Bone Age Assessment Empowered with Deep Learning: A Survey, Open Research Challenges and Future Directions
Abstract
1. Introduction
2. Typical Deep-Learning Models
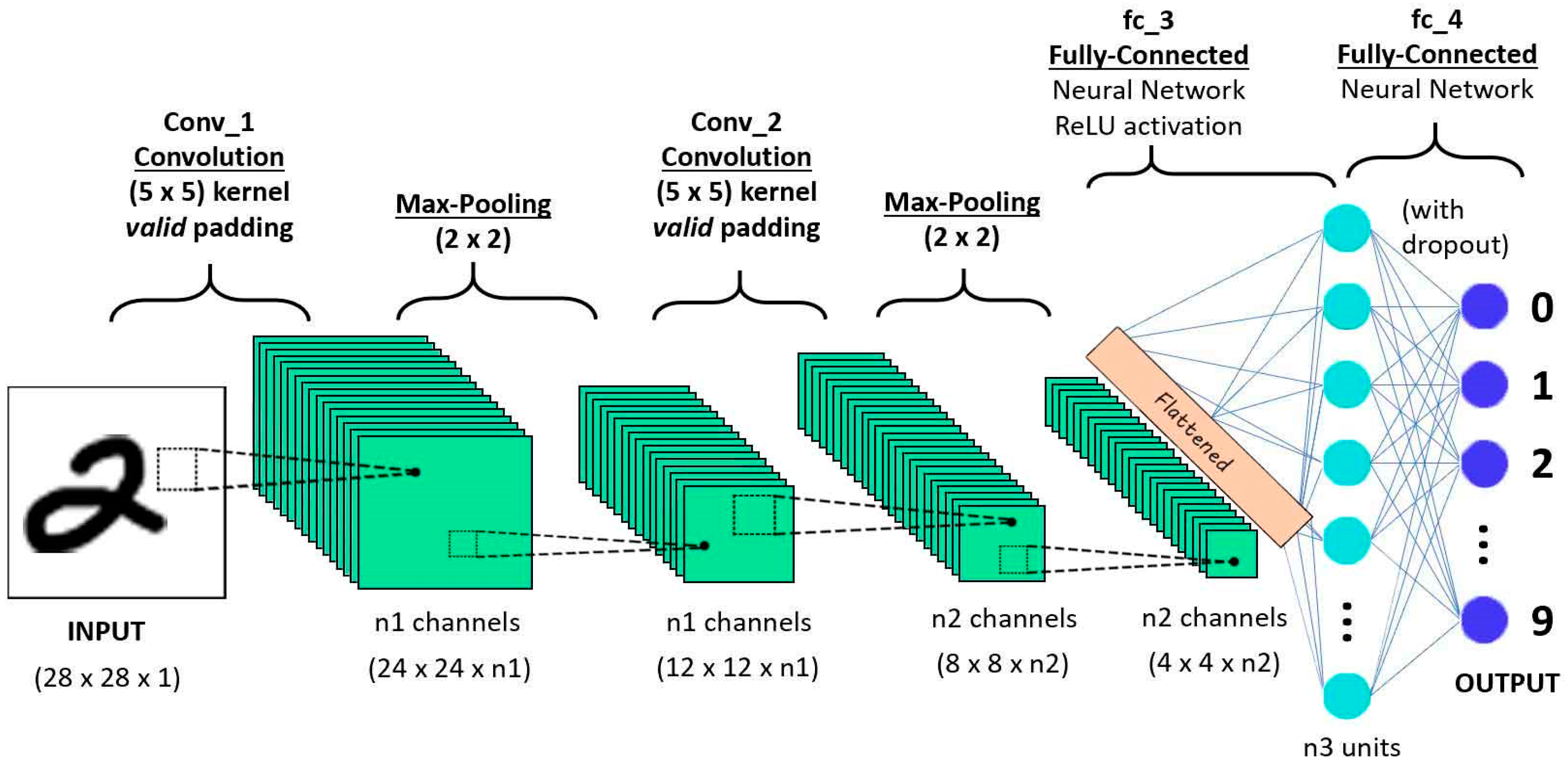
2.1. Convolutional Neural Network (CNN)
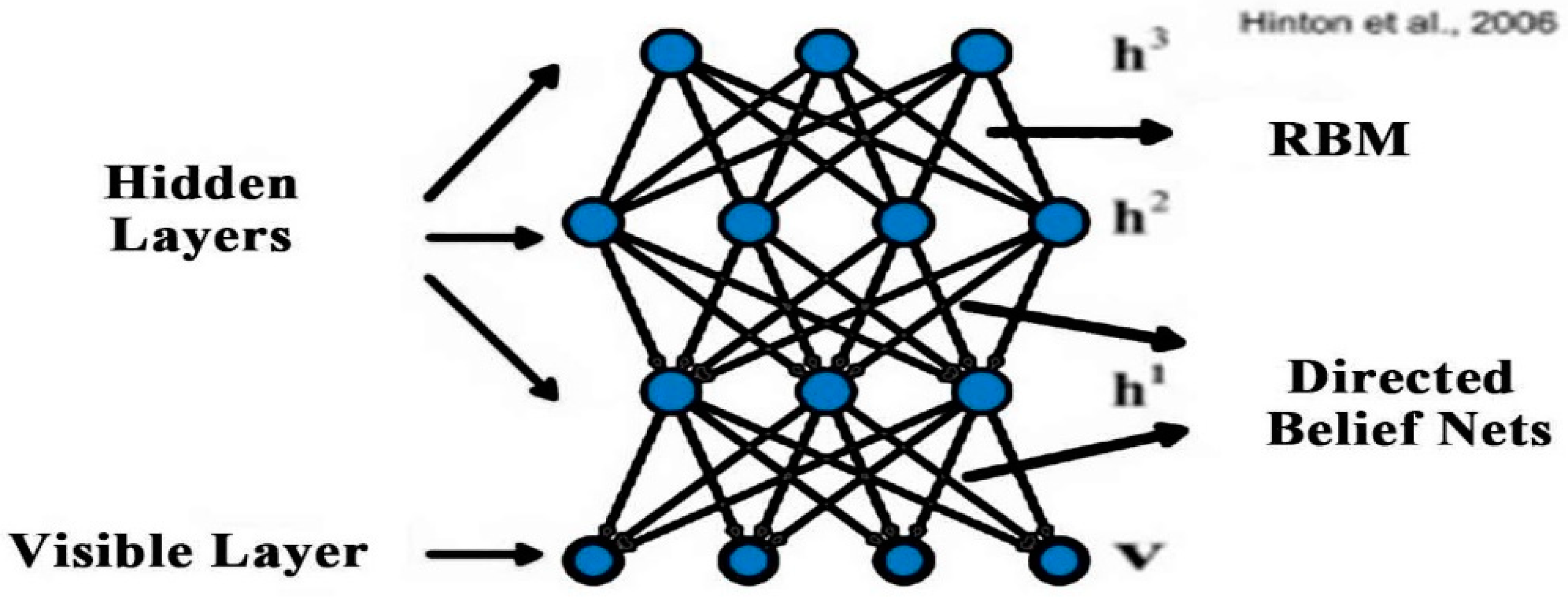
2.2. Deep Belief Network (DBN)
2.3. Recurrent Neural Network (RNN)
3. Deep-Learning Models for Bone Age Assessment
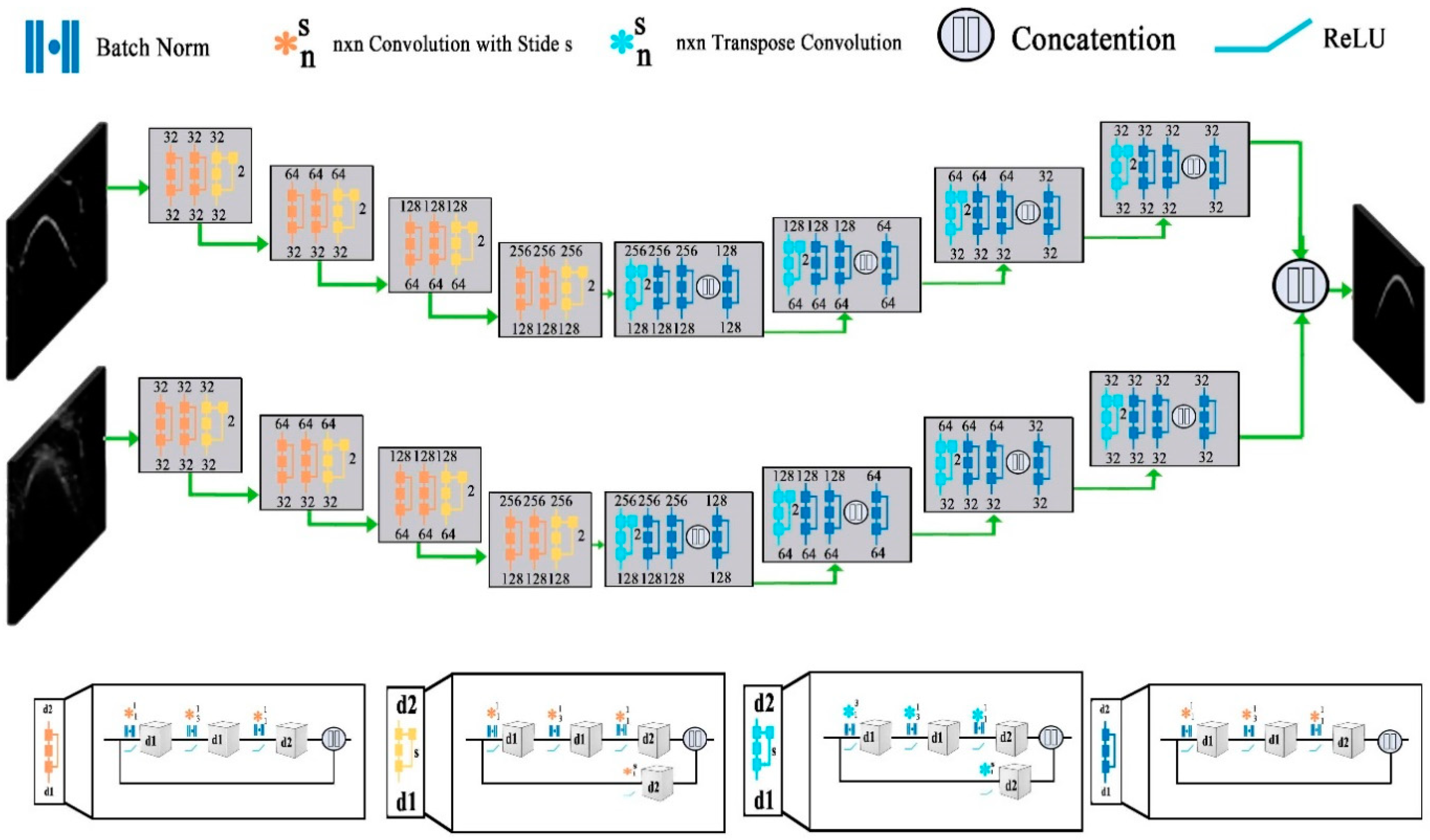
3.1. Deep-Learning Models for Bone Segmentation
3.2. Deep-Learning Models for Prediction of Bone Age
3.3. Deep-Learning Models for Classification
4. Discussion
4.1. Overview
4.2. Key Aspects of Successful Deep-Learning Models
4.3. Open Research Challenges, Limitations, and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Zhao, X.; Wu, Y.; Song, G.; Li, Z.; Zhang, Y.; Fan, Y. A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med. Image Anal. 2017, 43, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Jindal, A. Ultra sonogram images for thyroid segmentation and texture classification in diagnosis of malignant (cancerous) or benign (non-cancerous) nodules. Int. J. Eng. Innov. Technol. 2012, 1, 202–206. [Google Scholar]
- Christ, J.; Sivagowri, S.; Babu, G. Segmentation of Brain Tumors using Meta Heuristic Algorithms. Open J. Commun. Softw. 2014, 2014, 1–10. [Google Scholar] [CrossRef]
- White, H. Radiography of infants and children. JAMA 1963, 185, 223. [Google Scholar] [CrossRef]
- Carty, H. Assessment of skeletal maturity and prediction of adult height (TW3 method). J. Bone Jt. Surgery. Br. Vol. 2002, 310–311. [Google Scholar] [CrossRef]
- Poznanski, A.K.; Hernandez, R.J.; Guire, K.E.; Bereza, U.L.; Garn, S.M. Carpal Length in Children—A Useful Measurement in the Diagnosis of Rheumatoid Arthritis and Some Congenital Malformation Syndromes. Radiology 1978, 129, 661–668. [Google Scholar] [CrossRef]
- Berst, M.J.; Dolan, L.; Bogdanowicz, M.M.; Stevens, M.A.; Chow, S.; Brandser, E.A. Effect of Knowledge of Chronologic Age on the Variability of Pediatric Bone Age Determined Using the Greulich and Pyle Standards. Am. J. Roentgenol. 2001, 176, 507–510. [Google Scholar] [CrossRef]
- King, D.G.; Steventon, D.M.; O’Sullivan, M.P.; Cook, A.M.; Hornsby, V.P.L.; Jefferson, I.G.; King, P.R. Reproducibility of bone ages when performed by radiology registrars: An audit of Tanner and Whitehouse IIversusGreulich and Pyle methods. Br. J. Radiol. 1994, 67, 848–851. [Google Scholar] [CrossRef]
- Spampinato, C.; Palazzo, S.; Giordano, D.; Aldinucci, M.; Leonardi, R. Deep learning for automated skeletal bone age assessment in X-ray images. Med. Image Anal. 2017, 36, 41–51. [Google Scholar] [CrossRef]
- Eckstein, F.; Burstein, D.; Link, T.M. Quantitative MRI of cartilage and bone: Degenerative changes in osteoarthritis. NMR Biomed. 2006, 19, 822–854. [Google Scholar] [CrossRef]
- Daffner, R.; Lupetin, A.; Dash, N.; Deeb, Z.; Sefczek, R.; Schapiro, R. MRI in the detection of malignant infiltration of bone marrow. Am. J. Roentgenol. 1986, 146, 353–358. [Google Scholar] [CrossRef]
- Karampinos, D.C.; Ruschke, S.; Dieckmeyer, M.; Diefenbach, M.; Franz, D.; Gersing, A.S.; Krug, R.; Baum, T. Quantitative MRI and spectroscopy of bone marrow. J. Magn. Reson. Imaging 2017, 47, 332–353. [Google Scholar] [CrossRef]
- Jerban, S.; Ma, Y.; Wong, J.H.; Nazaran, A.; Searleman, A.; Wan, L.; Williams, J.; Du, J.; Chang, E.Y. Ultrashort echo time magnetic resonance imaging (UTE-MRI) of cortical bone correlates well with histomorphometric assessment of bone microstructure. Bone 2019, 123, 8–17. [Google Scholar] [CrossRef]
