Peptide Nucleic Acid (PNA)-Enhanced Specificity of a Dual-Target Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Assay for the Detection and Differentiation of SARS-CoV-2 from Related Viruses
Abstract
:1. Introduction
2. Materials and Methods
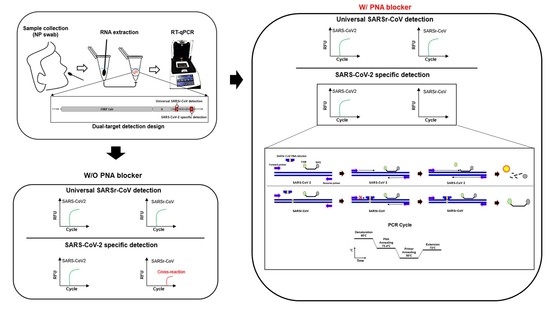
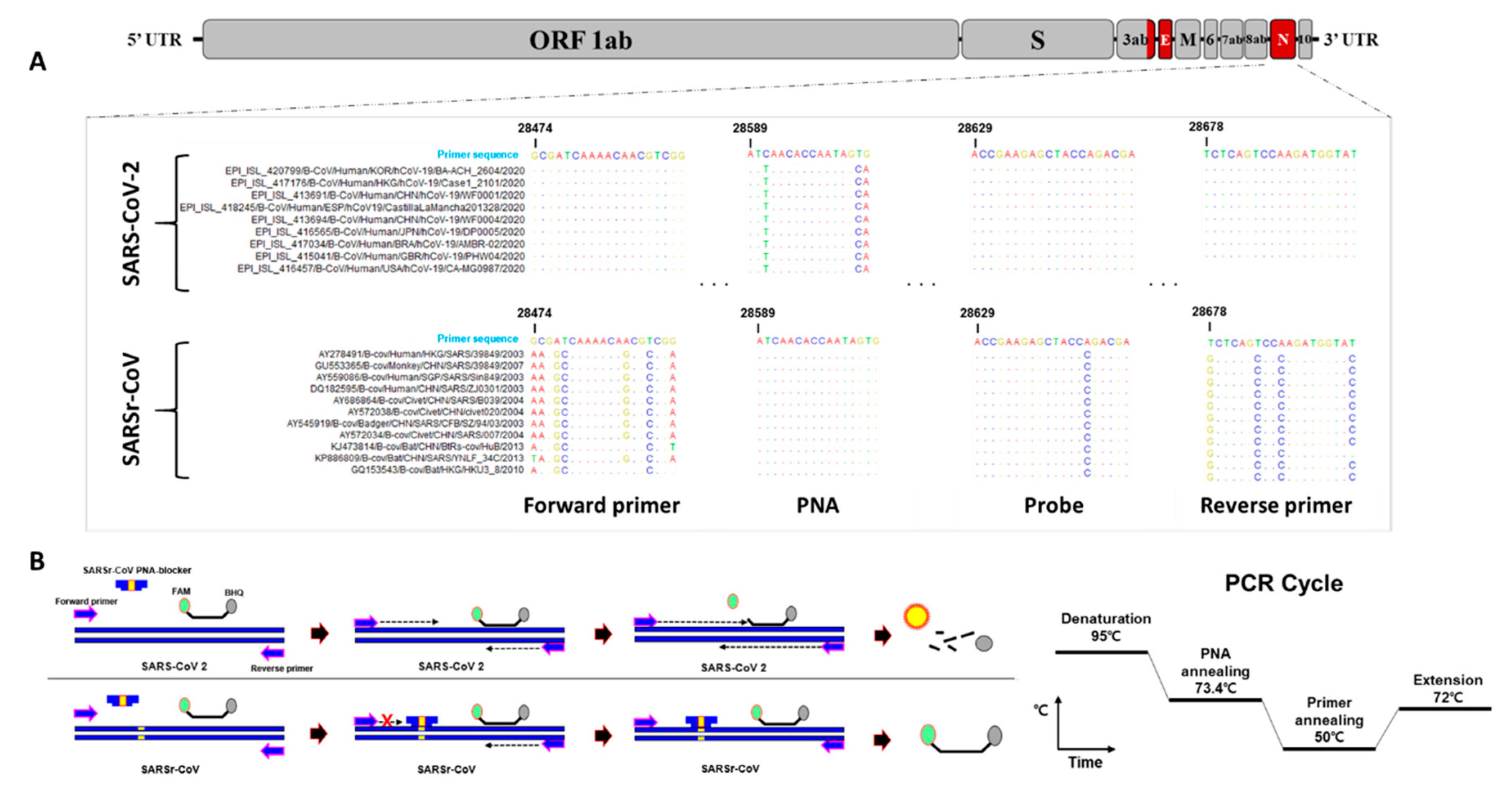
2.1. Primers, Probes, and PNA Blocker Design for Dual-Target Detection
2.2. Viral Propagation in Cell Lines and RNA Retrieval
2.3. SARS-CoV-2 Viruses and Clinical Specimens
2.4. Optimization of Primer and Probe Concentrations
2.5. RT-qPCR Condition Optimization
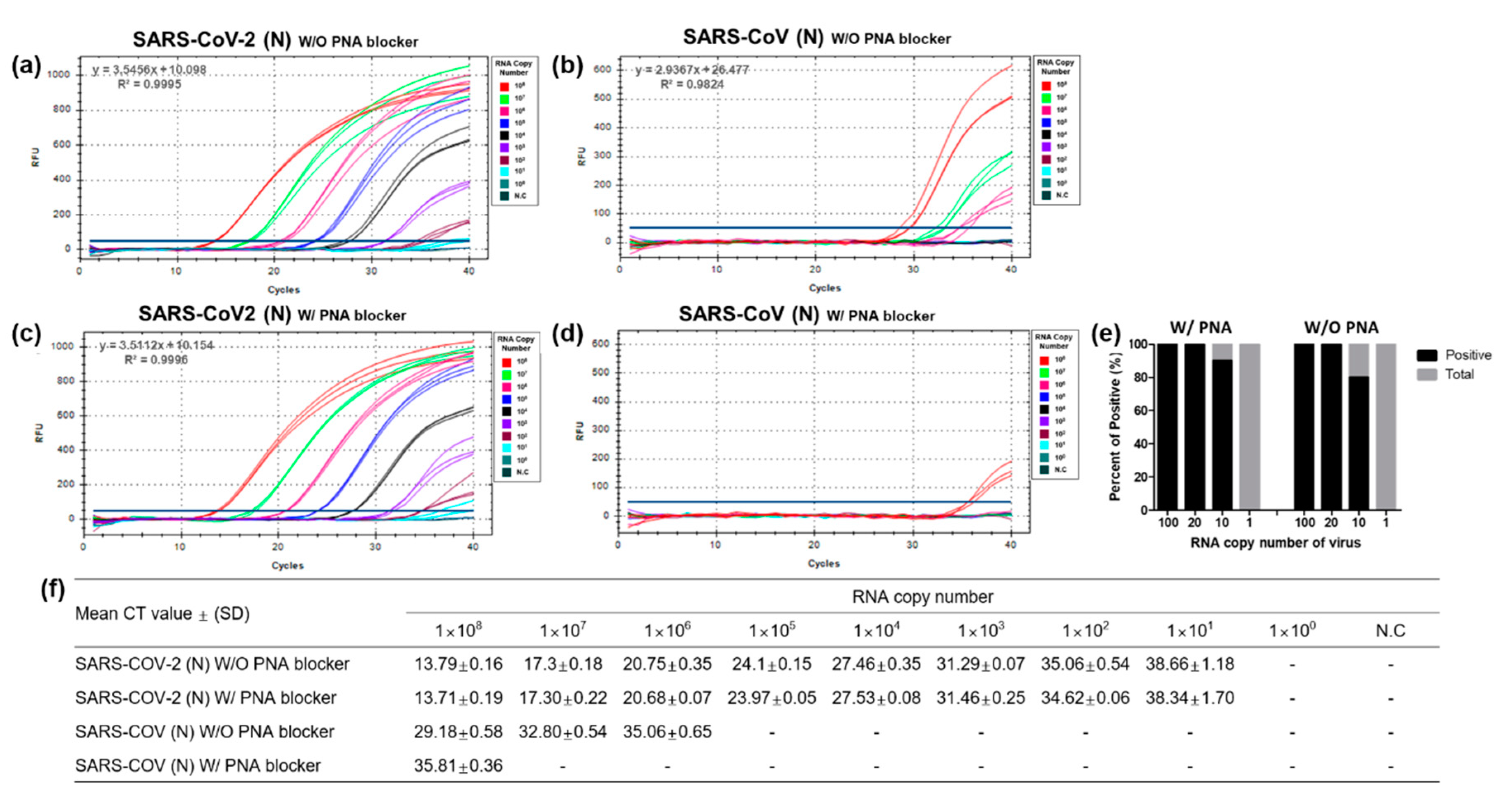
