Abstract
In this study we searched for correlations between polymorphic variants that determine sex hormone-binding globulin concentration (SHBGcon) and uterine fibroids (UFs). The work was performed on a sample of 1542 women (569 with UFs and 973 without UFs [control]), from whom we obtained experimental data on the distribution of nine single-nucleotide polymorphisms (SNPs) affecting the SHBGcon (data confirmed in genome-wide association studies [GWASs]). When searching for associations with UFs, both the independent effects of SNPs and the effects of their SNP–SNP interactions (SNP-SNPints) were taken into account during the “deep study” of the functionality of seven important UF loci and 115 strongly linked [r2 ≥ 0.80] variants (an in silico methodology was used). As the results show, two SHBGcon-related SNPs correlated with UF risk: rs3779195 [T/A] BAIAP2L1 (ORAA = 0.38; 95%CIAA = 0.20–0.91; pperm(AA) = 0.023) and rs440837 [A/G] ZBTB10 (ORGG = 1.93; 95%CIGG = 1.17–3.14; pperm(GG) = 0.010). At the same time, seven SHBGcon-related SNPs interacting with each other (four models of such SNP-SNPints [pperm ≤ 0.01)] were found to influence UF risk. These SHBGcon-related SNPs, determining susceptibility to UF, showed strong functional relevance and were involved in pathways of gene transcription regulation, interactions with hormone ligand-binding receptors, the content control of SHBG, testosterone, liver enzymes, lipids, etc. This study’s results demonstrate the effect of significant SHBGcon-related genetic determinants of UF risk.
1. Introduction
Uterine fibroids (UFs) are the most common pelvic tumors, affecting about 70% of women worldwide [1]. The disease is clinically manifested in at least 25–50% of patients [2,3]. The tumor reveals itself by severe menstrual bleeding, often leading to severe iron deficiency anemia, pelvic pain, and reproductive disorders, including infertility and pregnancy complications [1,3]. UFs often require surgical treatment [4,5]. UFs are the main cause of hysterectomies, accounting for at least one third of all hysterectomies [2]. The high UF prevalence has a significant economic impact on healthcare systems worldwide. Treatment costs for UF patients amount to hundreds of millions of dollars in countries such as Germany (USD 348 million) and France (USD 120 million) and reach tens of billions of dollars in the USA (USD 34.4 billion) [6]. It should be emphasized that the cost of treating women with UFs exceeds the cost of treating women with such common tumors as breast cancer and ovarian cancer [6]. In addition to direct healthcare costs, indirect costs due to temporary disability and the disability of women with uterine fibroids are estimated at USD 1.6–17.2 billion annually worldwide [7].
UFs are a genetically determined disorder, with a substantial hereditary contribution to UF development (26–69%, European twin studies data) [8,9]. The genetic determinants of UFs have been identified in more than ten GWASs (several dozen UF risk loci were detected, including genes “involved” in sex hormone pathways [ESR1, FSHB, etc.]) [10,11]; however, only a small part of UF heredity (≈13%) can be explained by the known GWAS (SNP) data [11], which confirms the relevance of further UF genetic studies aimed at detecting as-yet-unknown genetic risk factors for UFs.
The effect of sex hormones (such as estrogens and testosterone) on UFs is beyond doubt [12,13]. Estrogens are believed to stimulate the growth of myomatous nodes (interacting with estrogen α receptors and enhancing the expression of progesterone receptors, they increase the proliferative activity of uterine smooth muscle cells, etc.) [13]. The effect of testosterone on UF development is also significant (local transformation into estrogens under the action of aromatases, proapoptotic/antiproliferative effects, etc.); however, there is currently no definitive opinion in the literature on the direction of the effect of testosterone (risky/protective) on UF risk [12,13,14]. The pathophysiological effects of these hormones in the body (including UFs) will largely depend on SHBG [14]). SHBG, binding and transporting a major portion of testosterone (80%) [12] and estrogens (38%) [15], thus ensures the “regulation” of the level of the free fractions of these sex hormones in the organism (they account for 2% of estrogens and 1% of testosterone), which exhibit biological activity in the female (i.e., they are bioactive) [12,15]. Thus, the above data suggest that the modulation of testosterone and estrogen concentrations in the organism caused by SHBG (due to binding/release) (their low concentrations at high levels of SHBG and, conversely, high concentrations of testosterone/estrogen at low levels of SHBG) will be important in UF pathology. So, based on these literature materials, it can be noted that the main hypothetical biological mechanism by which SHBG levels can influence UF development is their significant effect on concentrations of bioavailable androgens/estrogens in the organism, which will be important in the UF formation.