- Carrion, C.; Guérin, E.; Gachard, N.; Le Guyader, A.; Giraut, S.; Feuillard, J. Adult Bone Marrow Three-Dimensional Phenotypic Landscape of B-Cell Differentiation. Cytom. Part. B Clin. Cytom. 2018, 96, 30–38. [Google Scholar] [CrossRef]
- Zimmermann, E.A.; Riedel, C.; Schmidt, F.N.; Stockhausen, K.E.; Chushkin, Y.; Schaible, E.; Gludovatz, B.; Vettorazzi, E.; Zontone, F.; Püschel, K.; et al. Mechanical Competence and Bone Quality Develop during Skeletal Growth. J. Bone Miner. Res. 2019, 34, 1461–1472. [Google Scholar] [CrossRef]
- Rafałko, G.; Borowska, M.; Szarmach, J. Statistical Analysis of Radiographic Textures Illustrating Healing Process after the Guided Bone Regeneration Surgery. In Proceedings of the Advances in Intelligent Systems and Computing; Springer Science and Business Media LLC: Berlin, Germany, 2018; pp. 217–226. [Google Scholar]
- Toghyani, S.; Nasseh, I.; Aoun, G.; Noujeim, M. Effect of Image Resolution and Compression on Fractal Analysis of the Periapical Bone. Acta Inform. Medica 2019, 27, 167–170. [Google Scholar] [CrossRef]
- Baltina, T.; Sachenkov, O.; Gerasimov, O.; Baltin, M.; Fedyanin, A.; Lavrov, I. The Influence of Hindlimb Unloading on the Bone Tissue’s Structure. BioNanoScience 2018, 8, 864–867. [Google Scholar] [CrossRef]
- Karamov, R.; Martulli, L.M.; Kerschbaum, M.; Sergeichev, I.; Swolfs, Y.; Lomov, S.V. Micro-CT based structure tensor analysis of fibre orientation in random fibre composites versus high-fidelity fibre identification methods. Compos. Struct. 2020, 235, 111818. [Google Scholar] [CrossRef]
- Gerasimov, O.; Yaikova, V.; Baltina, T.; Baltin, M.; Fedyanin, A.; Zamaliev, R.; Sachenkov, O. Modeling the change in the stiffness parameters of bone tissue under the influence of external loads. J. Phys. Conf. Ser. 2019, 1158, 022045. [Google Scholar] [CrossRef]
- Ren, Y.; Zhu, C.; Xiao, S. Small Object Detection in Optical Remote Sensing Images via Modified Faster R-CNN. Appl. Sci. 2018, 8, 813. [Google Scholar] [CrossRef]
- Li, S.; Song, W.; Fang, L.; Chen, Y.; Ghamisi, P.; Benediktsson, J.A. Deep Learning for Hyperspectral Image Classification: An Overview. IEEE Trans. Geosci. Remote. Sens. 2019, 57, 6690–6709. [Google Scholar] [CrossRef]
- Talo, M.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Application of deep transfer learning for automated brain abnormality classification using MR images. Cogn. Syst. Res. 2019, 54, 176–188. [Google Scholar] [CrossRef]
- Yang, X.; Ye, Y.; Li, X.; Lau, R.Y.; Zhang, X.; Huang, X. Hyperspectral Image Classification with Deep Learning Models. IEEE Trans. Geosci. Remote. Sens. 2018, 56, 5408–5423. [Google Scholar] [CrossRef]
- Sun, W.; Wang, R. Fully Convolutional Networks for Semantic Segmentation of Very High Resolution Remotely Sensed Images Combined With DSM. IEEE Geosci. Remote. Sens. Lett. 2018, 15, 474–478. [Google Scholar] [CrossRef]
- Monteiro, M.; Figueiredo, M.A.T.; Oliveira, A.L. Conditional Random Fields as Recurrent Neural Networks for 3D Medical Imaging Segmentation. arXiv 2018, arXiv:1807.07464. [Google Scholar]
- Yang, T.; Wu, Y.; Zhao, J.; Guan, L. Semantic segmentation via highly fused convolutional network with multiple soft cost functions. Cogn. Syst. Res. 2019, 53, 20–30. [Google Scholar] [CrossRef]
- Ambellan, F.; Tack, A.; Ehlke, M.; Zachow, S. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the Osteoarthritis Initiative. Med. Image Anal. 2019, 52, 109–118. [Google Scholar] [CrossRef]
