An Efficient and Streamlined System for In Vitro Regeneration and Genetic Transformation of Paper Mulberry (Broussonetia papyrifera)
Abstract
1. Introduction
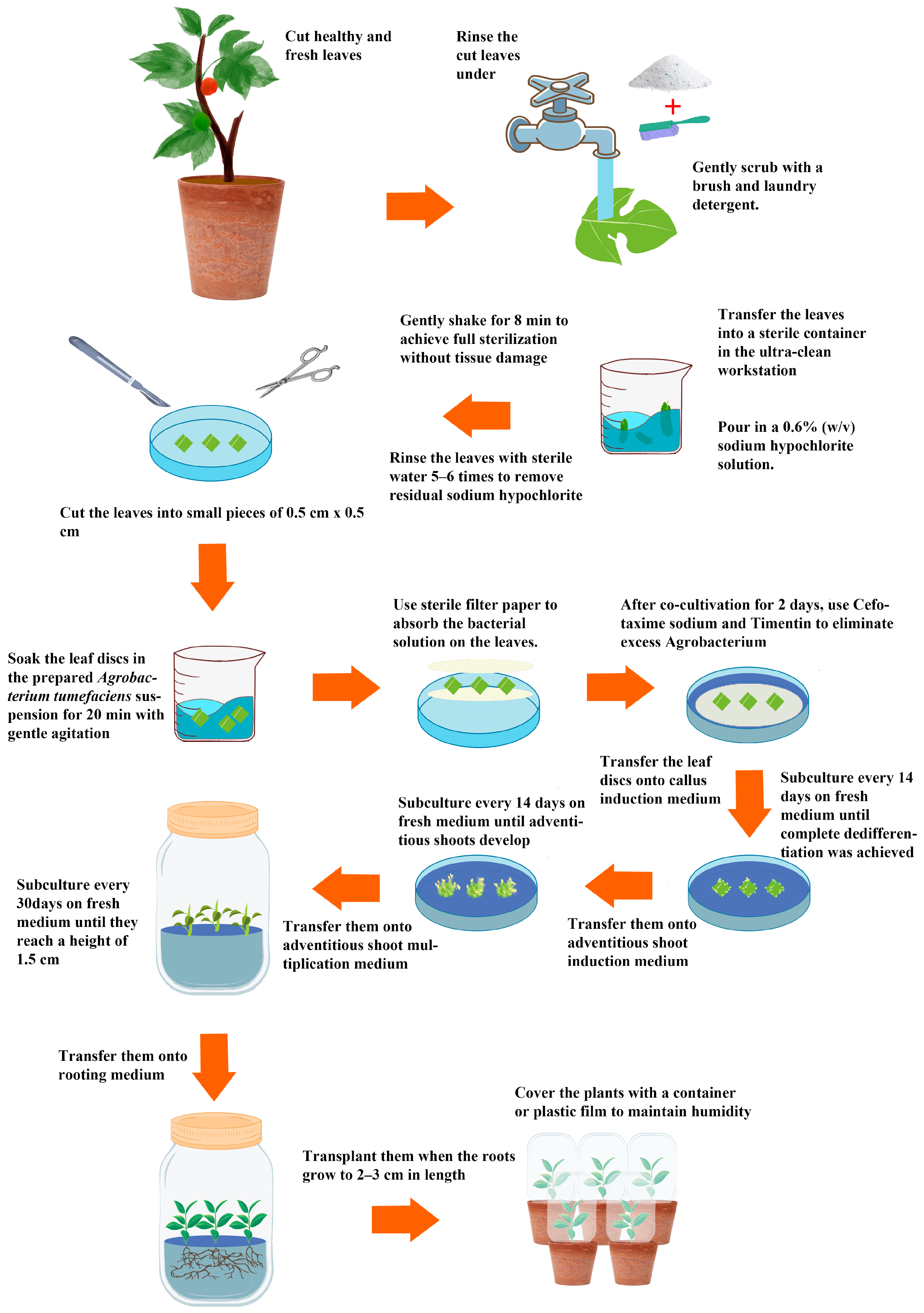
2. Materials and Methods
2.1. Plant Materials
2.2. Culture Media Optimization for Each Stage of B. papyrifera Tissue Culture
2.2.1. Evaluation of Sterilization Methods for Hybrid B. papyrifera Leaf Explants
2.2.2. Screening of Callus Induction Medium
