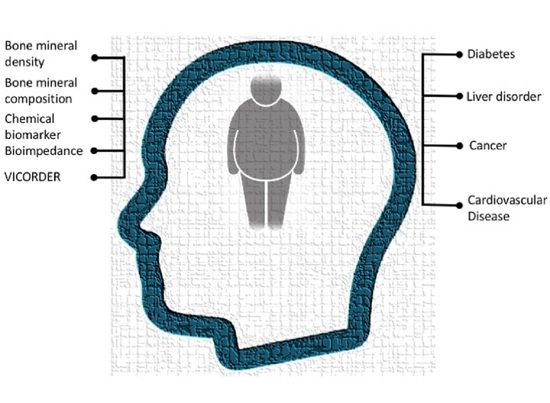
Obesity in Qatar: A Case-Control Study on the Identification of Associated Risk Factors
Abstract
1. Introduction
2. Methods
2.1. Ethical Approval
2.2. Cohort Description
2.3. Physio-Clinical Measurements
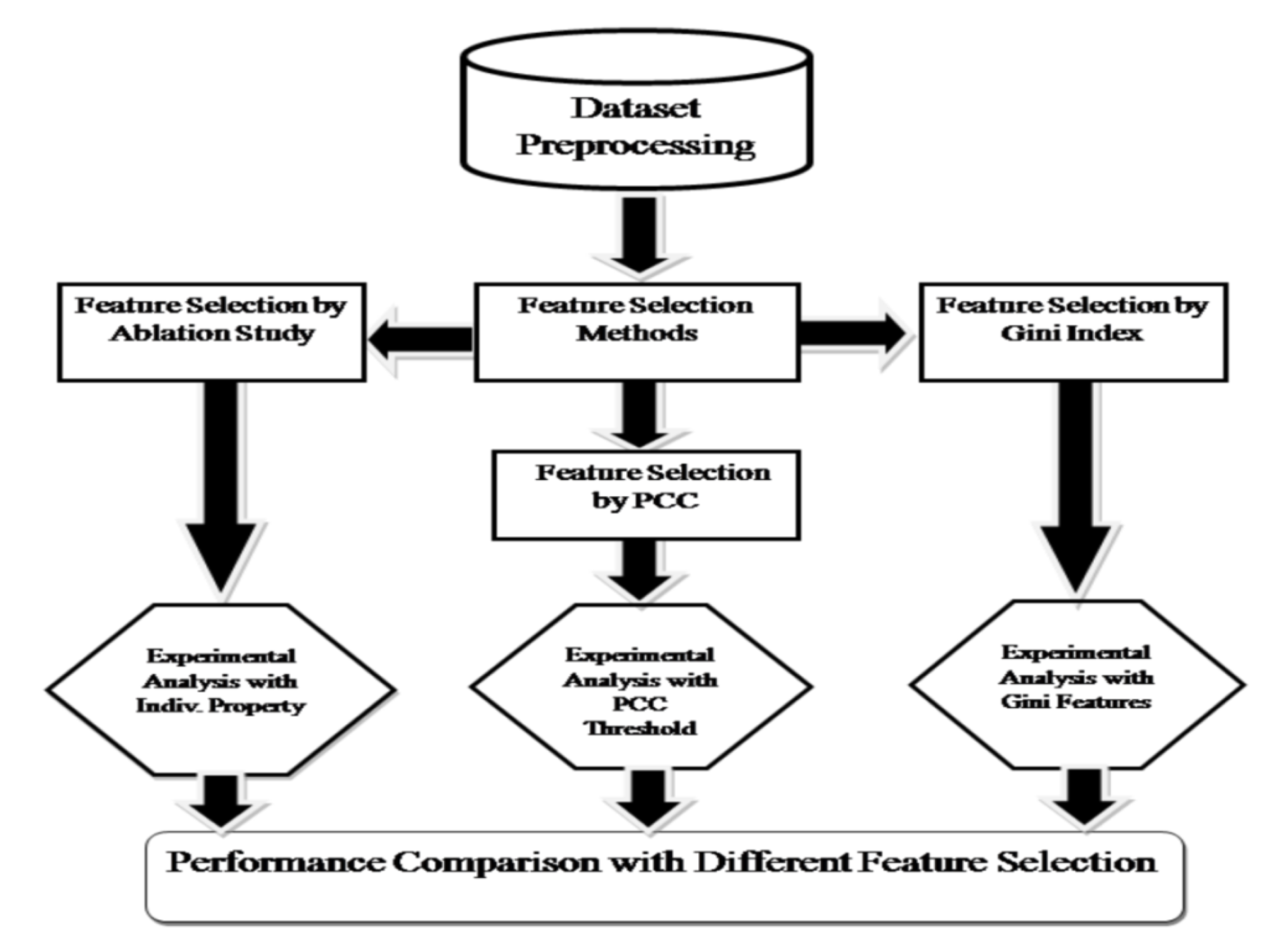
2.4. Data Pre-Processing
2.5. Feature Subset Selection
2.6. Statistical Significance of the Variables Selected by Feature Subset Selection Techniques
2.7. Machine Learning Model Development
2.8. Model Evaluation
3. Results
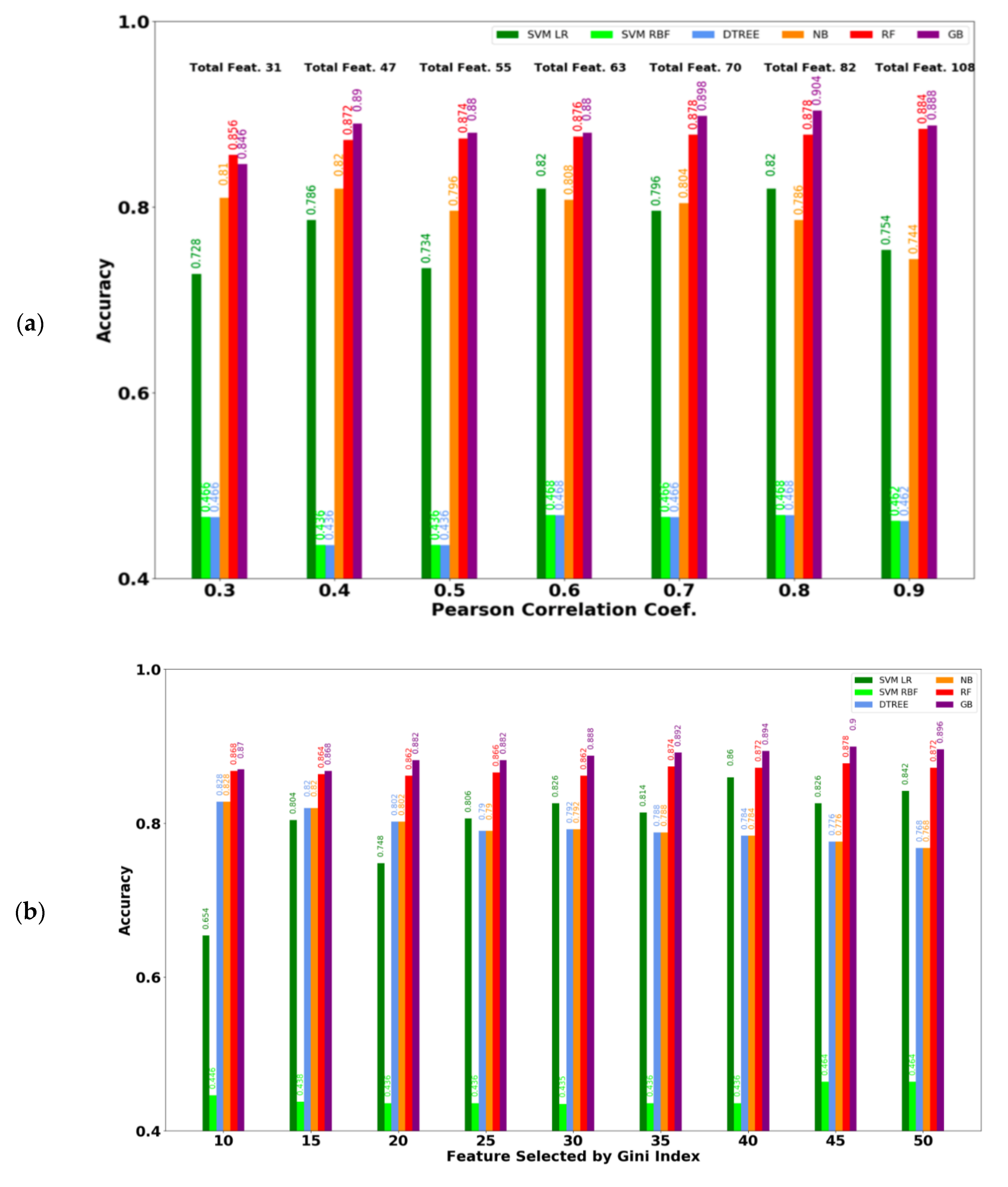
3.1. Performance of Machine Learning Models Based on Ablation Study
3.2. Performance of ML Models Considering the Selected Feature Subset
3.3. Performance of ML Models on Gender and Age Based Stratified Participants
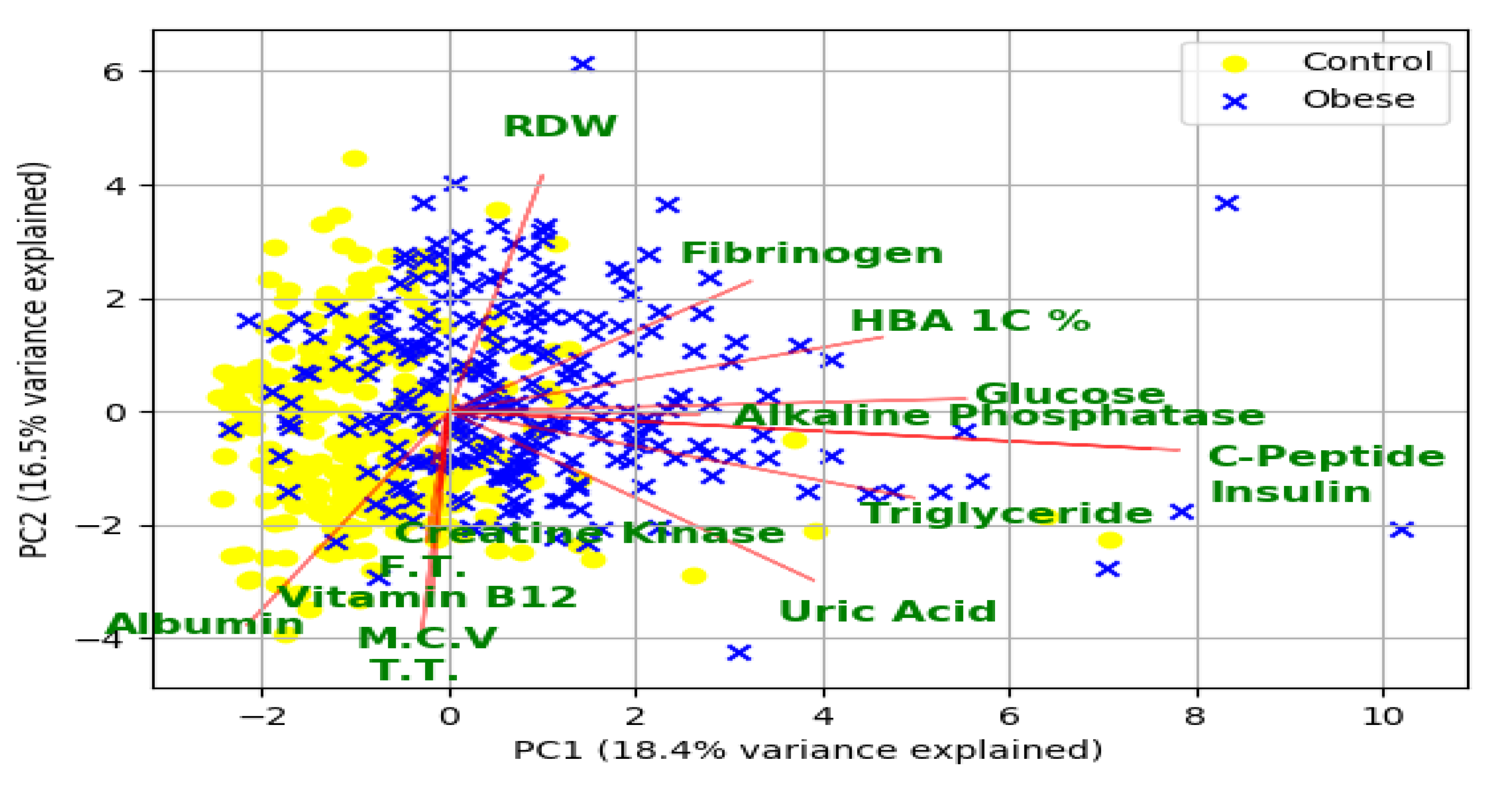
3.4. Statistical Significance of the Variables Selected by Feature Subset Selection Techniques
3.5. Potential Risk Factors for Obesity and Related Morbidities
3.6. Bone Mineral Density Associated Factors in Obesity
4. Discussion
4.1. Principal Findings
4.2. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kelly, T.; Yang, W.; Chen, C.-S.; Reynolds, K.; He, J. Global burden of obesity in 2005 and projections to 2030. Int. J. Obes. 2008, 32, 1431–1437. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Beydoun, M.A.; Liang, L.; Caballero, B.; Kumanyika, S.K. Will all Americans become overweight or obese? Estimating the progression and cost of the US obesity epidemic. Obesity 2008, 16, 2323–2330. [Google Scholar] [CrossRef]
- Ng, M.; Fleming, T.; Robinson, M.; Thomson, B.; Graetz, N.; Margono, C.; Mullany, E.C.; Biryukov, S.; Abbafati, C.; Abera, S.F.; et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014, 384, 766–781. [Google Scholar] [CrossRef]
- Smith, K.B.; Smith, M.S. Obesity Statistics. Primary Care: Clinics in Office Practice; Elsevier: Amsterdam, The Netherlands, 2016; Volume 43, pp. 121–135. [Google Scholar]