SHBGcon in the organism is genetically determined, as demonstrated by numerous GWASs [16,17,18,19,20,21,22]. However, the role of polymorphisms/genes that determine SHBGcon in UF development remains unexplored at the moment. To date, only one study by Wang et al. has investigated this association using Mendelian randomization (MR) of two cohorts, FinnGen and FibroGENE. While no significant association between genetic variants of SHBGcon and the risk of UF was found in the meta-analysis of the two studies, a cohort-specific association was identified in FibroGENE [23]. There are no other experimental genetic studies on this issue. So, the lack of knowledge on the role of the genetic determinants of SHBGcon in UF development dictates the need for research in this area, which is the subject of our study. The aim of the work is to examine correlations between polymorphic variations that determine the SHBGcon and UF.
2. Materials and Methods
2.1. Study Subjects
The phasing/design/ethical aspects of this study were reviewed and supported by the Ethics Committee (Medical Research) of Belgorod State National Research University. Each woman surveyed gave her consent (signed personally) to participate in the study. The overall number of the genetically studied group of women was n = 1542, including 569 with UF and 973 without UF [control]. All subjects were born in central Russia and were Europeans (Russian nationality) [24,25,26]. The UF diagnosis in all patients (ICD-10:code D25) was confirmed morphologically after hysterectomy in the gynecological department of the Belgorod Perinatal Center [27]. All women in the control group underwent a comprehensive medical evaluation (conducted at the Belgorod Perinatal Center), including a clinical examination, a medical history review, and pelvic ultrasound screening, specifically evaluated for proliferative disorders (UFs, endometriosis/adenomyosis, and endometrial hyperplasia). The control group included women who had no clinical, anamnestic, or ultrasound signs of proliferative diseases of the pelvic organs [27]. Despite the fact that ultrasound examination, used in the work to confirm the absence of UF in the control group, is the “gold standard” of uterine imaging and serves as the main method of diagnosing UFs, there is a possibility of non-detection of UFs (for example, small-sized UFs, cervical UFs, etc.) when it is used in the examination of women in the control group, which may be a limitation of the present study. Women with malignant diseases of the pelvic organs/breast, the severe pathology of the immune/vital system/organs were excluded from the study. The main characteristics of the UF/UF-free studied groups are shown in Table 1 (these materials were submitted in the previous genetic study of UFs [27]). The data in Table 1 point out the “UF vs. UF-free” differences—in terms of such indicators as age, weight, BMI, family history, a history of infertility, artificial abortions (and their number), chronic endometritis—which formed the basis for using these characteristics in “UF-SHBGcon-related SNP” association calculations as confounders.

Table 1.
The characteristics of participants from the case and control groups.
2.2. Experimental DNA Study (Selection/Genotyping of SHBGcon-Related Genetic Variants)
For the experimental study, DNA samples (isolated from peripheral blood using phenol, chloroform, and ethanol [28]) obtained during a previously conducted genetic study of UFs were used [27]. The DNA samples were stored in low-temperature (−80 °C) refrigerators (kelvinators), and had the required degree of purity (“260 nm/280 nm” ratio = 1.7–2.0) [29] and concentration (10–20 ng/mL) (the data on the DNA density/concentration were obtained on a NanoDrop™ 2000 (Thermo Fisher Scientific Inc., Waltham, MA, USA) [30]). Nine polymorphisms that showed associations with SHBGcon in previously performed GWASs were included in the work ([16,18,20,21,22]; detailed GWAS data on associations of the studied SNP with SHBGcon are presented in Table 2 and Supplementary Table S1). Also, when selecting polymorphisms, their potential functionality was taken into account (the assessment of the potential functionality of SNPs was carried out in silico using the bioinformatic online resource Haploreg—accessed 24 November 2024 [31]; the data obtained are presented in Supplementary Table S2). These are polymorphisms rs3779195 [T/A] BAIAP2L1, rs17496332 [A/G] PRMT6, rs8023580 [T/C] NR2F2, rs780093 [C/T] GCKR, rs440837 [A/G] ZBTB10, rs4149056 [T/C] SLCO1B1, rs10454142 [T/C] PPP1R21, rs12150660 [G/T] SHBG, and rs7910927 [G/T] JMJD1C. The SNP genotyping procedure was performed on CFX96 amplifiers [32]. The method of “blind” SNPs re-genotyping (for this purpose, we conducted repeated analyses for approximately 5% of the DNA samples [33]), which we employed to verify the accuracy of the genetic data, demonstrated a perfect match in the outcomes (specific genotypes) for 99% of the “blind” re-genotyped SNPs/samples, confirming the satisfactory quality of the genetic analysis.