2.2.3. Screening of Shoot Induction Medium
2.2.4. Screening of Shoot Multiplication Medium
2.2.5. Screening of Rooting Medium
2.3. Optimization of Genetic Transformation System for B. papyrifera
2.3.1. BpCHS8 Gene Cloning and Overexpression Vector Construction
2.3.2. Preparation of A. tumefaciens Suspension
2.3.3. Leaf-Disc Method for Infection and Transformation
2.4. Identification of Positive Transgenic Lines
2.4.1. Method for Evolutionary Analysis of BpCHS8
2.4.2. DNA-Level Identification of Transformants
2.4.3. qRT-PCR Analysis
2.5. Statistical Analyses
3. Results
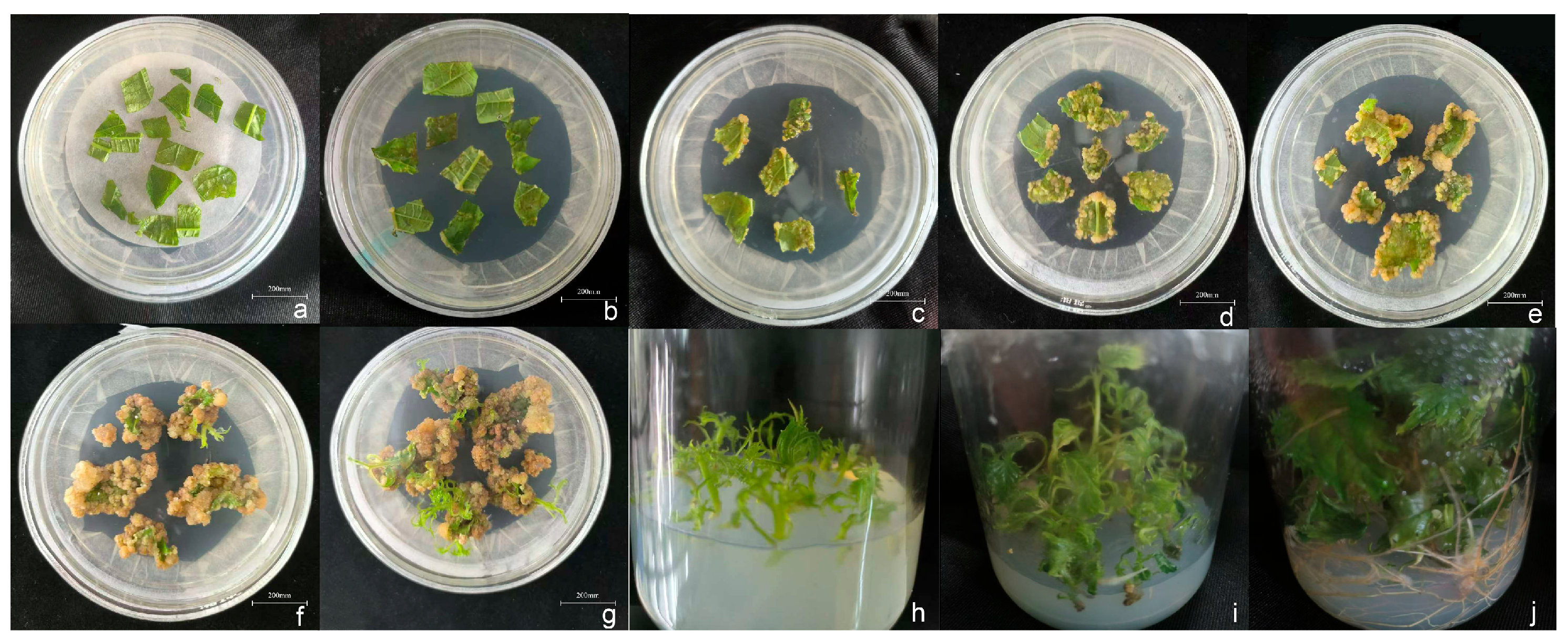
3.1. Optimization of In Vitro Regeneration of Hybrid B. papyrifera
3.1.1. Optimization of Leaf Explant Sterilization
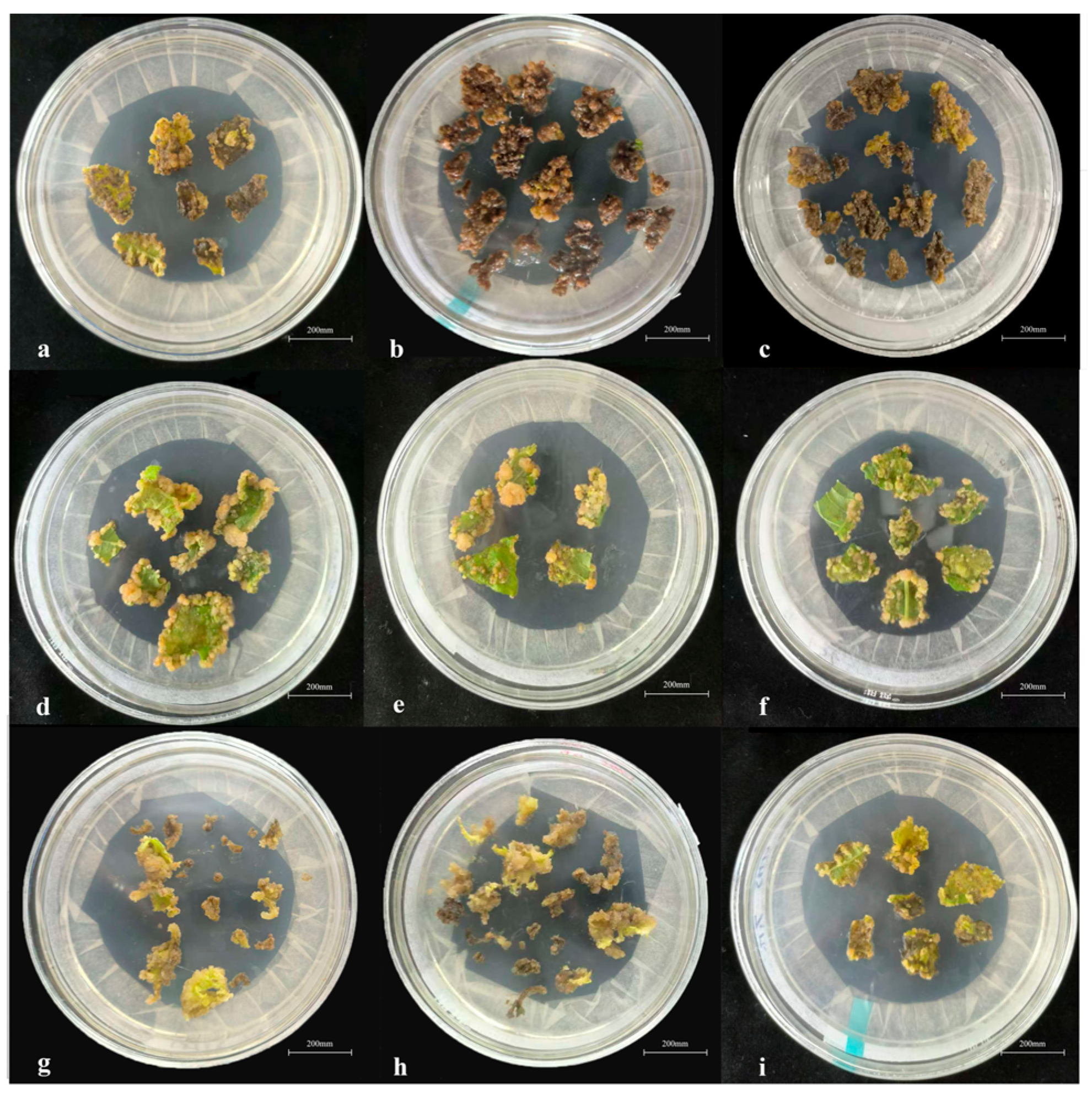
3.1.2. Callus Induction from Leaf Explants
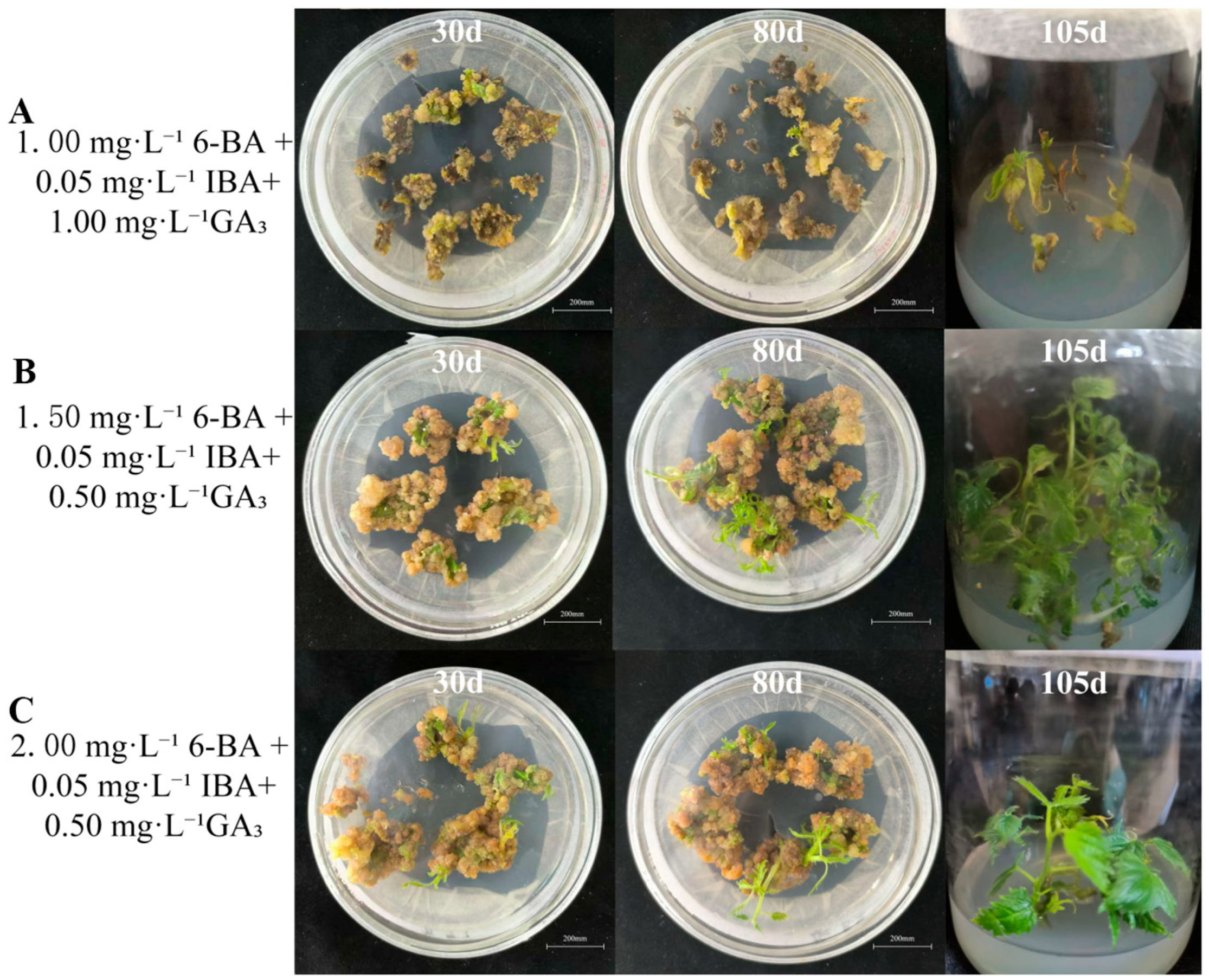
3.1.3. Adventitious Shoot Induction
3.1.4. Shoot Multiplication