2.6. Sensitivity and Specificity Confirmation of the PNA-Mediated Dual-Target RT-qPCR Assay
2.7. Ethics Statement
3. Results
3.1. Primer, Probe, and PNA Blocker Design
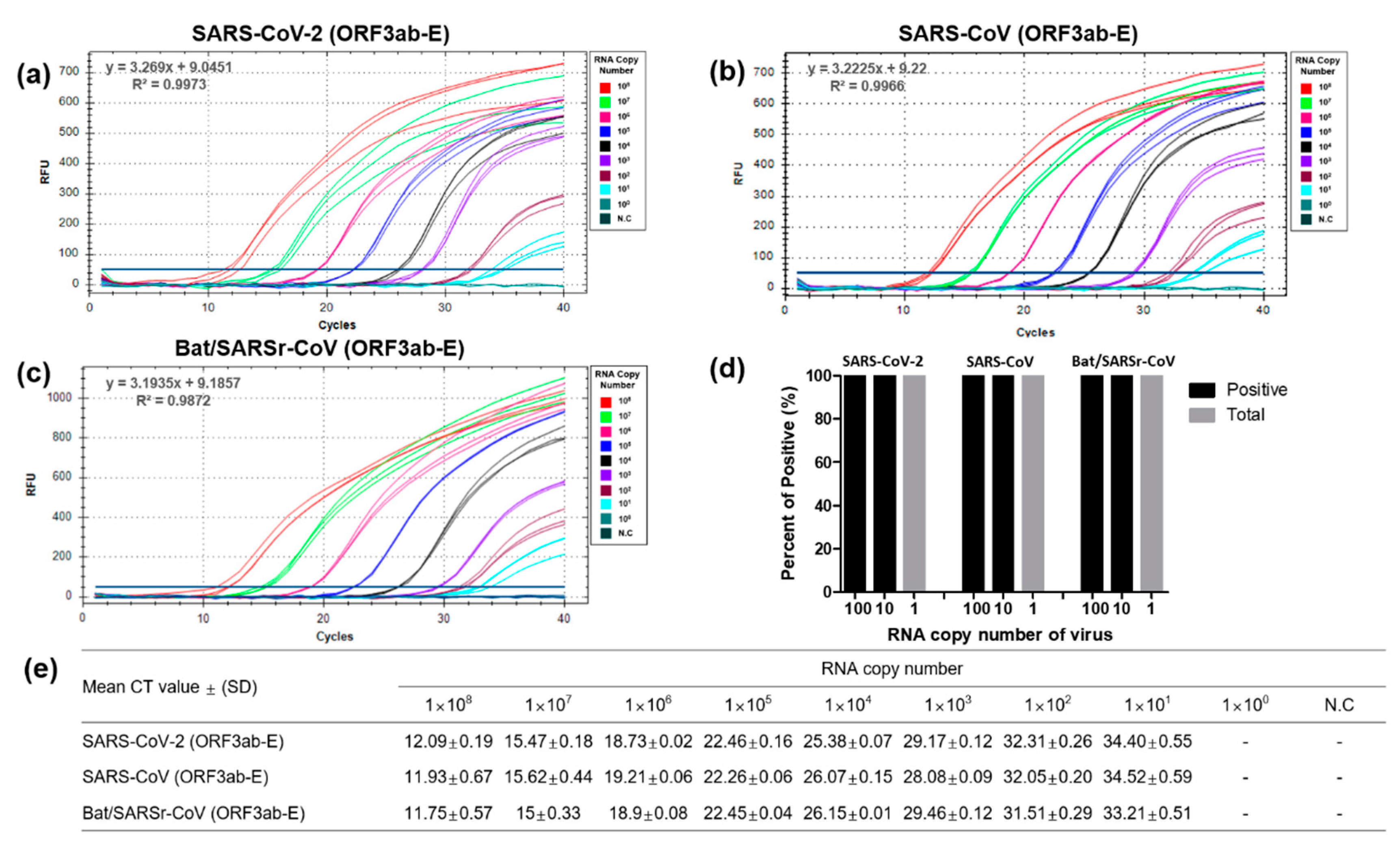
3.2. Optimization and Sensitivity Evaluation of the Universal RT-qPCR Assay for ORF3ab-E Gene Detection
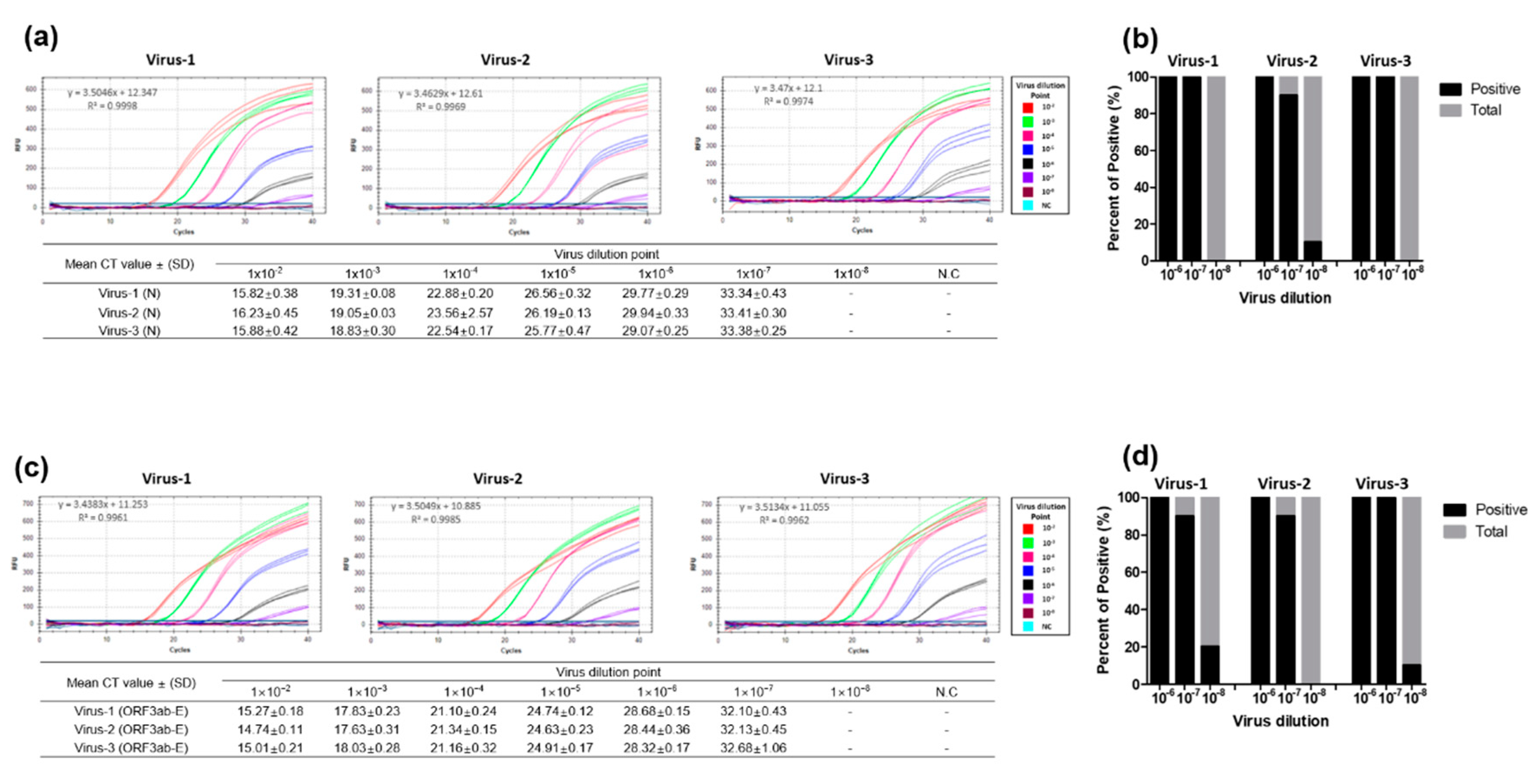
3.3. Performance Evaluation of the Dual-Target RT-qPCR Assay Using Intact Virus
3.4. Evaluation of the Dual-Target RT-qPCR Assay Using Clinical Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Han, Q.; Lin, Q.; Jin, S.; You, L. Coronavirus 2019-nCoV: A brief perspective from the front line. J. Infect. 