- Centers for Disease Control and Prevention. National Diabetes Fact Sheet: National Estimates and General Information on Diabetes and Prediabetes in the United States; US Department of Health and Human Services, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2011; Volume 201, pp. 2568–2569. [Google Scholar]
- Renehan, A.G.; Tyson, M.; Egger, M.; Heller, R.F.; Zwahlen, M. Body-mass index and incidence of cancer: A systematic review and meta-analysis of prospective observational studies. Lancet 2008, 371, 569–578. [Google Scholar] [CrossRef]
- Grotle, M.; Hagen, K.B.; Natvig, B.; Dahl, F.A.; Kvien, T.K. Obesity and osteoarthritis in knee, hip and/or hand: An epidemiological study in the general population with 10 years follow-up. BMC Musculoskelet. Disord. 2008, 9, 132. [Google Scholar] [CrossRef] [PubMed]
- Carman, W.J.; Sowers, M.; Hawthorne, V.M.; Weissfeld, L.A. Obesity as a risk factor for osteoarthritis of the hand and wrist: A prospective study. Am. J. Epidemiol. 1994, 139, 119–129. [Google Scholar] [CrossRef]
- Felson, D.T.; Zhang, Y.; Anthony, J.M.; Naimark, A.; Anderson, J.J. Weight loss reduces the risk for symptomatic knee osteoarthritis in women: The Framingham Study. Ann. Intern. Med. 1992, 116, 535–539. [Google Scholar] [CrossRef]
- Kurth, T.; Gaziano, J.M.; Berger, K.; Kase, C.S.; Rexrode, K.M.; Cook, N.R.; Buring, J.E.; Manson, J.E. Body Mass Index and the Risk of Stroke in Men. Arch. Intern. Med. 2002, 162, 2557–2562. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.J.; Parise, H.; Levy, D.; D’Agostino, R.B.; Wolf, P.A.; Vasan, R.S.; Benjamin, E.J. Obesity and the risk of new-onset atrial fibrillation. Jama 2004, 292, 2471–2477. [Google Scholar] [CrossRef] [PubMed]
- Poirier, P.; Giles, T.D.; Bray, G.A.; Hong, Y.; Stern, J.S.; Pi-Sunyer, F.X.; Eckel, R.H. Obesity and cardiovascular disease: Pathophysiology, evaluation, and effect of weight loss: An update of the 1997 American Heart Association Scientific Statement on Obesity and Heart Disease from the Obesity Committee of the Council on Nutrition, Physical Activity, and Metabolism. Circulation 2006, 113, 898–918. [Google Scholar]