3.1.5. Rooting of Regenerated Shoots
3.2. Optimization of Genetic Transformation for Hybrid B. papyrifera
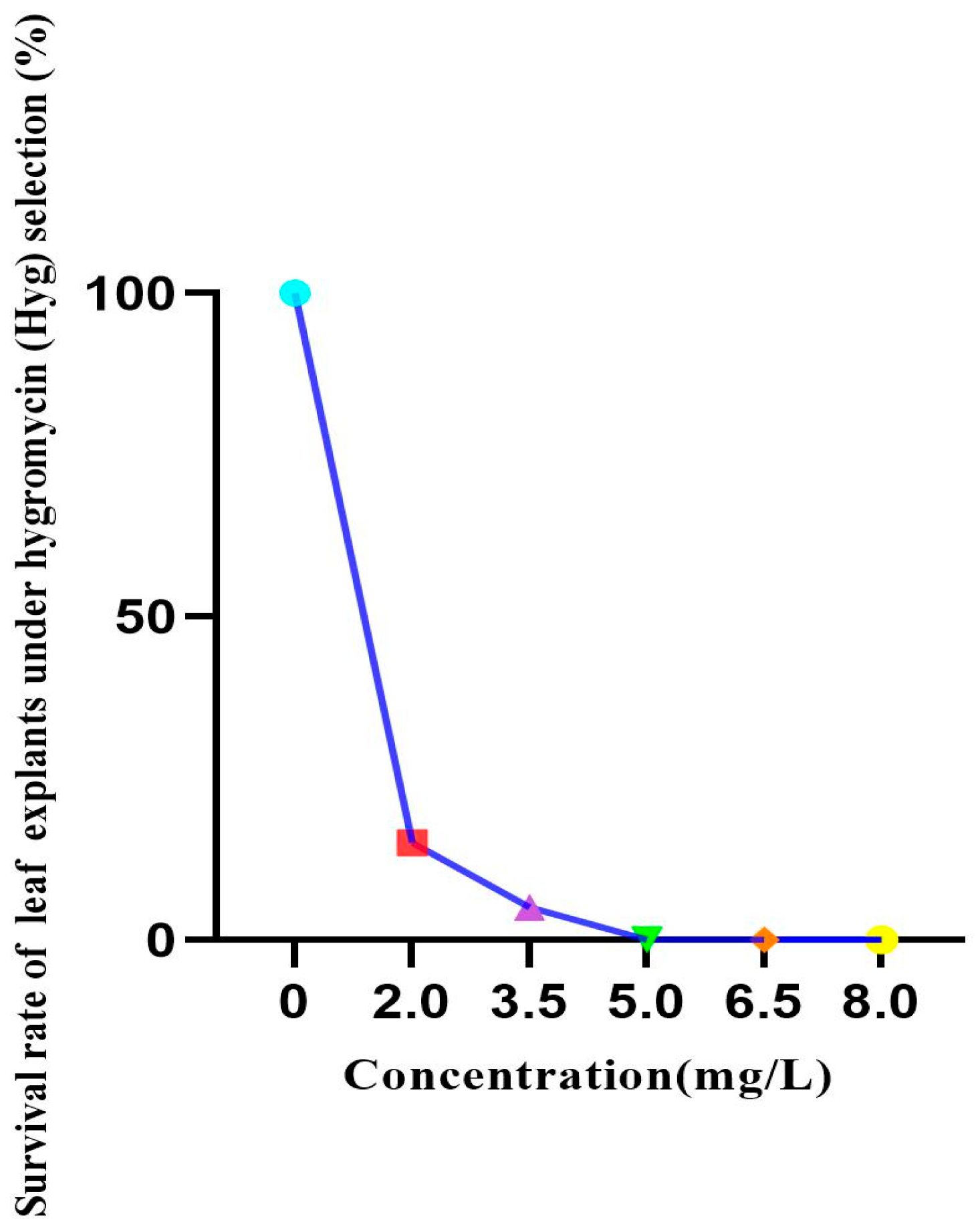
3.2.1. Determination of Hygromycin Selection Concentration
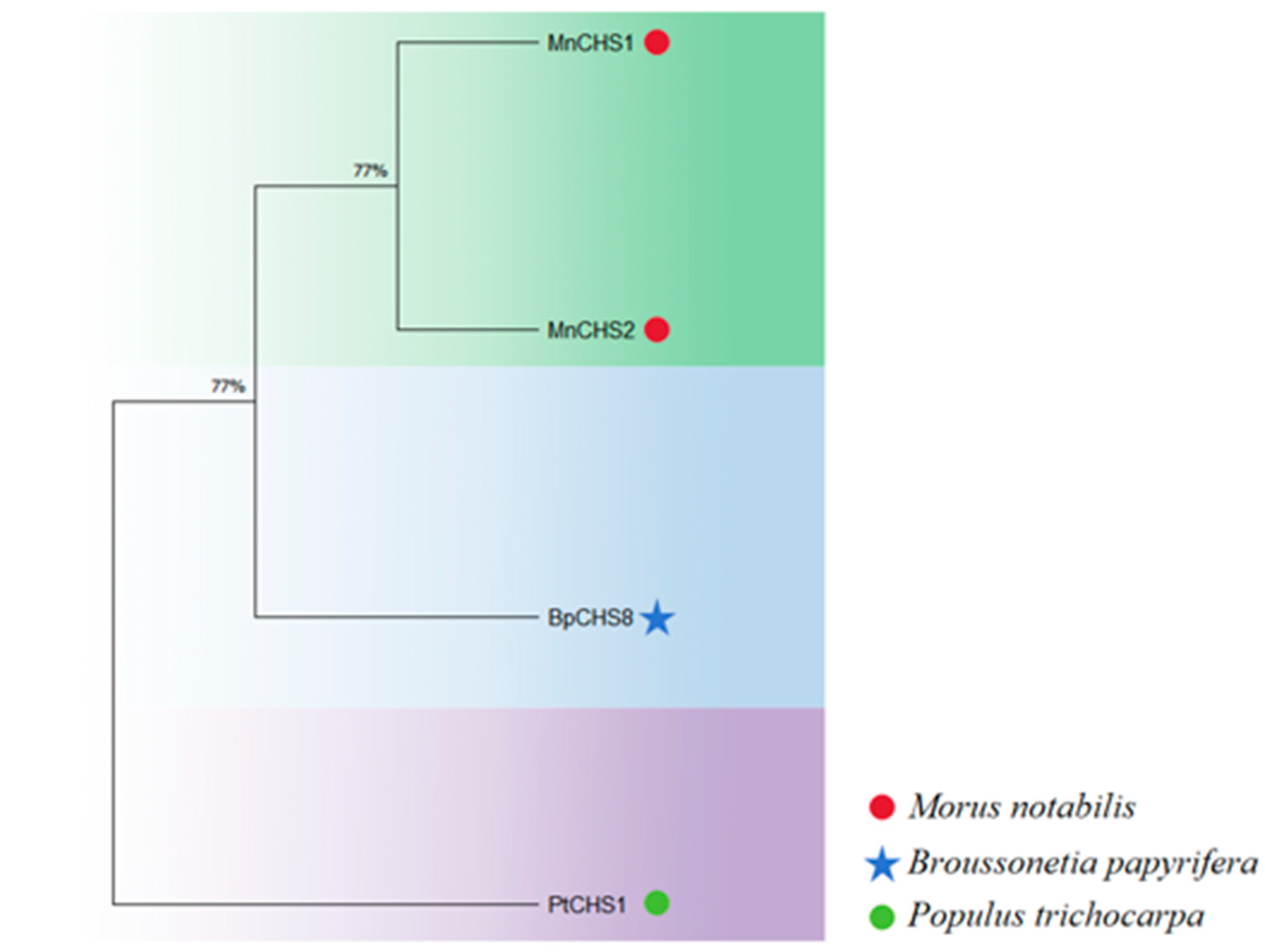
3.2.2. Evolutionary Analysis Results of BpCHS8
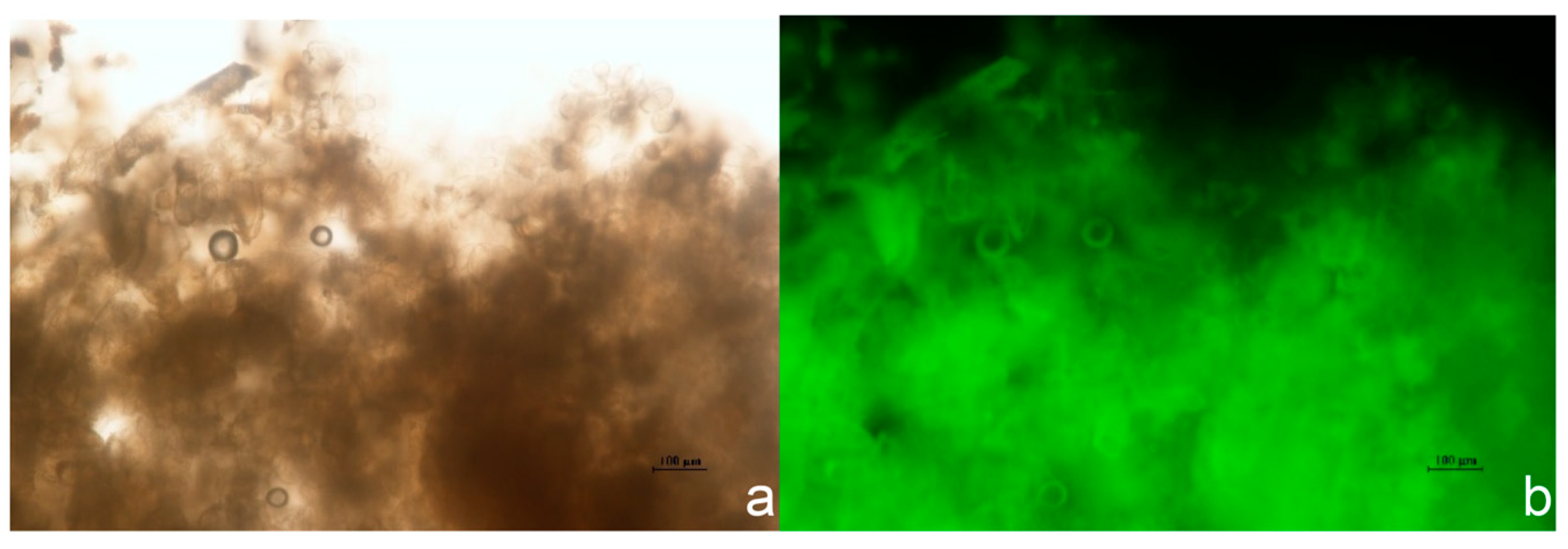
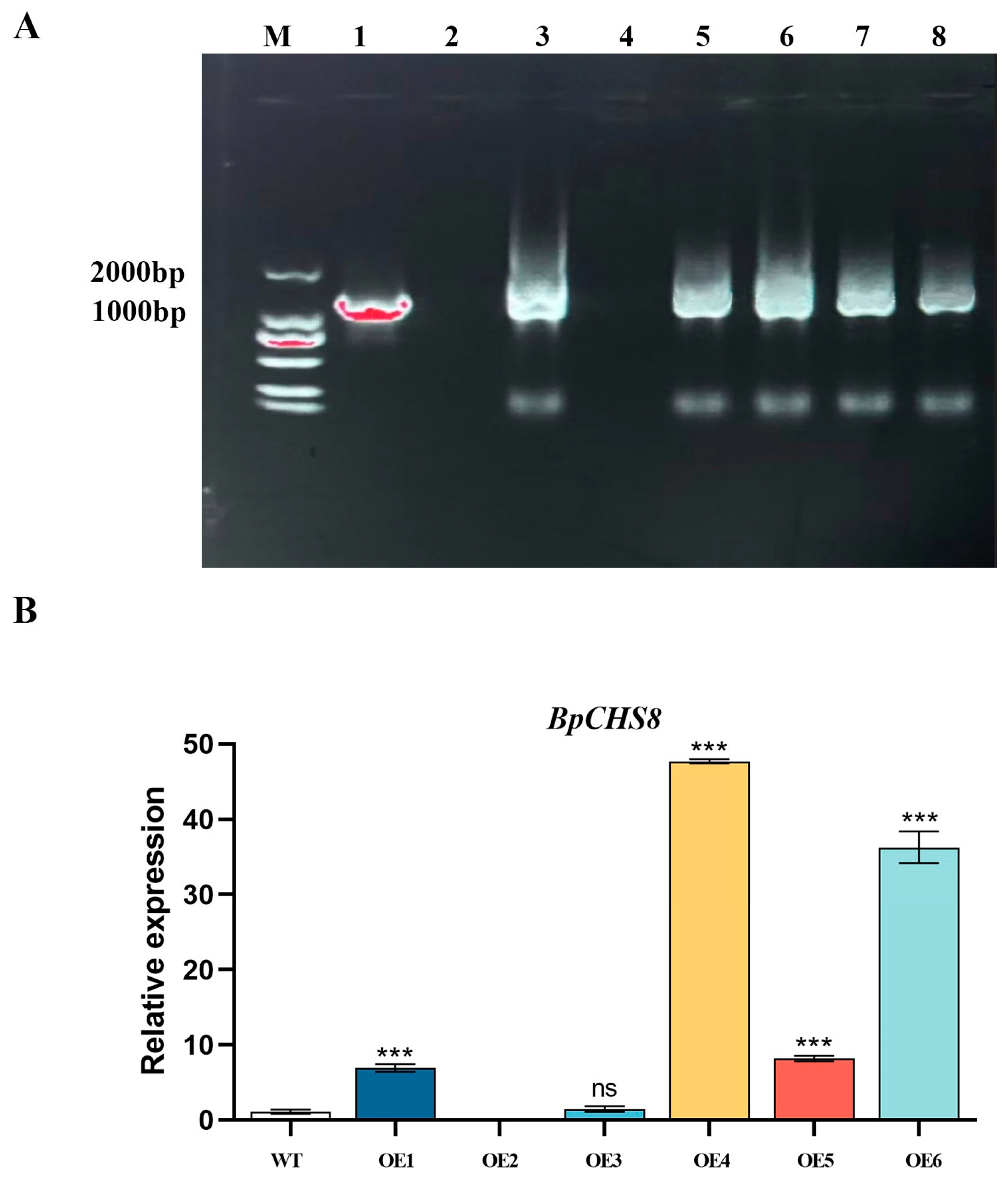
3.3. Genetic Transformation and Molecular Identification of BpCHS8::ps1300 Lines
3.3.1. Fluorescence Screening
3.3.2. PCR Confirmation of Transgene Integration
3.3.3. qRT-PCR Analysis of BpCHS8 Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, J.; Hu, B.; Fang, X. Comprehensive development and utilization of Broussonetia papyrifera (L.) L’Hér. ex Vent. Guizhou For. Sci. Technol. 2006, 2006, 61–64. Available online: http://dianda.cqvip.com/Qikan/Article/Detail?id=23607590 (accessed on 24 December 2025). (In Chinese).
