Unusual Morphological Changes of a Novel Wrinkled Bacterium Isolated from the Rice Rhizosphere Under Nutrient Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Bacterial Strain
2.2. Bacterial Culture Preparation
2.3. Bacterial Cell Morphology
2.4. Monitoring of Bacterial Cell Shape Under Nutrient Stress
2.5. Measurement of the Surface Area-to-Volume (S/V) Ratio of Bacterial Cells
2.6. Determination of Total Organic Carbon
2.7. Phylogenetic Analysis of 16S rRNA Gene Sequences
2.8. Physiological and Biochemical Tests
2.9. Chemotaxonomic Characterization
2.10. Whole-Genome Sequences of Strain YC6860T
2.11. Accession Numbers of 16S rRNA Gene and Genome Sequences of YC6860T
3. Results
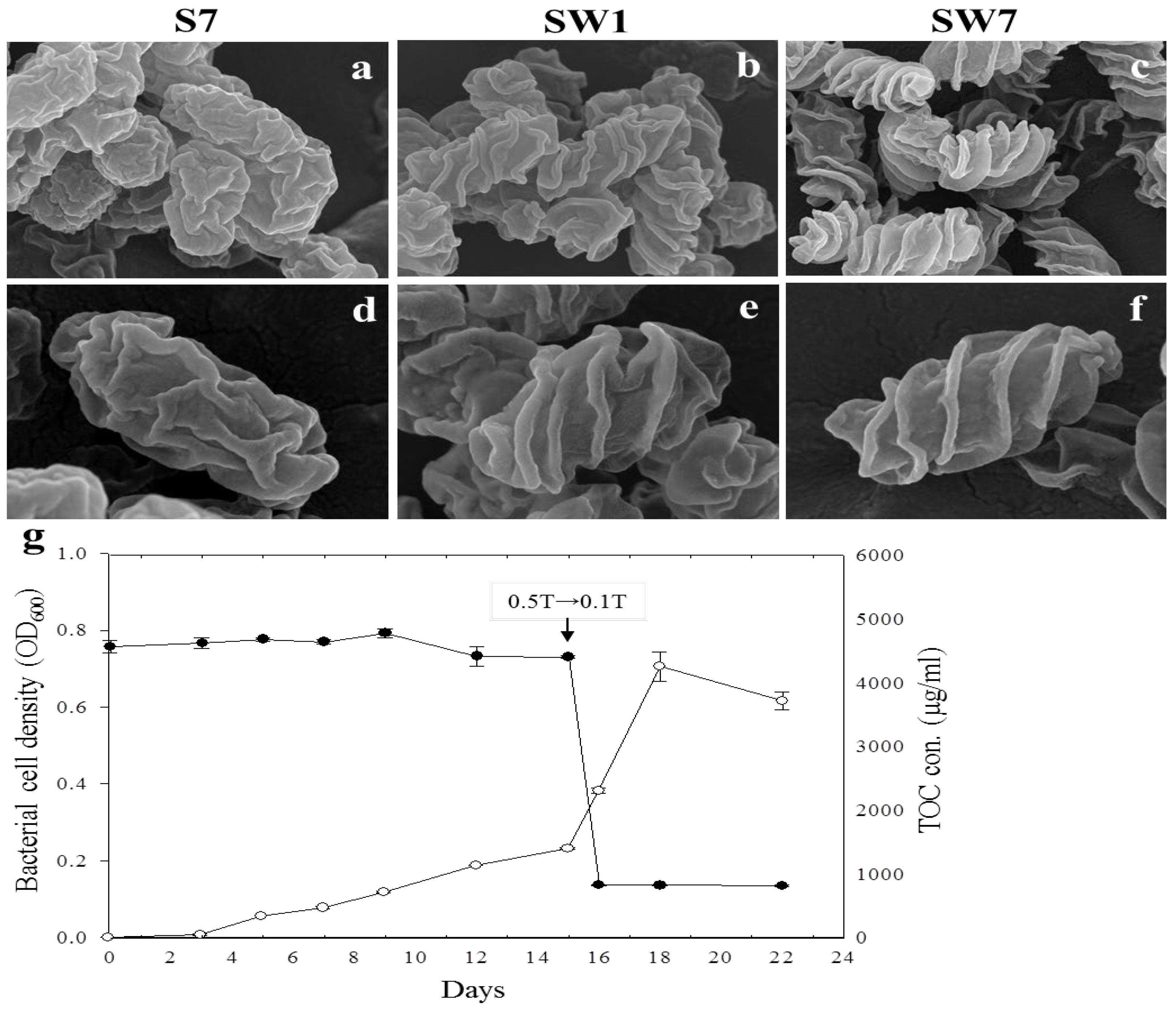
3.1. Growth and Morphology of the Strain YC6860T
3.2. Transition from Smooth to Wrinkled Shape
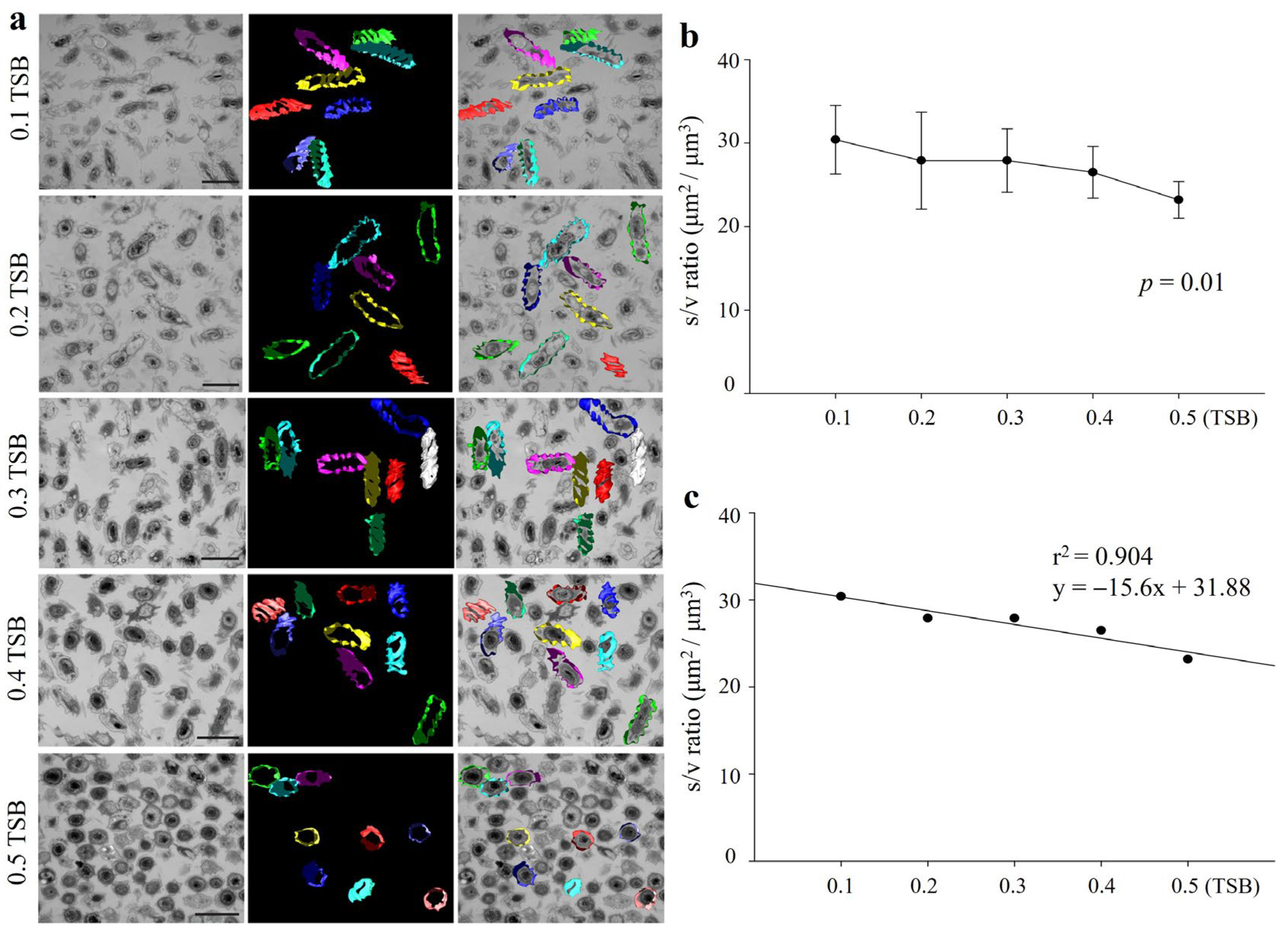
3.3. Morphological Transition Correlated with Surface Area-to-Volume (S/V) Ratio
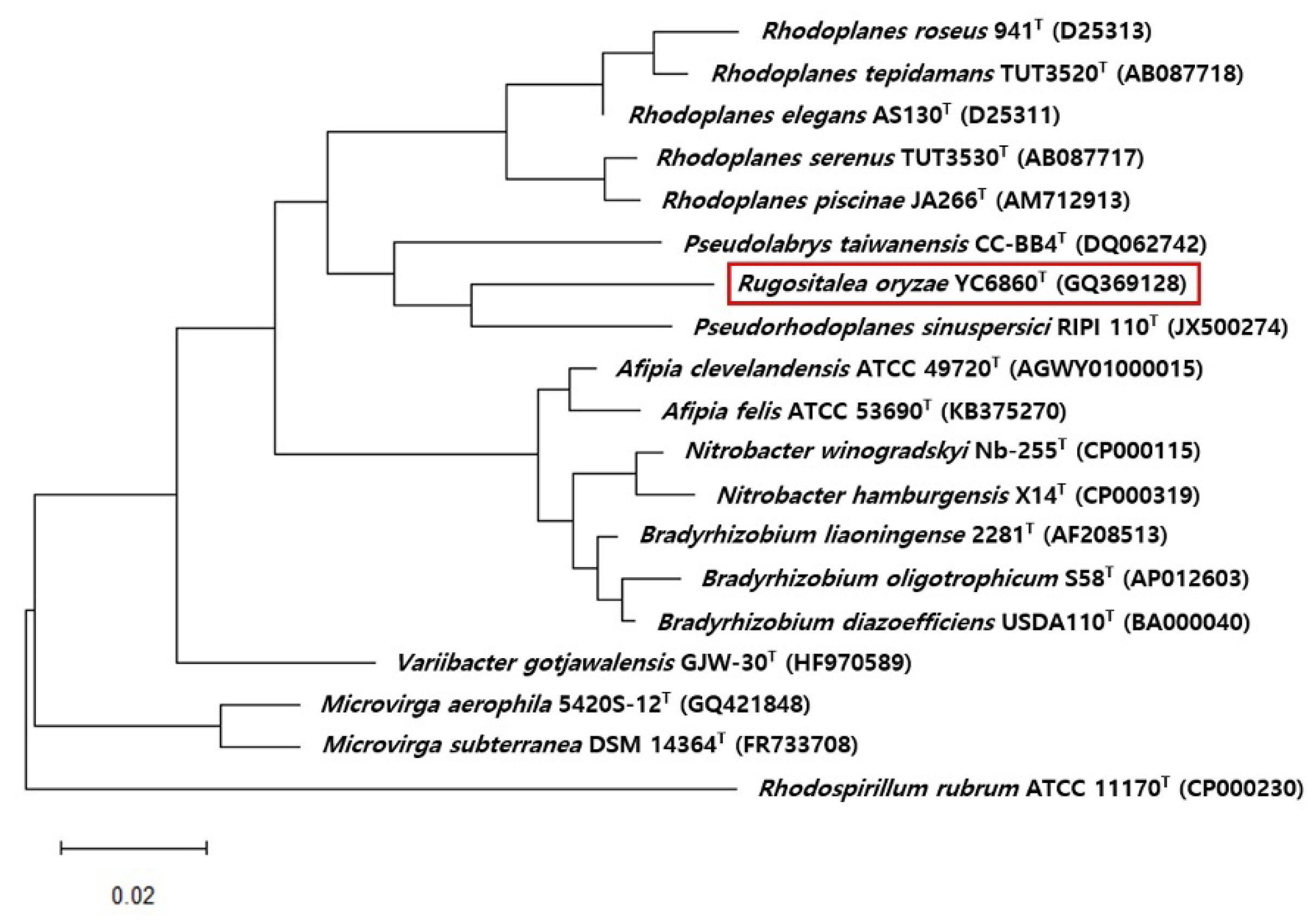
3.4. Phylogenetic Analysis
3.5. Phenotypic Characterization
3.6. Fatty Acid Analysis
3.7. Genome Description
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cabeen, M.T.; Jacobs-Wagner, C. Bacterial Cell Shape. Nat. Rev. Microbiol. 2005, 3, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Kysela, D.T.; Randich, A.M.; Caccamo, P.D.; Brun, Y.V. Diversity takes shape: Understanding the mechanistic and adaptive basis of bacterial morphology. PLoS Biol. 2016, 14, e1002565. [Google Scholar] [CrossRef] [PubMed]
- Typas, A.; Banzhaf, M.; Gross, C.A.; Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2012, 10, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.E.; Allegrini, M.; Iocoli, G.A.; Basualdo, J.; Villamil, M.B.; Zabaloy, M.C. Rhizospheric microbiome responses to cover crop suppression methods. Agronomy 2022, 12, 2246. [Google Scholar] [CrossRef]
- van Teeseling, M.C.F.; de Pedro, M.A.; Cava, F. Determinants of Bacterial Morphology: From Fundamentals to Possibilities for Antimicrobial Targeting. Front. Microbiol. 2017, 8, 1264. [Google Scholar] [CrossRef]