- Alsinan, A.Z.; Patel, V.M.; Hacihaliloglu, I. Automatic segmentation of bone surfaces from ultrasound using a filter-layer-guided CNN. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 775–783. [Google Scholar] [CrossRef]
- Fu, J.; Yang, Y.; Singhrao, K.; Ruan, D.; Low, D.A.; Lewis, J.H. Male pelvic synthetic CT generation from T1-weighted MRI using 2D and 3D convolutional neural networks. arXiv 2018, arXiv:1803.00131. [Google Scholar]
- Liu, F.; Zhou, Z.; Jang, H.; Samsonov, A.; Zhao, G.; Kijowski, R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn. Reson. Med. 2017, 79, 2379–2391. [Google Scholar] [CrossRef]
- Deniz, C.M.; Xiang, S.; Hallyburton, R.S.; Welbeck, A.; Babb, J.S.; Honig, S.; Cho, K.; Chang, G. Segmentation of the Proximal Femur from MR Images using Deep Convolutional Neural Networks. Sci. Rep. 2018, 8, 16485. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, S.; Nishio, M.; Yakami, M.; Nakagomi, K.; Togashi, K. Bone segmentation on whole-body CT using convolutional neural network with novel data augmentation techniques. Comput. Biol. Med. 2020, 121, 103767. [Google Scholar] [CrossRef] [PubMed]
- Ajayakumar, R.; Rajan, R. Predominant Instrument Recognition in Polyphonic Music Using GMM-DNN Framework. In Proceedings of the 2020 International Conference on Signal. Processing and Communications (SPCOM), Bangalore, India, 20–23 July 2020; pp. 1–5. [Google Scholar]
- Lunga, D.; Yang, H.L.; Reith, A.E.; Weaver, J.; Yuan, J.; Bhaduri, B.L. Domain-Adapted Convolutional Networks for Satellite Image Classification: A Large-Scale Interactive Learning Workflow. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2018, 11, 962–977. [Google Scholar] [CrossRef]
- Zhang, C.; Li, G.; Du, S. Multi-Scale Dense Networks for Hyperspectral Remote Sensing Image Classification. IEEE Trans. Geosci. Remote. Sens. 2019, 57, 9201–9222. [Google Scholar] [CrossRef]
- Karpathy, A.; Toderici, G.; Shetty, S.; Leung, T.; Sukthankar, R.; Fei-Fei, L.; Shetty, S.; Leung, T. Large-Scale Video Classification with Convolutional Neural Networks. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1725–1732. [Google Scholar]
- Tur, G.; Celikyilmaz, A.; He, X.; Hakkani-Tür, D.; Deng, L. Deep Learning in Conversational Language Understanding. In Deep Learning in Natural Language Processing; Springer Science and Business Media LLC: Singapore, 2018; pp. 23–48. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. Sep. 4, 2014. arXiv 2019, arXiv:1409.1556. [Google Scholar]
- Abdel-Hamid, O.; Mohamed, A.-R.; Jiang, H.; Deng, L.; Penn, G.; Yu, D. Convolutional Neural Networks for Speech Recognition. IEEE/ACM Trans. Audio Speech Lang. Process. 2014, 22, 1533–1545. [Google Scholar] [CrossRef]
- Qian, Y.; Bi, M.; Tan, T.; Yu, K. Very Deep Convolutional Neural Networks for Noise Robust Speech Recognition. IEEE/ACM Trans. Audio Speech Lang. Process. 2016, 24, 2263–2276. [Google Scholar] [CrossRef]
- Abdel-Hamid, O.; Mohamed, A.-R.; Jiang, H.; Penn, G. Applying Convolutional Neural Networks concepts to hybrid NN-HMM model for speech recognition. In Proceedings of the 2012 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Kyoto, Japan, 25–30 March 2012; pp. 4277–4280. [Google Scholar]
- Shi, H.; Ushio, T.; Endo, M.; Yamagami, K.; Horii, N. A multichannel convolutional neural network for cross-language dialog state tracking. In Proceedings of the 2016 IEEE Spoken Language Technology Workshop (SLT), San Diego, CA, USA, 13–16 December 2016; pp. 559–564. [Google Scholar] [CrossRef]
- Mane, D.T.; Kulkarni, U.V. A Survey on Supervised Convolutional Neural Network and Its Major Applications. In Deep Learning and Neural Networks; IGI Global: Hershey, PA, USA, 2020; pp. 1058–1071. [Google Scholar]