- Li, D.; Li, Y. The value and cultivation techniques of Broussonetia papyrifera. For. Sci. Technol. 2006, 2006, 41–42. (In Chinese) [Google Scholar] [CrossRef]
- Peng, X.; Shen, S. The paper mulberry: A novel model system for woody plant research. Chin. Bull. Bot. 2018, 53, 372–381. (In Chinese) [Google Scholar] [CrossRef]
- Saito, K.; Linquist, B.; Keobualapha, B.; Shiraiwa, T.; Horie, T. Broussonetia papyrifera (paper mulberry): Its growth, yield and potential as a fallow crop in slash-and-burn upland rice system of northern laos. Agrofor. Syst. 2009, 76, 525–532. [Google Scholar] [CrossRef]
- Zhang, M.; Fang, Y.; Ji, Y.; Jiang, Z.; Wang, L. Effects of salt stress on ion content, antioxidant enzymes and protein profile in different tissues of Broussonetia papyrifera. S. Afr. J. Bot. 2013, 85, 1–9. [Google Scholar] [CrossRef]
- Han, Z.; Guo, Z.; Zhang, Y.; Xiao, X.; Xu, Z.; Sun, Y. Pyrolysis characteristics of biomass impregnated with cadmium, copper and lead: Influence and distribution. Waste Biomass Valorization 2018, 9, 1223–1230. [Google Scholar] [CrossRef]
- Zeng, P.; Guo, C.; Xiao, X.; Peng, C.; Huang, P.; Xin, L. Effect of phytoremediation with Broussonetia papyrifera on the biological quality in soil contaminated with heavy metals. China Environ. Sci. 2018, 38, 2639–2645. (In Chinese) [Google Scholar] [CrossRef]
- Sun, J.; Peng, X.; Fan, W.; Tang, M.; Liu, J.; Shen, S. Functional analysis of BpDREB2 gene involved in salt and drought response from a woody plant Broussonetia papyrifera. Gene 2014, 535, 140–149. [Google Scholar] [CrossRef]
- Dou, Q.; Ru, H.; Wang, A.; Qin, X.; Ding, A. root characteristics of Hybrid Broussonetia papyrifera and their effects on soil physical properties. Agric. Sci.-Technol. Inf. 2022, 2022, 75–79. (In Chinese) [Google Scholar] [CrossRef]
- Yan, D.; Zhou, W.; Wang, X.; Ren, Y.; Yang, E.; Pian, R. Effects of different acid and alkali corrosion, temperature and light intensity on seed germination of Broussonetia papyrifera. J. South. Agric. 2019, 50, 1057–1063. (In Chinese) [Google Scholar] [CrossRef]
- Chen, T.; Gao, Z.; Cao, L.; Yan, H.; Xie, X.; Meng, Q.; Wang, Y.; Wang, P. Research progress on influencing factors of paper mulberry seedling raising. Guangdong Agric. Sci. 2021, 48, 72–80. (In Chinese) [Google Scholar] [CrossRef]
- Yang, X.; Wang, H. Propagation and cultivation of Broussonetia papyrifera. Trop. For. 2012, 40, 18–21. (In Chinese) [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; XU, G.; He, Z.; Lin, S. Breeding cultivation techniques of Broussonetia papyrifera and its application value. Chin. Hortic. Abstr. 2017, 33, 97–98. Available online: http://dianda.cqvip.com/Qikan/Article/Detail?id=672177893 (accessed on 24 December 2025). (In Chinese).
- Zhu, Y.; Zeng, L.; Zhang, H. Study on tissue culture breeding technology of Broussonetia papyrifera. For. Environ. Sci. 2014, 30, 18–21. Available online: http://dianda.cqvip.com/Qikan/Article/Detail?id=662100555 (accessed on 24 December 2025). (In Chinese).