- Cornforth, D.M.; Foster, K.R. Competition Sensing: The Social Side of Bacterial Stress Responses. Nat. Rev. Microbiol. 2013, 11, 285–293. [Google Scholar] [CrossRef]
- Young, K.D. The Selective Value of Bacterial Shape. Microbiol. Mol. Biol. Rev. 2006, 70, 660–703. [Google Scholar] [CrossRef]
- Young, K.D. Bacterial Morphology: Why Have Different Shapes? Curr. Opin. Microbiol. 2007, 10, 596–600. [Google Scholar] [CrossRef]
- Männik, J.; Driessen, R.; Galajda, P.; Keymer, J.E.; Dekker, C. Bacterial growth and motility in sub-micron constrictions. Proc. Natl. Acad. Sci. USA 2009, 106, 14861–14866. [Google Scholar] [CrossRef]
- Sun, J.; Rutherford, S.T.; Silhavy, T.J.; Huang, K.C. Physical properties of the bacterial outer membrane. Nat. Rev. Microbiol. 2022, 20, 236–248. [Google Scholar] [CrossRef]
- Ojkic, N.; Banerjee, S. Bacterial cell shape control by nutrient-dependent synthesis of cell division inhibitors. Biophys. J. 2021, 120, 2079–2084. [Google Scholar] [CrossRef]
- Kumar, P.; Libchaber, A. Pressure and temperature dependence of growth and morphology of Escherichia coli: Experiments and stochastic model. Biophys. J. 2013, 105, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Ghoul, M.; Mitri, S. The Ecology and Evolution of Microbial Competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.A.; Schlimpert, S.; Hughes, V.; Brun, Y.V.; Thanbichler, M.; Gitai, Z. Physiological role of stalk lengthening in Caulobacter crescentus. Commun. Integr. Biol. 2013, 6, e24561. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.C.; Blair, K.M.; Salama, N.R. Staying in Shape: The Impact of Cell Shape on Bacterial Survival in Diverse Environments. Microbiol. Mol. Biol. Rev. 2016, 80, 187–203. [Google Scholar] [CrossRef]
- Chien, A.C.; Hill, N.S.; Levin, P.A. Cell Size Control in Bacteria. Curr. Biol. 2012, 22, R340–R349. [Google Scholar] [CrossRef]
- Ratnayake, R.R.; Perera, B.M.; Rajapaksha, R.P.; Ekanayake, E.M.; Kumara, R.K.; Gunaratne, H.M. Soil carbon sequestration and nutrient status of tropical rice based cropping systems: Rice-Rice, Rice-Soya, Rice-Onion and Rice-Tobacco in Sri Lanka. Catena 2017, 150, 17–23. [Google Scholar] [CrossRef]