Table 2.
The GWAS data on associations of the studied polymorphisms of the candidate genes with the SHBGcon and testosterone concentrations.
2.3. Statistical Analysis (SNP–Multi-SNP Association Examined)
Previously, before analyzing the UF-SNP associations, the implementation of the Hardy–Weinberg (HW) rule by all SNPs among the UF/UF-free groups was considered [34,35].
We searched for an association of the SHBGcon-related polymorphisms with UFs in two directions [36]: (a) the relationship of individual loci with UFs was studied; (b) the contribution of SNP-SNPint (multi-SNP association) to exposure to UF was considered.
The association rates of individual SNPs with UFs (such as OR, 95%CI) [37]) were calculated using the logistic regression (the four most commonly used genetic-statistical models were used to evaluate various types of allelic variant interactions: dominant/additive/recessive/allelic [38]). The calculations were performed in gPLINK (a Java-related program [v.2.050]) [39] and took into account the effects of covariates (as indicated above, Table 1, these were age, weight, BMI, family history, a history of infertility, artificial abortions (and their number), and chronic endometritis), multiple comparisons (a permutation test was performed [adaptive version]) [40]) and the power of the associative connections (the Quanto software [v.1.2.4] was employed [41]). The values of “pperm” below 0.05 and “powers” above 80% were the basis for identifying reliable “UF-SNP” associations [42].
The MB-MDR (R-integrated) [43], GMDR (Java-integrated) [44,45], and MDR (Java-integrated) [46] programs were used to search/analyze the most UF-significant SNP-SNPint. When preparing data files for calculations in the MB-MDR, GMDR, and MDR programs, the missing genotype values were imputed using the MDR-Data Tool Software Overview (Java-integrated, v.3.0.2). All the UF-significant models we obtained took into account the necessary covariates (which are listed above) and were confirmed by permutation testing [40]. The permutation test was demonstrated to be efficient for the analysis of large massifs of GWAS data without reducing power [40]. As the most UF-significant SNP-SNPint models, we considered models that met the following requirements: (1) the level of their statistical significance after the 1st stage of MB-MDR analysis corresponded to a special threshold level “pborder” (determined based on the correction of the standard value “p = 0.05” by the number of genotype combinations considered at different SNP-SNPint levels/loci [i.e., the Bonferroni correction was introduced]), which corresponded to the values of pborder = 1.39 × 10−3 (0.05/36), pborder = 5.95 × 10−4 (0.05/84), and pborder = 3.97 × 10−4 (0.05/126) for SNP-SNPint models with 2, 3, and 4 loci, respectively [34]; (2) the level of their statistical significance after the 2nd stage of the MB-MDR analysis (the models were validated by permutation testing to minimize false positive results; 1000 permutations were performed) corresponded to the values of “pperm” < 0.01 (2- and 3-locus models) and < 0.001 (4-locus models). The two criteria mentioned above for identifying the most UF-significant SNP-SNPint models were met only by models of the 3rd and 4th levels (two models of each level), which were included in our work.
Next, we performed a cross-validation of these four most UF-significant SNP-SNPint models using the GMDR method/program (Java-integrated) [44,45]. The indicators of the cross validation consistency (CVC), testing balanced accuracy (TBA), sensitivity (Se), and specificity (Sp) of the models were calculated, taking into account the correction for necessary covariates. A correction for multiple comparisons was performed using a permutation test in Perl script (“perl GMDR_permutatin.pl”) GMDR software (v.1.0). In total, 1000 permutations (with 10 cross-validations) were performed, which ensures the level of statistical significance of the validated model for samples of more than 1000 individuals with at least pperm < 0.001.
The results derived by us in the study of “UF-SNP” associations were visualized (in the form of the SNP/SNP-SNPint contribution to the UF entropy [47]) on a graph (using the MDR program [46]).
2.4. The Evaluation of the Possible Functionality of UF-Correlated Variants: An In Silico Study
Following the identification the UF-associated loci, we performed the functional annotation of these variants and their LD proxies (r2 value: 0.80–1.00 [48,49]) to investigate the biological mechanisms of the observed association between SHBGcon-related SNPs and UF. To address this challenge, we used the following genomic/bioinformatic online resources [50,51]: (1) HaploReg_accessed_24 November 2024 [31]; (2) GTExportal_accessed_25 December 2024 [52]; and (3) STRING_accessed_08 December 2024 [53].