2020, 80, 373–377. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018, 100, 163–188. [Google Scholar] [CrossRef]
- Xiu, L.; Binder, R.A.; Alarja, N.A.; Kochek, K.; Coleman, K.K.; Than, S.T.; Bailey, E.S.; Bui, V.N.; Toh, T.H.; Erdman, D.D.; et al. A RT-PCR assay for the detection of coronaviruses from four genera. J. Clin. Virol. 2020, 128, 104391. [Google Scholar] [CrossRef]
- Corman, V.M.; Baldwin, H.J.; Tateno, A.F.; Zerbinati, R.M.; Annan, A.; Owusu, M.; Nkrumah, E.E.; Maganga, G.D.; Oppong, S.; Adu-Sarkodie, Y.; et al. Evidence for an Ancestral Association of Human Coronavirus 229E with Bats. J. Virol. 2015, 89, 11858–11870. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.; Shi, M.; Chommanard, C.; Queen, K.; Zhang, J.; Markotter, W.; Kuzmin, I.V.; Holmes, E.C.; Tong, S. Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huynh, J.; Li, S.; Yount, B.; Smith, A.; Sturges, L.; Olsen, J.C.; Nagel, J.; Johnson, J.B.; Agnihothram, S.; Gates, J.E.; et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012, 86, 12816–12825. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.F.; Lau, S.K.; To, K.K.; Cheng, V.C.; Woo, P.C.; Yuen, K.Y. Middle East respiratory syndrome coronavirus: Another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015, 28, 465–522. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef]
- Boley, P.A.; Alhamo, M.A.; Lossie, G.; Yadav, K.K.; Vasquez-Lee, M.; Saif, L.J.; Kenney, S.P. Porcine Deltacoronavirus Infection and Transmission in Poultry, United States (1). Emerg. Infect. Dis. 2020, 26, 255–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norz, D.; Fischer, N.; Schultze, A.; Kluge, S.; Mayer-Runge, U.; Aepfelbacher, M.; Pfefferle, S.; Lutgehetmann, M. Clinical evaluation of a SARS-CoV-2 RT-PCR assay on a fully automated system for rapid on-demand testing in the hospital setting. J. Clin. Virol. 2020, 128, 104390. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 4 August 2020).