- Cheng, Y.Q.; Yang, L.Z.; Cao, Z.H.; Ci, E.; Yin, S. Chronosequential changes of selected pedogenic properties in paddy soils as compared with non-paddy soils. Geoderma 2009, 151, 31–41. [Google Scholar] [CrossRef]
- Zhu, Z.; Ge, T.; Liu, S.; Hu, Y.; Ye, R.; Xiao, M.; Tong, C.; Kuzyakov, Y.; Wu, J. Rice rhizodeposits affect organic matter priming in paddy soil: The role of N fertilization and plant growth for enzyme activities, CO2 and CH4 emissions. Soil Biol. Biochem. 2018, 116, 369–377. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef]
- Lee, H.I.; Lee, J.H.; Park, K.H.; Sangurdekar, D.; Chang, W.S. Effect of soybean coumestrol on Bradyrhizobium japonicum nodulation ability, biofilm formation, and transcriptional profile. Appl. Environ. Microbiol. 2012, 78, 2896–2903. [Google Scholar] [CrossRef]
- Dheeman, S.; Tofazzal, I.M.; Egamberdieva, D.; Siddiqui, M.N. Soil Bacteria, Biofertilization and Soil Health; Springer: Berlin/Heidelberg, Germany, 2024. [Google Scholar]
- Carvalhais, L.C.; Dennis, P.G.; Fan, B.; Fedoseyenko, D.; Kierul, K.; Becker, A.; von Wiren, N.; Borriss, R. Linking plant nutritional status to plant-microbe interactions. PLoS ONE 2013, 8, e68555. [Google Scholar] [CrossRef]
- Dijkstra, F.A.; Carrillo, Y.; Pendall, E.; Morgan, J.A. Rhizosphere priming: A nutrient perspective. Front. Microbiol. 2013, 4, 216. [Google Scholar] [CrossRef]
- Narula, N.; Kothe, E.; Behl, R.K. Role of root exudates in plant-microbe Interactions. J. Appl. Bot. Food Qual. 2009, 82, 122–130. [Google Scholar]
- Megrian, D.; Taib, N.; Jaffe, A.L.; Banfield, J.F.; Gribaldo, S. Ancient origin and constrained evolution of the division and cell wall gene cluster in bacteria. Nat. Microbiol. 2022, 7, 2114–2127. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.; Huang, K.C. How and why cells grow as rods. BMC Biol. 2014, 12, 54. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Davis, R.M.; Kishony, R.; Kahne, D.; Ruiz, N. Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2012, 109, E2561–E2568. [Google Scholar] [CrossRef]