- Świętojański, P.; Ghoshal, A.; Renals, S. Convolutional Neural Networks for Distant Speech Recognition. IEEE Signal. Process. Lett. 2014, 21, 1120–1124. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the Dimensionality of Data with Neural Networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Hinton, G.E.; Osindero, S.; Teh, Y.-W. A Fast Learning Algorithm for Deep Belief Nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef]
- Larochelle, H.; Mandel, M.; Pascanu, R.; Bengio, y. Learning algorithms for the classification restricted boltzmann machine. J. Mach. Learn. Res. 2012, 13, 643–669. [Google Scholar]
- Salakhutdinov, R.; Mnih, A.; Hinton, G. Restricted Boltzmann machines for collaborative filtering. In Proceedings of the 24th International Conference on Machine learning, Corvalis, OR, USA, 20–24 June 2007; pp. 791–798. [Google Scholar]
- Cho, K.; Ilin, A.; Raiko, T. Improved Learning of Gaussian-Bernoulli Restricted Boltzmann Machines. In Proceedings of the Haptics: Science, Technology, Applications; Springer Science and Business Media LLC: Berlin, Germany, 2011; Volume 6791, pp. 10–17. [Google Scholar]
- Mohamed, A.-R.; Dahl, G.; Hinton, G. Acoustic Modeling Using Deep Belief Networks. IEEE Trans. Audio Speech Lang. Process. 2011, 20, 14–22. [Google Scholar] [CrossRef]
- Li, T.; Zhang, J.; Zhang, Y. Classification of hyperspectral image based on deep belief networks. In Proceedings of the 2014 IEEE International Conference on Image Processing (ICIP), Paris, France, 27–30 October 2014; pp. 5132–5136. [Google Scholar]
- Jie, N.; Xiongzhu, B.; Zhong, L.; Yao, W. An Improved Bilinear Deep Belief Network Algorithm for Image Classification. In Proceedings of the 2014 Tenth International Conference on Computational Intelligence and Security, Kunming, China, 15–16 November 2014; pp. 189–192. [Google Scholar]
- Huang, W.; Song, G.; Hong, H.; Xie, K. Deep Architecture for Traffic Flow Prediction: Deep Belief Networks with Multitask Learning. IEEE Trans. Intell. Transp. Syst. 2014, 15, 2191–2201. [Google Scholar] [CrossRef]
- Zhang, X.-L.; Wu, J. Deep Belief Networks Based Voice Activity Detection. IEEE Trans. Audio Speech Lang. Process. 2012, 21, 697–710. [Google Scholar] [CrossRef]
- Sarikaya, R.; Hinton, G.E.; Deoras, A. Application of Deep Belief Networks for Natural Language Understanding. IEEE/ACM Trans. Audio Speech Lang. Process. 2014, 22, 778–784. [Google Scholar] [CrossRef]
- Chung, J.; Gulcehre, C.; Cho, K.; Bengio, Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv 2014, arXiv:1412.3555. [Google Scholar]
- Gers, F.A.; Schraudolph, N.N.; Schmidhuber, J. Learning precise timing with LSTM recurrent networks. J. Mach. Learn. Res. 2002, 3, 115–143. [Google Scholar]
- Gers, F.A.; Schmidhuber, J.; Cummins, F. Learning to Forget: Continual Prediction with LSTM. Neural Comput. 2000, 12, 2451–2471. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Q.; Ding, M.; Li, J. High Precision Dimensional Measurement with Convolutional Neural Network and Bi-Directional Long Short-Term Memory (LSTM). Sensors 2019, 19, 5302. [Google Scholar] [CrossRef]
- Hermanto, A.; Adji, T.B.; Setiawan, N.A. Recurrent neural network language model for English-Indonesian Machine Translation: Experimental study. In Proceedings of the 2015 International Conference on Science in Information Technology (ICSITech), Yogyakarta, Indonesia, 27–28 October 2015; pp. 132–136. [Google Scholar]