3. Results
All analyzed loci demonstrated SNP distributions consistent with the Hardy–Weinberg equilibrium in both cohorts: UF (pHWE ≥ 0.024) and without UF (pHW ≥ 0.052) (Supplementary Table S3). We applied the Bonferroni correction based on the number of loci studied, pHW(Bonf) ≥ 0.006 [0.05/9]).
Two SHBG-related SNPs demonstrated significant correlations with UF risk: rs3779195 [T/A] BAIAP2L1 (ORAA = 0.38; 95%CIAA = 0.20–0.91; pperm(AA) = 0.023 [recessive genetic model]; powerAA = 81.84%) and rs440837 [A/G] ZBTB10 (ORGG = 1.93; 95%CIGG = 1.17–3.14; pperm(GG) = 0.010 [recessive genetic model]; power = 85.57%)) (Table 3).

Table 3.
Associations of the SHBG-significant gene polymorphisms with UF.
Also, seven SHBG-related SNPs [from the nine studied loci] such as rs3779195 [T/A] BAIAP2L1, rs17496332 [A/G] PRMT6, rs8023580 [T/C] NR2F2, rs780093 [C/T] GCKR, rs440837 [A/G] ZBTB10, rs10454142 [T/C] PPP1R21, and rs7910927 [G/T] JMJD1C, interacting amongst themselves (four models of such SNP-SNPint interactions [pperm ≤ 0.01] were found; the cross-validation parameters of these models were as follows: CVC = 10/10, testing balanced accuracy 50.72–56.68%, sensitivity 50.60–65.20%, and specificity 55.70–63.00%), influenced the UF risk (Table 3). Meanwhile, the effects of three loci (rs8023580 [T/C] NR2F2, rs780093 [C/T] GCKR, and rs10454142 [T/C] PPP1R21) were the most serious (75% of the SNP-SNPint models included each of these SNPs) (Table 4). After implementing the Bonferroni correction, the actual p-values for our models substantially exceeded the required thresholds: for three-locus interactions, we observed p = 4.88 × 10−5; against a threshold of 5.95 × 10−4; for four-locus interactions, p = 5.86 × 10−8; against a threshold of 3.97 × 10−4. This considerable margin between the observed values and threshold values confirms the high reliability of these interaction effects.

Table 4.
SNP × SNP interactions of SHBG-significant genes associated with UF.
As a result of the modeling procedure, 16 different UF-significant combinations of SNP-SNPint genotypes were identified (Supplementary Table S4), of which 81.25% (13/16) increased the UF risk, and correspondingly 18.75% (3/16) reduced the risk of UF. Three UF–risk combinations, such as rs8023580-TT NR2F2- rs10454142-TC PPP1R21- rs780093-TT GCKR- rs17496332-AA PRMT6 (β = 2.027), rs8023580-TT NR2F2-rs10454142-TC PPP1R21-rs780093-TT GCKR (β = 0.924), and rs8023580-TC NR2F2-rs7910927-GT JMJD1C-rs3779195-TA BAIAP2L1 (β = 2.669) have the greatest statistical significance (p = 0.0007, p = 0.006, and p = 0.007, respectively) (Supplementary Table S4).
The percentage of UF entropy, depending on the polymorphisms under consideration, was expected to be highest for the two loci rs3779195 [T/A] BAIAP2L1 (0.47%) and rs440837 [A/G] ZBTB10 (0.45%), which exhibit the main effect with respect to UF (Figure 1). The effect of the most remarkably paired SNP-SNPints of both antagonistic (−0.21–−0.26) and synergistic (0.26–0.28%) orientations on the UF risk was almost two times less than the independent exposures of the above two loci (Figure 1).
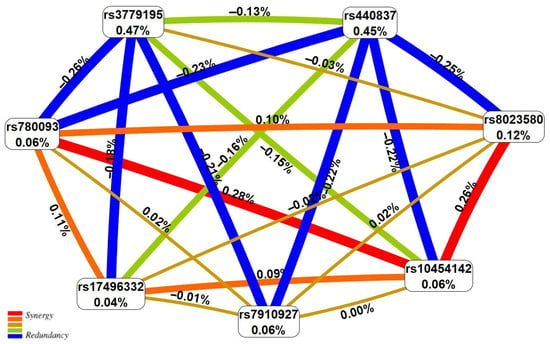
Figure 1.