- Harry, E.; Monahan, L.; Thomson, L. Bacterial cell division: The mechanism and its precision. Int. Rev. Cytol. 2006, 253, 27–94. [Google Scholar]
- Figge, R.M.; Divakaruni, A.V.; Gober, J.W. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 2004, 51, 1321–1332. [Google Scholar] [CrossRef]
- Weart, R.B.; Lee, A.H.; Chien, A.C.; Haeusser, D.P.; Hill, N.S.; Levin, P.A. A metabolic sensor governing cell size in bacteria. Cell 2007, 130, 335–347. [Google Scholar] [CrossRef]
- Jiang, C.; Caccamo, P.D.; Brun, Y.V. Mechanisms of bacterial morphogenesis: Evolutionary cell biology approaches provide new insights. BioEssays 2015, 37, 413–425. [Google Scholar] [CrossRef]
- Aslam, Z.; Yasir, M.; Yoon, H.S.; Jeon, C.O.; Chung, Y.R. Diversity of the bacterial community in the rice rhizosphere managed under conventional and no-tillage practices. J. Microbiol. 2013, 51, 747–756. [Google Scholar] [CrossRef]
- Giotis, E.S.; Blair, I.S.; McDowell, D.A. Morphological changes in Listeria monocytogenes subjected to sublethal alkaline stress. Int. J. Food Microbiol. 2007, 120, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.H. Electron microscopy and high-pressure freezing of Arabidopsis. Methods Cell Biol. 2010, 96, 259–283. [Google Scholar] [PubMed]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematic; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Maidak, B.L.; Olsen, G.J.; Larsen, N.; Overbeek, R.; McCaughey, M.J.; Woese, C.R. The RDP (Ribosomal Database Project). Nucleic Acids Res. 1997, 25, 109–110. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X Windows Interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kimura, M. The neutral theory of molecular evolution. Scient. Americ. 1979, 241, 98–129. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Fitch, W.M. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Biol. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Costilow, R.N. Biophysical factors in growth. In Manual of Methods for General Bacteriology; Gerhardt, P., Murray, R.G.E., Costilow, R.N., Nester, E.W., Wood, W.A., Krieg, N.R., Phillips, G.B., Eds.; American Society for Microbiology: Washington, D.C., USA, 1981; pp. 66–78. [Google Scholar]