- Fu, T.; Han, Y.; Li, X.; Liu, Y.; Wu, X. Integrating prosodic information into recurrent neural network language model for speech recognition. In Proceedings of the 2015 Asia-Pacific Signal and Information Processing Association Annual Summit and Conference (APSIPA), Hong Kong, China, 16–19 December 2015; pp. 1194–1197. [Google Scholar]
- Chen, X.; Liu, X.; Wang, Y.; Gales, M.J.F.; Woodland, P.C. Efficient Training and Evaluation of Recurrent Neural Network Language Models for Automatic Speech Recognition. IEEE/ACM Trans. Audio Speech Lang. Process. 2016, 24, 2146–2157. [Google Scholar] [CrossRef]
- Chien, J.-T.; Ku, Y.-C. Bayesian Recurrent Neural Network for Language Modeling. IEEE Trans. Neural Networks Learn. Syst. 2015, 27, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Y.; Xie, G.-S.; Liu, C.-L.; Bengio, Y. End-to-End Online Writer Identification with Recurrent Neural Network. IEEE Trans. Human-Mach. Syst. 2016, 47, 285–292. [Google Scholar] [CrossRef]
- Guo, H.; Song, S.; Wang, J.; Guo, M.; Cheng, Y.; Wang, Y.; Tamura, S. 3D surface voxel tracing corrector for accurate bone segmentation. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1549–1563. [Google Scholar] [CrossRef]
- Zaman, A.; Park, Y.; Park, C.; Park, I.; Joung, S. Deep Learning-based Bone Contour Segmentation from Ultrasound Images. 대한전자공학회 학술대회 2019, 1341–1342. [Google Scholar]
- Rehman, F.; Shah, S.I.A.; Riaz, M.N.; Gilani, S.O.; Faiza, R. A Region-Based Deep Level Set Formulation for Vertebral Bone Segmentation of Osteoporotic Fractures. J. Digit. Imaging 2019, 33, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: New York, NY, USA, 2015; pp. 234–241. [Google Scholar]
- Belal, S.L.; Sadik, M.; Kaboteh, R.; Enqvist, O.; Ulén, J.; Poulsen, M.H.; Simonsen, J.A.; Hoilund-Carlsen, P.F.; Edenbrandt, L.; Trägårdh, E. Deep learning for segmentation of 49 selected bones in CT scans: First step in automated PET/CT-based 3D quantification of skeletal metastases. Eur. J. Radiol. 2019, 113, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Villa, M.; Dardenne, G.; Nasan, M.; LeTissier, H.; Hamitouche, C.; Stindel, E. FCN Based Approach for the Automatic Segmentation of Bone Surfaces in Ultrasound Images. In Proceedings of the CAOS the 18th Annual Meeting of the International Society for Computer Assisted Orthopaedic Surgery, Beijing, China, 6–9 June 2018; Volume 13, pp. 1707–1716. [Google Scholar] [CrossRef]
- Wang, P.; Patel, V.M.; Hacihaliloglu, I. Simultaneous Segmentation and Classification of Bone Surfaces from Ultrasound Using a Multi-feature Guided CNN. In Proceedings of the Agreement Technologies; Springer Science and Business Media LLC: Berlin, Germany, 2018; pp. 134–142. [Google Scholar]
- Pham, D.D.; Dovletov, G.; Warwas, S.; Landgraeber, S.; Jager, M.; Pauli, J. Deep Learning with Anatomical Priors: Imitating Enhanced Autoencoders in Latent Space for Improved Pelvic Bone Segmentation in MRI. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1166–1169. [Google Scholar] [CrossRef]
- Hemke, R.; Buckless, C.G.; Tsao, A.; Wang, B.; Torriani, M. Deep learning for automated segmentation of pelvic muscles, fat, and bone from CT studies for body composition assessment. Skelet. Radiol. 2019, 49, 387–395. [Google Scholar] [CrossRef]
- Klein, A.; Warszawski, J.; Hillengaß, J.; Maier-Hein, K.H. Towards Whole-body CT Bone Segmentation. In Informatik Aktuell; Springer Science and Business Media LLC: Berlin, Germany, 2018; pp. 204–209. [Google Scholar]
- Greulich, W.W.; Pyle, S.I. Radiographic atlas of skeletal development of the hand and wrist. Am. J. Med. Sci. 1959, 238, 393. [Google Scholar] [CrossRef]