- Mandeya, J.; Kridli, S.A.-O. Childhood overweight and obesity in Qatar: A literature review. Avicenna 2014, 2014, 2. [Google Scholar] [CrossRef]
- Haj Bakri, A.; Al-Thani, A. Chronic Disease Risk Factor Surveillance: Qatar STEPS Report 2012; The Supreme Council of Health: Doha, Qatar, 2013. [Google Scholar]
- DeGregory, K.W.; Kuiper, P.; DeSilvio, T.; Pleuss, J.D.; Miller, R.; Roginski, J.W.; Fisher, C.B.; Harness, D.; Viswanath, S.; Heymsfield, S.B.; et al. A review of machine learning in obesity. Obes. Rev. 2018, 19, 668–685. [Google Scholar] [CrossRef]
- Abdel-Aal, R.; Mangoud, A. Modeling obesity using abductive networks. Comput. Biomed. Res. 1997, 30, 451–471. [Google Scholar] [CrossRef] [PubMed]
- Dugan, T.M.; Mukhopadhyay, S.; Carroll, A.; Downs, S. Machine learning techniques for prediction of early childhood obesity. Appl. Clin. Inform. 2015, 6, 506–520. [Google Scholar]
- Ergün, U. The classification of obesity disease in logistic regression and neural network methods. J. Med Syst. 2009, 33, 67. [Google Scholar] [CrossRef] [PubMed]
- ALNohair, S. Obesity in gulf countries. Int. J. Health Sci. 2014, 8, 79. [Google Scholar] [CrossRef]
- Al-Thani, M.H.; Al-Thani, A.; Al-Chetachi, W.F.; Khalifa, S.A.; Akram, H.; Poovelil, B.V.; Almalki, B.A.; Bakri, A.H.; Arora, P.; Badawi, A. Dietary and nutritional factors influencing obesity in Qatari adults and the modifying effect of physical activity. J. Obes. Weight-Loss Med. 2015, 1. [Google Scholar] [CrossRef]
- Bener, A. Prevalence of obesity, overweight, and underweight in Qatari adolescents. Food Nutr. Bull. 2006, 27, 39–45. [Google Scholar] [CrossRef]
- Ullah, E.; Mall, R.; Rawi, R.; Moustaid-Moussa, N.; Butt, A.A.; Bensmail, H. Harnessing Qatar Biobank to understand type 2 diabetes and obesity in adult Qataris from the First Qatar Biobank Project. J. Transl. Med. 2018, 16, 99. [Google Scholar] [CrossRef]