- Cappuccino, J.; Sherman, N. Microbiology: A Laboratory Manual, 6th ed.; Benjamin Cummings Pearson Education: San Francisco, CA, USA, 2002; pp. 15–224. [Google Scholar]
- Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Current Protocols in Molecular Biology; Janssen, K., Ed.; Wiley: New York, NY, USA, 1994; Volume 1. [Google Scholar]
- Mesbah, M.; Premachandran, U.; Whitman, W.B. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 1989, 39, 159–167. [Google Scholar] [CrossRef]
- Tindall, B.J. A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst. Appl. Microbiol. 1990, 13, 128–130. [Google Scholar] [CrossRef]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; Technical Note; MIDI Inc.: Newark, DE, USA, 1990. [Google Scholar]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A Program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile Hidden Markov Models. Bioinforma. Oxf. Engl. 1998, 14, 755–763. [Google Scholar]
- Overbeek, R.; Begley, T.; Butler, R.M.; Choudhuri, J.V.; Chuang, H.-Y.; Cohoon, M.; de Crécy-Lagard, V.; Diaz, N.; Disz, T.; Edwards, R. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005, 33, 5691–5702. [Google Scholar] [CrossRef]
- Yu, C.; Zavaljevski, N.; Desai, V.; Reifman, J. Genome-wide enzyme annotation with precision control: Catalytic families (CatFam) databases. Proteins Struct. Funct. Bioinforma. 2009, 74, 449–460. [Google Scholar] [CrossRef]
- Sheffield, J.B. ImageJ, a Useful Tool for Biological Image processing and analysis. Microsc. Microanal. 2007, 13, 200–201. [Google Scholar] [CrossRef]
- Tirandaz, H.; Dastgheib, S.M.M.; Amoozegar, M.A.; Shavandi, M.; De La Haba, R.R.; Ventosa, A. Pseudorhodoplanes sinuspersici Gen. Nov., Sp. Nov., isolated from oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 4743–4748. [Google Scholar] [CrossRef]
- Hiraishi, A.; Okamura, K. Proposal of Rhodoplanes tepidamans Sp. Nov. to accommodate the thermotolerant phototrophic bacterium previously referred to as “Rhodoplanes (Rhodopseudomonas) cryptolactis”. Int. J. Syst. Evol. Microbiol. 2017, 67, 1540–1545. [Google Scholar] [CrossRef] [PubMed]