- Jiang, X.; Luo, M.; Zou, Z.; Wang, X.; Chen, C.; Qiu, J. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J. Med. Virol. 2020. [Google Scholar] [CrossRef]
- Chia, P.Y.; Coleman, K.K.; Tan, Y.K.; Ong, S.W.X.; Gum, M.; Lau, S.K.; Lim, X.F.; Lim, A.S.; Sutjipto, S.; Lee, P.H.; et al. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat. Commun. 2020, 11, 2800. [Google Scholar] [CrossRef]
- WHO. Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV). Available online: https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov) (accessed on 19 June 2020).
- Eigner, U.; Reucher, S.; Hefner, N.; Staffa-Peichl, S.; Kolb, M.; Betz, U.; Holfelder, M.; Spier, G.; Pfefferle, S.; Lutgehetmann, M. Clinical evaluation of multiplex RT-PCR assays for the detection of influenza A/B and respiratory syncytial virus using a high throughput system. J. Virol. Methods 2019, 269, 49–54. [Google Scholar] [CrossRef]
- Greub, G.; Sahli, R.; Brouillet, R.; Jaton, K. Ten years of R&D and full automation in molecular diagnosis. Future Microbiol. 2016, 11, 403–425. [Google Scholar] [CrossRef] [Green Version]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brunink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eur. Surveill. 2020, 25. [Google Scholar] [CrossRef] [Green Version]
- Baek, Y.H.; Um, J.; Antigua, K.J.C.; Park, J.H.; Kim, Y.; Oh, S.; Kim, Y.I.; Choi, W.S.; Kim, S.G.; Jeong, J.H.; et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 998–1007. [Google Scholar] [CrossRef] [Green Version]
- Park, G.S.; Ku, K.; Baek, S.H.; Kim, S.J.; Kim, S.I.; Kim, B.T.; Maeng, J.S. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 2020, 22, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, C. Fast, portable tests come online to curb coronavirus pandemic. Nat. Biotechnol. 2020, 38, 515–518. [Google Scholar] [CrossRef] [PubMed]
- WHO. Laboratory Testing for 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases. Available online: https://www.who.int/publications/i/item/laboratory-testing-of-2019-novel-coronavirus-(-2019-ncov)-in-suspected-human-cases-interim-guidance-17-january-2020 (accessed on 19 June 2020).
- Shen, K.; Yang, Y.; Wang, T.; Zhao, D.; Jiang, Y.; Jin, R.; Zheng, Y.; Xu, B.; Xie, Z.; Lin, L.; et al. Diagnosis, treatment, and prevention of 2019 novel coronavirus infection in children: Experts’ consensus statement. World J. Pediatr. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfefferle, S.; Reucher, S.; Norz, D.; Lutgehetmann, M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Eur. Surveill. 2020, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [Green Version]
- Porcheddu, A.; Giacomelli, G. Peptide nucleic acids (PNAs), a chemical overview. Curr. Med. Chem. 2005, 12, 2561–2599. [Google Scholar] [CrossRef]
- Machnik, G.; Skudrzyk, E.; Buldak, L.; Labuzek, K.; Ruczynski, J.; Alenowicz, M.; Rekowski, P.; Nowak, P.J.; Okopien, B. A Novel, Highly Selective RT-QPCR Method for Quantification of MSRV Using PNA Clamping Syncytin-1 (ERVWE1). Mol. Biotechnol. 2015, 57, 801–813. [Google Scholar] [CrossRef] [Green Version]
- Pellestor, F.; Paulasova, P. The peptide nucleic acids (PNAs), powerful tools for molecular genetics and cytogenetics. Eur. J. Hum. Genet. 2004, 12, 694–700. [Google Scholar] [CrossRef]
- Pellestor, F.; Paulasova, P. The peptide nucleic acids, efficient tools for molecular diagnosis (Review). Int. J. Mol. Med. 2004, 13, 521–525. [Google Scholar] [CrossRef]
- Nao, N.; Shirato, K.; Katano, H.; Matsuyama, S.; Takeda, M. Detection of Second Case of 2019-nCoV Infection in Japan. Available online: https://www.niid.go.jp/niid/images/vir3/nCoV/method-niid-20200123-2_erratum.pdf (accessed on 19 June 2020).