- Al Kuwari, H.; Al Thani, A.; Al Marri, A.; Al Kaabi, A.; Abderrahim, H.; Afifi, N.; Qafoud, F.; Chan, Q.; Tzoulaki, I.; Downey, P.; et al. The Qatar Biobank: Background and methods. BMC Public Health 2015, 15, 1208. [Google Scholar] [CrossRef] [PubMed]
- Al Thani, A.; Fthenou, E.; Paparrodopoulos, S.; Al Marri, A.; Shi, Z.; Qafoud, F.; Afifi, N. Qatar Biobank cohort study: Study design and first results. Am. J. Epidemiol. 2019, 188, 1420–1433. [Google Scholar] [CrossRef]
- Elliott, P.; Vergnaud, A.-C.; Singh, D.; Neasham, D.; Spear, J.; Heard, A. The Airwave Health Monitoring Study of police officers and staff in Great Britain: Rationale, design and methods. Environ. Res. 2014, 134, 280–285. [Google Scholar] [CrossRef]
- Cleutjens, F.A.; Spruit, M.A.; Ponds, R.W.; Dijkstra, J.B.; Franssen, F.M.; Wouters, E.F.; Janssen, D.J. Cognitive functioning in obstructive lung disease: Results from the United Kingdom biobank. J. Am. Med. Dir. Assoc. 2014, 15, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Keehn, L.; Milne, L.; McNeill, K.; Chowienczyk, P.; Sinha, M.D. Measurement of pulse wave velocity in children: Comparison of volumetric and tonometric sensors, brachial-femoral and carotid-femoral pathways. J. Hypertens. 2014, 32, 1464. [Google Scholar] [CrossRef] [PubMed]
- Blake, G.M.; Fogelman, I. An update on dual-energy x-ray absorptiometry. In Seminars in Nuclear Medicine; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Anderson, T.W.; Darling, D.A. A test of goodness of fit. J. Am. Stat. Assoc. 1954, 49, 765–769. [Google Scholar] [CrossRef]
- Student. The probable error of a mean. Biometrika 1908, 6, 1–25. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Fan, R.-E.; Chang, K.-W.; Hsieh, C.-J.; Wang, X.-R.; Lin, C.-J. LIBLINEAR: A library for large linear classification. J. Mach. Learn. Res. 2008, 9, 1871–1874. [Google Scholar]