- Hiraishi, A.; Ueda, Y. Rhodoplanes Gen. Nov., A new genus of phototrophic bacteria including Rhodopseudomonas rosea as Rhodoplanes roseus Comb. Nov. and Rhodoplanes elegans Sp. Nov. Int. J. Syst. Bacteriol. 1994, 44, 665–673. [Google Scholar] [CrossRef]
- Kämpfer, P.; Young, C.C.; Arun, A.B.; Shen, F.T.; Jäckel, U.; Rosselló-Mora, R.; Lai, W.A.; Rekha, P.D. Pseudolabrys taiwanensis Gen. Nov., Sp. Nov., an Alphaproteobacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2006, 56, 2469–2472. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Wagner, J.K.; Setayeshgar, S.; Sharon, L.A.; Reilly, J.P.; Brun, Y.V. A nutrient uptake role for bacterial cell envelope extensions. Proc. Natl. Acad. Sci. USA 2006, 103, 11772–11777. [Google Scholar] [CrossRef]
- Kuykendall, L.D. Order VI. Rhizobiales Ord. Nov. Bergey’s Man. Syst. Bacteriol. 2005, 324–568. [Google Scholar]
- Diomandé, S.E.; Nguyen-The, C.; Guinebretière, M.H.; Broussolle, V.; Brillard, J. Role of fatty acids in Bacillus environmental adaptation. Front. Microbiol. 2015, 6, 813. [Google Scholar] [CrossRef]
- Bae, H.C.; Cota-Robles, E.H.; Casida, L.E. Microflora of soil as viewed by transmission electron microscopy. Appl. Microbiol. 1972, 23, 637–648. [Google Scholar] [CrossRef]
- Carballido-López, R. The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. 2006, 70, 888–909. [Google Scholar] [CrossRef]
- Hett, E.C.; Rubin, E.J. Bacterial Growth and Cell Division: A mycobacterial perspective. Microbiol. Mol. Biol. Rev. 2008, 72, 126–156. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | Rugositalea oryzae YC6860T | Pseudorhodoplanes sinuspersici RIPI 110T | Rhodoplanes tepidamans TUT3520T | Rhodoplanes elegans AS130T | Pseudolabrys taiwanensis CC-BB4T |
|---|---|---|---|---|---|
| Isolation source | Rice rhizosphere | Oil-contaminated Soil site | Hot springs | Sludge | Soil |
| Gram-staining | Negative | Negative | Negative | Negative | Negative |
| Motility | Motile | Non-motile | Motile | Motile | Non-motile |
| NaCl requirement | 0–1% | 0–3.5% | 0% | <1% | ND |
| Oxidase/Catalase | +/+ | +/+ | ND | ND | +/+ |