- Garn, S.M.; Tanner, J.M. Assessment of Skeletal Maturity and Prediction of Adult Height (TW2 Method). Man 1986, 21, 142. [Google Scholar] [CrossRef]
- Nadeem, M.W.; Al Ghamdi, M.A.; Hussain, M.; Khan, M.A.; Masood, K.; AlMotiri, S.H.; Butt, S.A. Brain Tumor Analysis Empowered with Deep Learning: A Review, Taxonomy, and Future Challenges. Brain Sci. 2020, 10, 118. [Google Scholar] [CrossRef]
- Lee, H.; Tajmir, S.; Lee, J.; Zissen, M.; Yeshiwas, B.A.; Alkasab, T.K.; Choy, G.; Do, S. Fully Automated Deep Learning System for Bone Age Assessment. J. Digit. Imaging 2017, 30, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Liang, B.; Li, J.; Zheng, Z. A Deep Automated Skeletal Bone Age Assessment Model with Heterogeneous Features Learning. J. Med. Syst. 2018, 42, 249. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, J.; Zhang, Y.; Lu, Y.; Liu, S.-Y. Automatic feature extraction in X-ray image based on deep learning approach for determination of bone age. Futur. Gener. Comput. Syst. 2020, 110, 795–801. [Google Scholar] [CrossRef]
- Kim, J.R.; Yoon, H.M.; Hong, S.H.; Kim, S.; Shim, W.H.; Lee, J.S.; Cho, Y.A. Computerized Bone Age Estimation Using Deep Learning Based Program: Evaluation of the Accuracy and Efficiency. Am. J. Roentgenol. 2017, 209, 1374–1380. [Google Scholar] [CrossRef]
- Van Steenkiste, T.; Ruyssinck, J.; Janssens, O.; Vandersmissen, B.; Vandecasteele, F.; Devolder, P.; Achten, E.; Van Hoecke, S.; Deschrijver, D.; Dhaene, T. Automated Assessment of Bone Age Using Deep Learning and Gaussian Process Regression. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018; pp. 674–677. [Google Scholar]
- Lee, J.H.; Kim, K.G. Applying Deep Learning in Medical Images: The Case of Bone Age Estimation. Health Inform. Res. 2018, 24, 86–92. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Q.; Han, J.; Jia, Y. Application of Deep learning in Bone age assessment. IOP Conf. Series: Earth Environ. Sci. 2018, 199, 032012. [Google Scholar]
- Wang, S.; Summers, R.M. Machine learning and radiology. Med. Image Anal. 2012, 16, 933–951. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.C.; Okoye, M.I.; Kiple, D.; Kimmerle, E.H.; Reinhard, K.J. Three-dimensional (3-D) imaging in post-mortem examinations: Elucidation and identification of cranial and facial fractures in victims of homicide utilizing 3-D computerized imaging reconstruction techniques. Int. J. Leg. Med. 1999, 113, 33–37. [Google Scholar] [CrossRef]
- Heimer, J.; Thali, M.J.; Ebert, L. Classification based on the presence of skull fractures on curved maximum intensity skull projections by means of deep learning. J. Forensic Radiol. Imaging 2018, 14, 16–20. [Google Scholar] [CrossRef]
- Connor, S.; Tan, G.; Fernando, R.; Chaudhury, N. Computed tomography pseudofractures of the mid face and skull base. Clin. Radiol. 2005, 60, 1268–1279. [Google Scholar] [CrossRef] [PubMed]
- Mutasa, S.; Chang, P.D.; Ruzal-Shapiro, C.; Ayyala, R. MABAL: A Novel Deep-Learning Architecture for Machine-Assisted Bone Age Labeling. J. Digit. Imaging 2018, 31, 513–519. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the Thirty-first AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Bui, T.D.; Lee, J.-J.; Shin, J. Incorporated region detection and classification using deep convolutional networks for bone age assessment. Artif. Intell. Med. 2019, 97, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Khalid, H.; Hussain, M.; Al Ghamdi, M.A.; Khalid, T.; Khalid, K.; Khan, M.A.; Fatima, K.; Masood, K.; AlMotiri, S.H.; Farooq, M.S.; et al. A Comparative Systematic Literature Review on Knee Bone Reports from MRI, X-rays and CT Scans Using Deep Learning and Machine Learning Methodologies. Diagnostics 2020, 10, 518. [Google Scholar] [CrossRef]