- Amari, S.-I.; Wu, S. Improving support vector machine classifiers by modifying kernel functions. Neural Netw. 1999, 12, 783–789. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Stone, C.J.; Olshen, R.A. Classification and Regression Trees; CRC Press: Boca Raton, FL, USA, 1984. [Google Scholar]
- McCallum, A.; Nigam, K. A comparison of event models for naive bayes text classification. In AAAI-98 Workshop on Learning for Text Categorization; Citeseer: Madison, WI, USA, 1998. [Google Scholar]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Dunstan, D.W.; Zimmet, P.Z.; Welborn, T.A.; De Courten, M.P.; Cameron, A.J.; Sicree, R.A.; Dwyer, T.; Colagiuri, S.; Jolley, D.; Knuiman, M.; et al. The rising prevalence of diabetes and impaired glucose tolerance: The Australian Diabetes, Obesity and Lifestyle Study. Diabetes Care 2002, 25, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Rohlfing, C.L.; Wiedmeyer, H.M.; Little, R.R.; England, J.D.; Tennill, A.; Goldstein, D.E. Defining the relationship between plasma glucose and HbA(1c): Analysis of glucose profiles and HbA(1c) in the Diabetes Control and Complications Trial. Diabetes Care 2002, 25, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Zimmet, P.; Magliano, D.; Matsuzawa, Y.; Alberti, G.; Shaw, J. The metabolic syndrome: A global public health problem and a new definition. J. Atheroscler. Thromb. 2005, 12, 295–300. [Google Scholar] [CrossRef]
- Dai, X.; Yuan, J.; Yao, P.; Yang, B.; Gui, L.; Zhang, X.; Guo, H.; Wang, Y.; Chen, W.; Wei, S.; et al. Association between serum uric acid and the metabolic syndrome among a middle- and old-age Chinese population. Eur. J. Epidemiol. 2013, 28, 669–676. [Google Scholar] [CrossRef]
- Tsutsumi, V.; Nakamura, T.; Ueno, T.; Torimura, T.; Aguirre-García, J. Structure and Ultrastructure of the Normal and Diseased Liver. In Liver Pathophysiology; Elsevier: Amsterdam, The Netherlands, 2017; pp. 23–44. [Google Scholar]
- Díez, J.; Iglesias, P. Relationship between thyrotropin and body mass index in euthyroid subjects. Exp. Clin. Endocrinol. Diabetes 2011, 119, 144–150. [Google Scholar] [CrossRef]