| Temperature (°C) | 16–30 | 15–35 | 40 | 30–35 (optimal) | 15–36 |
| pH | 6.0–10.0 | 5.5–8.0 | 6.8–7.5 | 6–8.5 | 4.2–8.5 |
| Aerobic/Anaerobic condition | +/- | +/+ | +/+ | +/+ | +/- |
| Alkaline phosphatase | - | ND | ND | + | + |
| Lipase (C14) | - | ND | ND | - | + |
| DNA G + C content (%) | 63.5 | 59.4 | 69.9 | 69.7 | 67.0 |
| Major quinone | Q-10 | Q-10 | Q-10, R-10 | Q-10, RQ-10 | ND |
| Fatty Acids (%) | 0.1 TSB | 0.5 TSB |
|---|---|---|
| Saturated | ||
| C10:0 | - | 4.02 |
| C12:0 | 1.37 | 4.62 |
| C14:0 | 0.78 | 0.98 |
| C16:0 | 18.93 | 19.78 |
| C17:0 | 0.93 | 0.77 |
| C18:0 | 1.38 | 1.41 |
| Unsaturated | ||
| C17:1w8c | 0.73 | - |
| C18:1w5c | - | 0.1 |
| C18:1w7c Hydroxy | 55.18 | 58.33 |
| C16:1 2OH | 0.17 | - |
| C18:0 3OH | 0.56 | 0.41 |
| Cyclopropane | ||
| C17:0 CYCLO | 1.33 | 0.51 |
| C19:0 CYCLO w8c | 12.2 | 4.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chung, Y.R.; Lee, J.E.; Aslam, Z.; Chung, E.J.; Lee, K.H.; Kang, B.H.; Khan, A.; Niraula, S.; Chang, W.-S. Unusual Morphological Changes of a Novel Wrinkled Bacterium Isolated from the Rice Rhizosphere Under Nutrient Stress. Life 2025, 15, 1337. https://doi.org/10.3390/life15091337
Chung YR, Lee JE, Aslam Z, Chung EJ, Lee KH, Kang BH, Khan A, Niraula S, Chang W-S. Unusual Morphological Changes of a Novel Wrinkled Bacterium Isolated from the Rice Rhizosphere Under Nutrient Stress. Life. 2025; 15(9):1337. https://doi.org/10.3390/life15091337
Chicago/Turabian StyleChung, Young Ryun, Jung Eun Lee, Zubair Aslam, Eu Jin Chung, Kwang Hee Lee, Byung Ho Kang, Ajmal Khan, Sarbjeet Niraula, and Woo-Suk Chang. 2025. "Unusual Morphological Changes of a Novel Wrinkled Bacterium Isolated from the Rice Rhizosphere Under Nutrient Stress" Life 15, no. 9: 1337. https://doi.org/10.3390/life15091337
APA StyleChung, Y. R., Lee, J. E., Aslam, Z., Chung, E. J., Lee, K. H., Kang, B. H., Khan, A., Niraula, S., & Chang, W.-S. (2025). Unusual Morphological Changes of a Novel Wrinkled Bacterium Isolated from the Rice Rhizosphere Under Nutrient Stress. Life, 15(9), 1337. https://doi.org/10.3390/life15091337