- Jung, Y.; Park, G.S.; Moon, J.H.; Ku, K.; Beak, S.H.; Lee, C.S.; Kim, S.; Park, E.C.; Park, D.; Lee, J.H.; et al. Comparative Analysis of Primer-Probe Sets for RT-qPCR of COVID-19 Causative Virus (SARS-CoV-2). ACS Infect. Dis. 2020, 6, 2513–2523. [Google Scholar] [CrossRef]
- Xiu, L.; Zhang, C.; Wu, Z.; Peng, J. Establishment and Application of a Universal Coronavirus Screening Method Using MALDI-TOF Mass Spectrometry. Front. Microbiol. 2017, 8, 1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Wu, R.; Xing, Y.; Du, Q.; Xue, Z.; Xi, Y.; Yang, Y.; Deng, Y.; Han, Y.; Li, K.; et al. Influence of Different Inactivation Methods on Severe Acute Respiratory Syndrome Coronavirus 2 RNA Copy Number. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef] [PubMed]
- Sakai, J.; Tarumoto, N.; Orihara, Y.; Kawamura, R.; Kodana, M.; Matsuzaki, N.; Matsumura, R.; Okane, K.; Kawamura, T.; Takeuchi, S.; et al. Evaluation of a high-speed but low-throughput RT-qPCR system for SARS-CoV-2 detection. J. Hosp. Infect. 2020. [Google Scholar] [CrossRef] [PubMed]
- Poon, L.L.; Chan, K.H.; Wong, O.K.; Cheung, T.K.; Ng, I.; Zheng, B.; Seto, W.H.; Yuen, K.Y.; Guan, Y.; Peiris, J.S. Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clin. Chem. 2004, 50, 67–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.K.; Chen, S.Y.; Liu, I.J.; Chen, Y.C.; Chen, H.L.; Yang, C.F.; Chen, P.J.; Yeh, S.H.; Kao, C.L.; Huang, L.M.; et al. Detection of SARS-associated coronavirus in throat wash and saliva in early diagnosis. Emerg. Infect. Dis. 2004, 10, 1213–1219. [Google Scholar] [CrossRef]
- Poljak, M.; Korva, M.; Knap Gasper, N.; Fujs Komlos, K.; Sagadin, M.; Ursic, T.; Avsic Zupanc, T.; Petrovec, M. Clinical Evaluation of the cobas SARS-CoV-2 Test and a Diagnostic Platform Switch during 48 Hours in the Midst of the COVID-19 Pandemic. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef] [Green Version]

| Assay Signature | Target Region | Primer Position | Primer/Probe | Sequence (5′ to 3′) | Concentration (Per Reaction) |
|---|---|---|---|---|---|
| PNA-mediated SARS-CoV-2 detection | Nucleocapsid | 28371–28389 a | Forward primer | GCGATCAAAACAACGTCGG | 250 nM |
| Nucleocapsid | 28575–28593 a | Reverse primer | ATACCATCTTGGACTGAGA | 250 nM | |
| Nucleocapsid | 28526–28545 a | Probe | FAM-ACCGAAGAGCTACCAGACGA-BHQ | 250 nM | |
| Nucleocapsid | 28486–28502 a | PNA blocker | ATCAACACCAATAGTGGc | 500 nM | |
| 28342–28358 b | |||||
| Universal SARSr-CoV detection | ORF3ab gene | 26184–26201 a | Forward primer | CCGACGACGACTACTAGC | 250 nM |
| Envelope protein | 26365–26386 a | Reverse primer | CTCACGTTAACAATATTGCAGC | 250 nM | |
| Envelope protein | 26329–26384 a | Probe | FAM-TAGCCATCCTTACTGCGCTT-BHQ | 250 nM |