- Zhou, J.; Li, Z.; Zhi, W.; Liang, B.; Moses, D.; Dawes, L. Using Convolutional Neural Networks and Transfer Learning for Bone Age Classification. In Proceedings of the 2017 International Conference on Digital Image Computing: Techniques and Applications (DICTA), Sydney, Australia, 29 November–1 December 2017; pp. 1–6. [Google Scholar]


| Study | Data Set | Method/Model | Proposed Solution | Languages/Libraries/Frameworks/Tools/Software’s for Implementation and Simulation | Evaluation |
|---|---|---|---|---|---|
| Faisal Rehman et al. [68] | CSI 2014 and xVertSeg.v1 | U-Net | FU-Net based Model | Python using Tensor flow on windows desktop system Intel-(R) i-7 Central Processing Unit (CPU) and 1080 GTX graphics card with GPU memory 8 GB. | Dice Score = 96.4 ± 0.8 ASD (mm) = 0.1 ± 0.05 |
| Ahmed Z. Alsinan et al. [29] | B-mode US images ClariusC3 | Convolutional Neural Network | Filter-Layer-Guided CNN | Keras framework and Tensor flow, Intel Xeon CPU at 3.00 GHz, and Nvidia Titan-X GPU with 8 GB of memory, Sonix-Touch | F-Score = 95% Bone Surface Localization Error = 0.2 mm |
| Robert Hemke et al. [74] | Custom Developed (200 Samples) | Convolutional Neural Network | Deep Convolutional Neural Network-based Model | Python 3.7, Keras library (V2.2.4, https://keras.io), Tensorflow 1.13.1, Multi-GPU (4× NVIDIA Titan Xp units) | Dice score = 0.92 Mean Time to segment one CT image = 0.07 s (GPU), 2.51 s (CPU) |
| Asaduz Zaman et al. [67] | Custom Developed (1950 Samples) | U-Net | U-Net based Encoder–Decoder Model | Not Mentioned | Dice Score = 0.692 ± 0.011 |
| Sarah Lindgren Belala et al. [70] | Custom Developed at Sahlgrenska University Hospital, Goteborg, Sweden (100 Samples) | CNN | Fully Convolutional Neural Network | Not Mentioned | Sorensen-Dice index (SDI) for Sacrum bone = 0.88% |
| D. D. Pham et al. [73] | Custom Developed | U-Net | 2D Encoder-Decoder based U-Net Model | Tensor flow, GTX 1080 GPU | Dice Score = 73.45 ± 5.93 |
| Haoyan Guo et al. [66] | Custom Developed (212 Samples) | Not Mention | Gaussian Standard Deviation (GSD) | C++, Ubuntu platform, PC with a 2.33 GHz Intel quad-core processor, 8 GB RAM | Dice Overlap Coefficient = 98.06 ± 0.58% |
| M. Villa et al. [71] | Custom developed US images based (3692 Samples) | Fully Convolutional Networks (FCN) | FCN based Model | Python, Caffe framework | RMSE = 1.32 ± 3.70 mm Mean Recall = 62% Precision = 64% F1 Score = 57% Accuracy = 80% Specificity = 83% |
| Andre Klein et al. [75] | Custom-developed (6000 Samples) | U-Net | U-Net with Padded Convolutions based Model | MITK, NVIDIA Titan X GPU. | Dice Score = 0.96 |
| Puyang Wang et al. [72] | Custom developed US images based (519 Samples) | CNN | Multi-Feature Guided Convolutional Neural Network (CNN) | MATLAB | Recall = 0.97 Precision = 0.965 F-score = 0.968 |
| Study | Data Set | Method/Model | Proposed Solution | Languages/Libraries/ Frameworks/ Tools/Software’s Used for Implementation and Simulation | Evaluation |
|---|---|---|---|---|---|
| Xu Chen et al. [81] | Custom Developed at Shengjing Hospital of China Medical University (Samples) | Convolutional Neural Network (CNN) | Depth Neural Network, Local Binary Patterns (LBP) features and Glutamate Cysteine Ligase Modifier subunit (GCLM) | Tensor flow | Average Absolute Error = 0.455 |