- Milionis, A.; Milionis, C. Correlation Between Body Mass Index and Thyroid Function in Euthyroid Individuals in Greece; ISRN Biomarkers: London, UK, 2013; Volume 2013. [Google Scholar]
- Rhee, M.C.; Ahmadi, S.-F.; Kalantar-Zadeh, K. The dual roles of obesity in chronic kidney disease: A review of the current literature. Curr. Opin. Nephrol. Hypertens. 2016, 25, 208. [Google Scholar] [CrossRef]
- Salamat, M.R.; Salamat, A.H.; Janghorbani, M. Association between obesity and bone mineral density by gender and menopausal status. Endocrinol. Metab. 2016, 31, 547–558. [Google Scholar] [CrossRef]
- López-Gómez, J.J.; Castrillón, J.L.P.; de Luis Román, D.A. Impact of obesity on bone metabolism. Endocrinol. Nutr. 2016, 63, 551–559. (In English) [Google Scholar] [CrossRef]
- Shapses, S.A.; Pop, L.C.; Wang, Y. Obesity is a concern for bone health with aging. Nutr. Res. 2017. 39, 1–13. [CrossRef]
- De Laet, C.; Kanis, J.A.; Odén, A.; Johanson, H.; Johnell, O.; Delmas, P.; Eisman, J.A.; Kroger, H.; Fujiwara, S.; Garnero, P.; et al. Body mass index as a predictor of fracture risk: A meta-analysis. Osteoporos. Int. 2005, 16, 1330–1338. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Lee, J.Y.; Sung, J. Obesity and Bone Health Revisited: A Mendelian Randomization Study for Koreans. J. Bone Miner. Res. 2019, 34, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.; Schneider, G.; Ertel, N.; Worton, E. Obesity, androgens, estrogens, and cancer risk. Cancer Res. 1982, 42, 3281s–s3285. [Google Scholar] [PubMed]
- Johansson, H.; Kanis, J.A.; Odén, A.; McCloskey, E.; Chapurlat, R.D.; Christiansen, C.; Cummings, S.R.; Diez-Perez, A.; Eisman, J.A.; Fujiwara, S.; et al. A meta-analysis of the association of fracture risk and body mass index in women. J. Bone Miner. Res. 2014, 29, 223–233. [Google Scholar] [CrossRef] [PubMed]
| Features | Size | Examples |
|---|---|---|
| All | 236 | Spirometry, Physio-clinical Biomarkers, VICORDER, DXA |