| Virus | Specimen Type | Number of Specimen | Detection Percentage (%) | Ct Value a | ||
|---|---|---|---|---|---|---|
| PNA-N Gene | ORF3ab-E Gene | |||||
| Coronavirus | SARS-CoV-2 | Clinical | 23 b | 82.6 | 100 | 18.17–37.93 c |
| MERS-CoV | Spike | 1 | ND | ND | 15.9 | |
| 229E | Clinical/Spike | 17 | ND | ND | 17.17–40.61 | |
| NL63 | Clinical/Spike | 13 | ND | ND | 17.18–40.91 | |
| OC43 | Clinical/Spike | 17 | ND | ND | 17.62–40.17 | |
| Influenza virus | Type B | Clinical/Spike | 58 | ND | ND | 16.05–41.0 |
| H1N1 | Clinical/Spike | 17 | ND | ND | 15.84–41.15 | |
| H3N2 | Clinical/Spike | 68 | ND | ND | 9.16–41.18 | |
| HPAI H5NX | Spike | 8 | ND | ND | 22.2–24.54 | |
| H7N9 | Spike | 1 | ND | ND | 21.25 | |
| Other respiratory viruses | MPV | Clinical | 11 | ND | ND | 19.10–39.49 |
| RSV A | Clinical | 3 | ND | ND | 21.07–30.8 | |
| RSV B | Clinical | 46 | ND | ND | 16.77–41.97 | |
| PIV | Clinical | 8 | ND | ND | 27.93–39.75 | |
| AdV | Clinical | 3 | ND | ND | 27.96–40.21 | |
| HRV | Clinical | 37 | ND | ND | 15.00–40.00 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, W.-S.; Jeong, J.H.; Nicolas, H.D.G.; Oh, S.; Antigua, K.J.C.; Park, J.-H.; Kim, B.; Yoon, S.-W.; Shin, K.S.; Choi, Y.K.; et al. Peptide Nucleic Acid (PNA)-Enhanced Specificity of a Dual-Target Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Assay for the Detection and Differentiation of SARS-CoV-2 from Related Viruses. Diagnostics 2020, 10, 775. https://doi.org/10.3390/diagnostics10100775
Choi W-S, Jeong JH, Nicolas HDG, Oh S, Antigua KJC, Park J-H, Kim B, Yoon S-W, Shin KS, Choi YK, et al. Peptide Nucleic Acid (PNA)-Enhanced Specificity of a Dual-Target Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Assay for the Detection and Differentiation of SARS-CoV-2 from Related Viruses. Diagnostics. 2020; 10(10):775. https://doi.org/10.3390/diagnostics10100775
Chicago/Turabian StyleChoi, Won-Suk, Ju Hwan Jeong, Halcyon Dawn G. Nicolas, Sol Oh, Khristine Joy C. Antigua, Ji-Hyun Park, Beomkyu Kim, Sun-Woo Yoon, Kyeong Seob Shin, Young Ki Choi, and et al. 2020. "Peptide Nucleic Acid (PNA)-Enhanced Specificity of a Dual-Target Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Assay for the Detection and Differentiation of SARS-CoV-2 from Related Viruses" Diagnostics 10, no. 10: 775. https://doi.org/10.3390/diagnostics10100775
APA StyleChoi, W.-S., Jeong, J. H., Nicolas, H. D. G., Oh, S., Antigua, K. J. C., Park, J.-H., Kim, B., Yoon, S.-W., Shin, K. S., Choi, Y. K., Baek, Y. H., & Song, M.-S. (2020). Peptide Nucleic Acid (PNA)-Enhanced Specificity of a Dual-Target Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Assay for the Detection and Differentiation of SARS-CoV-2 from Related Viruses. Diagnostics, 10(10), 775. https://doi.org/10.3390/diagnostics10100775