| Chao Tong et al. [80] | Public Database Digital Hand-Atlas (Samples) | Convolutional Neural Networks (CNNs) | Convolutional Neural Networks (CNNs) and Support Vector Regression (SVR) based Model | Matlab, Keras framework with Tensor Flow | Mean Absolute Error (MAE) = 0.547 |
| Jang Hyung Lee et al. [84] | Radiological Society of North America (RSNA) challenge dataset | CNN | CNN and CaffeNet based Model | Caffe, Tensorflow, Keras, Theano and Torch, Linux Ubuntu OS, NVIDIA GTX 1060 GPU, CUDA library, and CUDNN library. | Concordance Correlation Coefficient = 0.78 |
| Tom Van Steenkiste et al. [83] | Radiological Society of North America challenge dataset (Samples) | Visual Geometry Group (VGG16)echanical Competence and Bone Quality Deve | Visual Geometry Group (VGG16) and Gaussian Process Regression (GPR) based Model | Not Mention | Mean Absolute Difference = 6.80 (−0.94) |
| Hyunkwang Lee et al. [79] | Custom-developed using open-source software OsiriX and DICOM images (Samples) | CNN | ImageNet pre-trained, fine-tuned convolutional neural network (CNN) | GoogLeNet and Caffe Zoo | Accuracy = 98.56% |
| Jeong Rye Kim et al. [82] | Custom Developed (Samples) | Deep Neural Network | Greulich-Pyle and Deep Neural Network-Based Model | Not Mention | Root Mean Square Error = 0.42 |
| Study | Data Set | Method/Model | Proposed Solution | Languages/Libraries/ Frameworks/ Tools/Software’s Used for Implementation and Simulation | Evaluation |
|---|---|---|---|---|---|
| Jakob Heime et al. [89] | Custom-developed (150 Samples) | Deep Neural Network | Deep Neural Network-based Customized Model | VIDI, COGNEX, Natick, MA, USA | Accuracy = 0.965% Threshold = 0.79% Sensitivity = 91.4% Specificity = 87.5% |
| Simukayi Mutasa et al. [91] | Digital Hand-Atlas (Samples) | Convolutional Neural Networks (CNNs) | Customized Convolutional Neural Networks with Inception Block | Python Tensor Flow v1.1 library, Ubuntu 16.04 workstation, NVIDIA TITAN X Pascal GPU. | Mean Absolute Error (MAE) = 0.536 |
| Yagang WANG et al. [85] | GoogLeNet | Matlab | Accuracy = 94.4% | ||
| Toan Duc Bui et al. [95] | Public Dataset Digital Hand-Atlas (DHA) (1375 Samples) | Deep Convolutional Networks (DNNs) | Deep Convolutional Networks (DNNs) and Tanner Whitehouse (TW3) based Model | Not Mentioned | MAE = 0.59 RMS = 0.76 |
| Jianlong Zhou et al. [97] | Digital Hand-Atlas | Convolutional Neural Networks | Convolutional Neural Networks and Transfer Learning Based Model | Not Mentioned | MAE = 0.72 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nadeem, M.W.; Goh, H.G.; Ali, A.; Hussain, M.; Khan, M.A.; Ponnusamy, V.a/p. Bone Age Assessment Empowered with Deep Learning: A Survey, Open Research Challenges and Future Directions. Diagnostics 2020, 10, 781. https://doi.org/10.3390/diagnostics10100781
Nadeem MW, Goh HG, Ali A, Hussain M, Khan MA, Ponnusamy Va/p. Bone Age Assessment Empowered with Deep Learning: A Survey, Open Research Challenges and Future Directions. Diagnostics. 2020; 10(10):781. https://doi.org/10.3390/diagnostics10100781
Chicago/Turabian StyleNadeem, Muhammad Waqas, Hock Guan Goh, Abid Ali, Muzammil Hussain, Muhammad Adnan Khan, and Vasaki a/p Ponnusamy. 2020. "Bone Age Assessment Empowered with Deep Learning: A Survey, Open Research Challenges and Future Directions" Diagnostics 10, no. 10: 781. https://doi.org/10.3390/diagnostics10100781
APA StyleNadeem, M. W., Goh, H. G., Ali, A., Hussain, M., Khan, M. A., & Ponnusamy, V. a/p. (2020). Bone Age Assessment Empowered with Deep Learning: A Survey, Open Research Challenges and Future Directions. Diagnostics, 10(10), 781. https://doi.org/10.3390/diagnostics10100781