| Spirometry | 35 | Forced expiratory time (FET), Forced expiratory flow (FEF), Forced Vital Capacity (FVC), etc. |
| Physio-clinical Biomarkers | 70 | Hemoglobin, red blood cells, white blood cells, Lymphocyte, Platelet count, Sodium, Urea, Glucose, Cholesterol, Systolic blood pressure, Diastolic blood pressure, hip waist circumference, etc. |
| VICORDER | 5 | Heart Beats, Heart rate, PPI (Pulse Pressure Index), Pulse Wave (PWV) |
| DXA body composition | 5 | CT bone mass android, CT bone mass android visceral, CT bone mass arms, CT bone mass gynoid, CT bone mass total |
| DXA Densitometry | 121 | DT area arms, DT area head, DT area pelvis, DT area spine, DT area total, etc. |
| Obese | Control | |||||
|---|---|---|---|---|---|---|
| Unit | Mean | Standard Deviation | Mean | Standard Deviation | p-Value | |
| Age | year | 35.89 | 9.898 | 30.28 | 8.32 | 1.54 × 10−11 |
| BMI | kg/m2 | 34.60 | 4.08 | 23.29 | 2.81 | 1.10 × 10−83 |
| Weight | kg | 94.22 | 15.15 | 63.99 | 10.74 | 9.38 × 10−72 |
| Z-FMI | - | 2.65 | 1.13 | −0.24 | 0.708 | 1.99 × 10−83 |
| Waist circumference | cm | 97.45 | 10.71 | 75.02 | 8.58 | 8.85 × 10−94 |
| Hip circumference | cm | 116.66 | 8.80 | 97.16 | 6.75 | 2.30 × 10−77 |
| waist-to-hip ratio (WHR) | - | 0.84 | 0.087 | 0.77 | 0.076 | 5.27 × 10−16 |
| Property | Evaluation Parameter | SVM (linear) | SVM (rbf) | Decision Tree | Naïve Bayes | RF | GB |
|---|---|---|---|---|---|---|---|
| All | Accuracy | 0.806 | 0.47 | 0.812 | 0.688 | 0.868 | 0.882 |
| Precision | 0.874 | 0.305 | 0.803 | 0.701 | 0.85 | 0.881 | |
| Recall | 0.748 | 0.58 | 0.813 | 0.678 | 0.891 | 0.879 | |
| MCC | 0.635 | 0.068 | 0.616 | 0.379 | 0.739 | 0.763 | |
| Spirometry | Accuracy | 0.543 | 0.518 | 0.557 | 0.543 | 0.587 | 0.628 |
| Precision | 0.354 | 0.578 | 0.557 | 0.522 | 0.582 | 0.618 | |
| Recall | 0.313 | 0.146 | 0.521 | 0.750 | 0.578 | 0.611 | |
| MCC | 0.045 | 0.12 | 0.112 | 0.099 | 0.178 | 0.256 | |
| VICORDER | Accuracy | 0.525 | 0.565 | 0.583 | 0.593 | 0.633 | 0.64 |
| Precision | 0.597 | 0.545 | 0.579 | 0.746 | 0.635 | 0.645 | |