- Su, J.; Xian, K.; Fu, C.; He, J.; Liu, B.; Huang, N. Selection of suitable reference genes in Paulownia Fortunei (Seem.) Hemsl. Under different tissues and abiotic stresses for qPCR normalization. Czech J. Genet. Plant Breed. 2023, 59, 205–218. [Google Scholar] [CrossRef]
- Zhou, T.; Lin, Y.; Lin, Y.; Luo, J.; Ding, J. Regeneration and Agrobacterium-mediated genetic transformation of twelve Eucalyptus species. For. Res. 2022, 2, 15. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Guan, L.; Li, Z.; Wang, H.; Luo, J. Optimization of high-efficiency tissue culture regeneration systems in gray poplar. Life 2023, 13, 1896. [Google Scholar] [CrossRef]
- Wei, H.; Liu, Z.; Ma, L.; Wan, W.; Sheng, D. Callus induction and plant regeneration from leaves of Hybrid Broussonetia papyrifera. J. Beijing For. Univ. 2010, 32, 116–120. (In Chinese) [Google Scholar] [CrossRef]
- Li, A.; Deng, H.; Yang, L.; Liu, J. Establishment of regrowing system for Japanese Broussonetia kazinoki. Hubei For. Sci. Technol. 2007, 36, 14–17. (In Chinese) [Google Scholar] [CrossRef]
- Wang, W. Estalishiment of Tissue Culture Regeneration System and Salt-Tolerance of Hybrid Broussonetia papyrifera. Master’s Thesis, Beijing Forestry University, Beijing, China, 2010; pp. 2–3. (In Chinese). [Google Scholar]
- Yan, H.; Hao, Z.; Chen, T.; Cao, L.; Wang, P. Callus induction of Hybrid Broussonetia papyrifera leaves. Henan Sci. 2020, 38, 1776–1780. (In Chinese) [Google Scholar] [CrossRef]
- Liu, X. Minamata disease in Japan: More than just corporate responsibility. China Ecol. Civiliz. 2021, 6, 86. (In Chinese) [Google Scholar]
- Zhong, J. Preliminary Study on Improving Hybrid Broussonetia papyrifera via EMS Mutagenesis and Agrobacterium-Mediated Genetic Transformation. Master’s Thesis, South China Agricultural University, Guangzhou, China, 2011; pp. 3–8. (In Chinese). [Google Scholar] [CrossRef]
- Peng, X.; Liu, H.; Chen, P.; Tang, F.; Hu, Y.; Wang, F.; Pi, Z.; Zhao, M.; Chen, N.; Chen, H.; et al. A chromosome-scale genome assembly of paper mulberry (Broussonetia papyrifera) provides new insights into its forage and papermaking usage. Mol. Plant 2019, 12, 661–677. [Google Scholar] [CrossRef]
- Zhang, W. Cadmium Resistance and Regulation Pathway of BpTT2-Overexpressed Broussonetia papyrifera. Doctoral Dissertation, Central South University of Forestry and Technology, Changsha, China, 2022; pp. 53–60. (In Chinese). [Google Scholar] [CrossRef]
- Li, M.; Li, Y.; Li, H.; Wu, G. Overexpression of AtNHX5 improves tolerance to both salt and drought stress in Broussonetia papyrifera (l.) Vent. Tree Physiol. 2011, 31, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Hajdukiewicz, P.; Svab, Z.; Maliga, P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 1994, 25, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Glazebrook, J. Transformation of Agrobacterium using the freeze-thaw method. Cold Spring Harb. Protoc. 2006, pdb.prot4666. [Google Scholar] [CrossRef]
- Han, Y.; Ding, T.; Su, B.; Jiang, H. Genome-wide identification, characterization and expression analysis of the chalcone synthase family in maize. Int. J. Mol. Sci. 2016, 17, 161. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lan, Z.; Wang, H.; Xu, C.; Zhou, Z.; Cao, J.; Liu, Y.; Sun, Z.; Mu, D.; Han, J.; et al. Genome-wide identification of chalcone synthase (CHS) family members and their expression patterns at the sprouting stage of common bean (Phaseolus vulgaris) under abiotic stress. Sci. Hortic. 2024, 334, 113309. [Google Scholar] [CrossRef]
- Yu, M.; Shi, G.; Lv, Q.; Yang, L.; Tang, R.; Zhang, H. Invitroculture and shoot regeneration from leaves and petioles of Broussonetia papyrifera. For. Res. 2006, 19, 253–256. (In Chinese) [Google Scholar] [CrossRef]
- Li, Y. Somatic Embryo Induction and Artificial Seed Preparation of Hybrid Broussonetia papyrifera. Master’s Thesis, Dalian Polytechnic University, Dalian, China, 2015; pp. 33–36. (In Chinese). [Google Scholar]
- Zhang, C.; Zhang, Z.; Chen, Y.; Li, Y. Study on somatic embryo induction of Hybrid Broussonetia papyrifera. J. Henan Agric. Sci. 2016, 45, 127–131. (In Chinese) [Google Scholar] [CrossRef]
- Tian, R.; Huang, R.; Lu, S.; Xu, A.; Dai, Y.; Gong, Y.; Qin, Y.; Qin, Z. Establishment of tissue culture system of Hybrid Broussonetia papyrifera (L.) Vent. Hubei Agric. Sci. 2019, 58, 120–123. (In Chinese) [Google Scholar] [CrossRef]
- Lin, J.; Zhang, B.; Zou, J.; Luo, Z.; Yang, H.; Zhou, P.; Chen, X.; Zhou, W. Induction of tetraploids in paper mulberry (Broussonetia papyrifera (l.) L’hér. Ex vent.) By colchicine. BMC Plant Biol. 2023, 23, 574. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. Studies on the Tolerance of Broussonetia papyrifera to Cadmium and Manganese Under the Condition of the Tissue Culture. Master’s Thesis, Central South University of Forestry and Technology, Changsha, China, 2021; pp. 15–25. (In Chinese). [Google Scholar] [CrossRef]
