 1 | Bladder | 1.00 | 0.90 | 0.10 | 1.00 | 0.92 | 0.08 | 1.00 | 0.91 | 0.09 |
| Prostate | 0.92 | 0.82 | 0.10 | 0.99 | 0.83 | 0.17 | 1.00 | 0.86 | 0.14 | |
| Lymph nodes | 0.88 | 0.74 | 0.14 | 0.89 | 0.78 | 0.12 | 0.95 | 0.81 | 0.14 | |
| Penile bulb | 0.82 | 0.56 | 0.26 | 0.87 | 0.71 | 0.15 | 0.80 | 0.66 | 0.14 | |
| Rectum | 0.97 | 0.74 | 0.23 | 0.97 | 0.77 | 0.19 | 0.98 | 0.82 | 0.16 | |
| Seminal vesicles | 0.84 | 0.78 | 0.06 | 0.97 | 0.75 | 0.22 | 0.97 | 0.74 | 0.24 | |
| Oncologist 2 | Bladder | 1.00 | 0.91 | 0.08 | 1.00 | 0.92 | 0.08 | |||
| Prostate | 0.92 | 0.86 | 0.06 | 0.92 | 0.80 | 0.12 | ||||
| Lymph nodes | 0.87 | 0.78 | 0.10 | 0.91 | 0.72 | 0.19 | ||||
| Penile bulb | 0.82 | 0.57 | 0.24 | 0.90 | 0.61 | 0.29 | ||||
| Rectum | 0.99 | 0.64 | 0.35 | 0.99 | 0.76 | 0.23 | ||||
| Seminal vesicles | 0.84 | 0.79 | 0.05 | 0.83 | 0.79 | 0.04 | ||||
| Oncologist 3 | Bladder | 1.00 | 0.92 | 0.08 | ||||||
| Prostate | 0.99 | 0.82 | 0.18 | |||||||
| Lymph nodes | 0.92 | 0.74 | 0.18 | |||||||
| Penile bulb | 0.80 | 0.74 | 0.06 | |||||||
| Rectum | 0.99 | 0.77 | 0.22 | |||||||
| Seminal vesicles | 0.97 | 0.77 | 0.20 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiljunen, T.; Akram, S.; Niemelä, J.; Löyttyniemi, E.; Seppälä, J.; Heikkilä, J.; Vuolukka, K.; Kääriäinen, O.-S.; Heikkilä, V.-P.; Lehtiö, K.; et al. A Deep Learning-Based Automated CT Segmentation of Prostate Cancer Anatomy for Radiation Therapy Planning-A Retrospective Multicenter Study. Diagnostics 2020, 10, 959. https://doi.org/10.3390/diagnostics10110959
Kiljunen T, Akram S, Niemelä J, Löyttyniemi E, Seppälä J, Heikkilä J, Vuolukka K, Kääriäinen O-S, Heikkilä V-P, Lehtiö K, et al. A Deep Learning-Based Automated CT Segmentation of Prostate Cancer Anatomy for Radiation Therapy Planning-A Retrospective Multicenter Study. Diagnostics. 2020; 10(11):959. https://doi.org/10.3390/diagnostics10110959
Chicago/Turabian StyleKiljunen, Timo, Saad Akram, Jarkko Niemelä, Eliisa Löyttyniemi, Jan Seppälä, Janne Heikkilä, Kristiina Vuolukka, Okko-Sakari Kääriäinen, Vesa-Pekka Heikkilä, Kaisa Lehtiö, and et al. 2020. "A Deep Learning-Based Automated CT Segmentation of Prostate Cancer Anatomy for Radiation Therapy Planning-A Retrospective Multicenter Study" Diagnostics 10, no. 11: 959. https://doi.org/10.3390/diagnostics10110959
APA StyleKiljunen, T., Akram, S., Niemelä, J., Löyttyniemi, E., Seppälä, J., Heikkilä, J., Vuolukka, K., Kääriäinen, O.-S., Heikkilä, V.-P., Lehtiö, K., Nikkinen, J., Gershkevitsh, E., Borkvel, A., Adamson, M., Zolotuhhin, D., Kolk, K., Pang, E. P. P., Tuan, J. K. L., Master, Z., ... Keyriläinen, J. (2020). A Deep Learning-Based Automated CT Segmentation of Prostate Cancer Anatomy for Radiation Therapy Planning-A Retrospective Multicenter Study. Diagnostics, 10(11), 959. https://doi.org/10.3390/diagnostics10110959