| Recall | 0.441 | 0.574 | 0.587 | 0.246 | 0.603 | 0.603 | |
| MCC | 0.090 | 0.126 | 0.17 | 0.23 | 0.266 | 0.28 | |
| Physio-clinical Biomarker | Accuracy | 0.679 | 0.489 | 0.645 | 0.75 | 0.802 | 0.807 |
| Precision | 0.729 | 0.04 | 0.644 | 0.767 | 0.814 | 0.821 | |
| Recall | 0.657 | 0.1 | 0.634 | 0.692 | 0.772 | 0.786 | |
| MCC | 0.408 | 0.0 | 0.293 | 0.502 | 0.609 | 0.621 | |
| DXA Body Composition | Accuracy | 0.754 | 0.438 | 0.792 | 0.736 | 0.832 | 0.83 |
| Precision | 0.833 | 0.269 | 0.794 | 0.755 | 0.84 | 0.841 | |
| Recall | 0.717 | 0.6 | 0.799 | 0.697 | 0.823 | 0.817 | |
| MCC | 0.545 | 0.012 | 0.581 | 0.473 | 0.665 | 0.661 | |
| DXA Densitometry | Accuracy | 0.68 | 0.436 | 0.682 | 0.626 | 0.758 | 0.79 |
| Precision | 0.834 | 0.27 | 0.690 | 0.643 | 0.734 | 0.785 | |
| Recall | 0.565 | 0.6 | 0.661 | 0.597 | 0.813 | 0.803 | |
| MCC | 0.422 | 0.0 | 0.368 | 0.26 | 0.523 | 0.586 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khondaker, M.T.I.; Khan, J.Y.; Refaee, M.A.; Hajj, N.E.; Rahman, M.S.; Alam, T. Obesity in Qatar: A Case-Control Study on the Identification of Associated Risk Factors. Diagnostics 2020, 10, 883. https://doi.org/10.3390/diagnostics10110883
Khondaker MTI, Khan JY, Refaee MA, Hajj NE, Rahman MS, Alam T. Obesity in Qatar: A Case-Control Study on the Identification of Associated Risk Factors. Diagnostics. 2020; 10(11):883. https://doi.org/10.3390/diagnostics10110883
Chicago/Turabian StyleKhondaker, Md. Tawkat Islam, Junaed Younus Khan, Mahmoud Ahmed Refaee, Nady El Hajj, M. Sohel Rahman, and Tanvir Alam. 2020. "Obesity in Qatar: A Case-Control Study on the Identification of Associated Risk Factors" Diagnostics 10, no. 11: 883. https://doi.org/10.3390/diagnostics10110883
APA StyleKhondaker, M. T. I., Khan, J. Y., Refaee, M. A., Hajj, N. E., Rahman, M. S., & Alam, T. (2020). Obesity in Qatar: A Case-Control Study on the Identification of Associated Risk Factors. Diagnostics, 10(11), 883. https://doi.org/10.3390/diagnostics10110883