- Liu, S.; Li, X.; Xu, L.; Zhang, G. Hormone functions in adventitious root formation during cutting propagation of woody plants. J. Plant Res. 2025, 138, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, H.; Jiang, H.; Wu, G. Establishment of a highly efficient Agrobacterium tumefaciens—Mediated leaf disc transformation method for Broussonetia papyrifera. Plant Cell Tissue Organ Cult. 2008, 93, 249–255. [Google Scholar] [CrossRef]
- Yi, J.; Derynck, M.R.; Li, X.; Telmer, P.; Marsolais, F.; Dhaubhadel, S. A single-repeat MYB transcription factor, GmMYB176, regulates CHS8 gene expression and affects isoflavonoid biosynthesis in soybean. Plant J. 2010, 62, 1019–1034. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Hu, J.; Xu, G.; Song, H.; Zhang, L.; Cao, Y. An efficient transformation system for fast production of VcCHS transgenic blueberry callus and its expressional analysis. Plants 2023, 12, 2905. [Google Scholar] [CrossRef] [PubMed]
| Disinfection Method | Disinfection Time/min | Number of Explants/Plant | Average Contamination Rate/% | Average Browning Rate/% | Average Explant Survival Rate/% |
|---|---|---|---|---|---|
| 0.6% NaClO | 8 | 30 | |||
| 27 | |||||
| 29 | 35.84 ± 11.39 a | 31.70 ± 8.72 c | 33.6 ± 9.59 a | ||
| 15 | 30 | ||||
| 29 | |||||
| 28 | 28.80 ± 13.34 a | 50.53 ± 4.39 ab | 23.04 ± 7.31 ab | ||
| 0.3% HgCl2 | 8 | 30 | |||
| 30 | |||||
| 30 | 37.78 ± 13.47 a | 42.22 ± 6.94 b | 20.00 ± 10.00 ab | ||
| 15 | 30 | ||||
| 29 | |||||
| 27 | 40.83 ± 3.91 a | 41.77 ± 5.83 b | 17.39 ± 6.57 ab | ||
| 10% H2O2 | 8 | 30 | |||
| 29 | |||||
| 30 | 39.43 ± 9.15 a | 39.31 ± 1.20 b | 21.26 ± 8.18 ab | ||
| 15 | 28 | ||||
| 30 | |||||
| 30 | 26.27 ± 6.08 a | 58.73 ± 15.27 a | 15.00 ± 9.28 b |
| Treatment | Concentration of 6-BA/(mg·L−1) | Concentration of IBA/(mg·L−1) | Number of Leaves/Piece | Callus Rate/% | Growth Potential |
|---|---|---|---|---|---|
| CIM1 | 1.00 | 0.05 | 40 | 38/40 = 95 | ** |
| CIM2 | 1.00 | 0.10 | 40 | 38/40 = 95 | * |
| CIM3 | 1.00 | 0.15 | 40 | 30/40 = 75 | * |
| CIM4 | 1.50 | 0.05 | 40 | 38/40 = 95 | *** |
| CIM5 | 1.50 | 0.10 | 40 | 36/40 = 90 | *** |
| CIM6 | 1.50 | 0.15 | 40 | 39/40 = 97.5 | *** |
| CIM7 | 2.00 | 0.05 | 40 | 36/40 = 90 | *** |
| CIM8 | 2.00 | 0.10 | 40 | 32/40 = 80 | ** |
| CIM9 | 2.00 | 0.15 | 40 | 30/40 = 75 | ** |
| Treatment | Concentration of 6-BA/(mg·L−1) | Concentration of IBA/(mg·L−1) | Concentration of GA3/(mg·L−1) | Number of Inoculated Explants/Plant | Callus Induction Rate of Adventitious Bud/% | Shoot Conversion Rate % |
|---|---|---|---|---|---|---|
| SIM1 | 1.00 | 0.05 | 0.50 | 30 | 50.00 ± 3.30 f | 0.33 ± 0.58 e |
| SIM2 | 1.00 | 0.05 | 1.00 | 30 | 65.57 ± 1.96 de | 12.11 ± 3.20 d |
| SIM3 | 1.00 | 0.05 | 1.50 | 30 | 60.00 ± 3.33 e | 0.00 ± 0.00 e |
| SIM4 | 1.50 | 0.05 | 0.50 | 30 | 90.00 ± 3.33 a | 83.89 ± 2.71 a |
| SIM5 | 1.50 | 0.05 | 1.00 | 30 | 86.67 ± 3.34 a | 74.29 ± 3.07 b |
| SIM6 | 1.50 | 0.05 | 1.50 | 30 | 82.22 ± 5.09 ab | 60.57 ± 7.96 c |
| SIM7 | 2.00 | 0.05 | 0.50 | 30 | 88.89 ± 1.92 a | 68.76 ± 1.89 bc |
| SIM8 | 2.00 | 0.05 | 1.00 | 30 | 70.00 ± 3.33 cd | 66.62 ± 1.59 bc |
| SIM9 | 2.00 | 0.05 | 1.50 | 30 | 75.55 ± 3.85 bc | 63.26 ± 0.66 c |
| Treatment | Concentration of 6-BA/(mg·L−1) | Concentration of IBA/(mg·L−1) | Adventitious Shoots/Plant | Shoot Multiplication Coefficient |
|---|---|---|---|---|
| SMM1 | 0.15 | 0.05 | 20.00 | 5.70 ± 0.10 a |
| SMM2 | 0.15 | 0.10 | 20.00 | 5.27 ± 0.40 a |
| SMM3 | 0.15 | 0.15 | 20.00 | 5.47 ± 0.76 a |
| SMM4 | 0.50 | 0.05 | 20.00 | 3.03 ± 0.61 c |
| SMM5 | 0.50 | 0.10 | 20.00 | 2.33 ± 0.61 cd |
| SMM6 | 0.50 | 0.15 | 20.00 | 4.13 ± 0.71 b |
| SMM7 | 1.00 | 0.05 | 20.00 | 2.07 ± 0.76 cd |
| SMM8 | 1.00 | 0.10 | 20.00 | 1.73 ± 0.71 ef |
| SMM9 | 1.00 | 0.15 | 20.00 | 0.93 ± 0.61 f |
| Treatment | Concentration of IBA/(mg·L−1) | Number of Leaves/Piece | Rooting Percentage/% |
|---|---|---|---|
| RM1 | 0.05 | 40 | 89.17 ± 3.82 a |
| RM2 | 0.10 | 40 | 85.00 ± 2.50 ab |
| RM3 | 0.15 | 40 | 72.50 ± 4.33 bc |
| RM4 | 0.20 | 40 | 64.17 ± 8.04 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Ye, F.; Ke, T.; Deng, S.; Pan, L.; Tang, M.; Hu, W. An Efficient and Streamlined System for In Vitro Regeneration and Genetic Transformation of Paper Mulberry (Broussonetia papyrifera). Life 2026, 16, 78. https://doi.org/10.3390/life16010078
Ye F, Ke T, Deng S, Pan L, Tang M, Hu W. An Efficient and Streamlined System for In Vitro Regeneration and Genetic Transformation of Paper Mulberry (Broussonetia papyrifera). Life. 2026; 16(1):78. https://doi.org/10.3390/life16010078
Chicago/Turabian StyleYe, Fangyu, Tong Ke, Shuiqing Deng, Lan Pan, Ming Tang, and Wentao Hu. 2026. "An Efficient and Streamlined System for In Vitro Regeneration and Genetic Transformation of Paper Mulberry (Broussonetia papyrifera)" Life 16, no. 1: 78. https://doi.org/10.3390/life16010078
APA StyleYe, F., Ke, T., Deng, S., Pan, L., Tang, M., & Hu, W. (2026). An Efficient and Streamlined System for In Vitro Regeneration and Genetic Transformation of Paper Mulberry (Broussonetia papyrifera). Life, 16(1), 78. https://doi.org/10.3390/life16010078