The entropy graph of the UF-associated SNP × SNP interactions (based on the MDR analysis). Positive values of entropy indicate synergistic interactions while the negative values indicate redundancy. The red and orange colors denote strong and moderate synergism, respectively, and the brown color denotes the independent effect; green and blue denote moderate and strong antagonism.
3.1. Potential Functionality of the UF-Associated Polymorphisms
Having identified the associations of SHBG-related polymorphisms with UF, we further assessed the potential functionality of UF-associated loci (and strongly linked SNPs [r2 ≥ 0.80]) in the organism (using the in silico approach for this), due to which these polymorphisms may be involved in a susceptibility to UF. We conducted this analysis in two directions: firstly, we examined the functionality of two UF-causal loci (independently associated with UF), rs440837 [A/G] ZBTB10 (together with 5 proxy variants) and rs3779195 [T/A] BAIAP2L1 (together with 20 proxy variants); secondly, we evaluated the functionality of all seven UF-associated polymorphisms related with SHBG (both independently and during SNP-SNPint) (together with 115 proxy variants).
3.1.1. The Characterization of the Functionality of the Two UF-Causal Loci
SNP rs440837 [A/G] ZBTB10
The variant rs440837 [A/G] ZBTB10 and its five proxy loci are located within functionally significant regulatory regions near ZBTB10/RP11-48B3.3/RP11-48B3.4 genes that were involved in interaction with 22 transcription factors (TFs). In the liver, both rs440837 [A/G] ZBTB10 and its proxy locus, rs7013042 RP11-48B3.4, were posted in a DNA region where epigenetic modifications of histones (acetylation/methylation) marking potential promoters/enhancers [H3K4me3/H3K4me1] occur (including active promoters/enhancers [H3K9ac/H3K27ac]) (Table 5, Supplementary Table S5).

Table 5.
The potential functionality of the UF-causal SNPs rs440837 [A/G] ZBTB10 and rs3779195 [T/A] BAIAP2L1 and their proxy variants (r ≥ 0.80) in organism as a whole and in the liver (SHBG synthesis place) (in silico data).
SNP rs3779195 [T/A] BAIAP2L1
rs3779195 [T/A] BAIAP2L1 and its 20 LD SNPs were extremely significant for the DNA (in the BAIAP2L1/BRI3 gene region) collaboration regulation with 68 TFs (17 SNPs) and 18 regulatory proteins (4 SNPs) (Table 5, Supplementary Table S5), and affect the transcription of 15 genes (Table 4, Supplementary Table S6, Supplementary Table S7) and the splicing of 3 genes (BRI3, TECPR1, BAIAP2L1) (Table 5, Supplementary Table S8, Supplementary Table S9) including both organs of the female reproductive system (uterus [eQTL-RP11-307C18.1], ovary [eQTL-RP11-307C18.1]) and other organs directly related to the UF pathophysiology: adipose tissue [eQTL-BRI3, RP11-307C18.1; sQTL-BRI3], musculoskeletal tissue [eQTL-BRI3, RP11-307C18.1, ASNS, BAIAP2L1; sQTL-BRI3], the brain [eQTL-BHLHA15, RP11-307C18.1; sQTL-TECPR1, BRI3], adrenal glands [eQTL-RP11-307C18.1], the thyroid gland [eQTL-BAIAP2L1, RP11-307C18.1, TECPR1, BHLHA15, LMTK2; sQTL-BRI3], the blood [eQTL-TECPR1, RP11-307C18.1], etc. In the liver, both rs3779195 [T/A] BAIAP2L1 and several of its proxy SNPs were located in genomic regions where various histones’ epigenetic modifications (acetylation/methylation) were present, such as H3K4me3 (labeling the promoters [3 SNPs]), H3K4me1 (marking the enhancers [10 SNPs]), H3K9ac (labeling the active promoters [6 SNPs]), and H3K27ac (marking the active enhancers [8 SNPs]) (Table 5, Supplementary Table S5). Also, in the liver, rs3779195 [T/A] BAIAP2L1 and 17 proxy SNPs were linked with the expression of two genes (BRI3 and RP11-307C18.1). Interestingly, UF-protective allele A rs3779195 [T/A] BAIAP2L1 was associated with high RP11-307C18.1 transcription (NES:−0.54) and low BRI3 expression (NES:0.87) in the liver (Supplementary Table S6).
After characterizing the functionality of the two UF-causal loci and their proxy SNPs, we then evaluated the protein–protein communications (PPcs) linked with these loci in the STRING program. For example, the PPc of 22 TFs and three proteins encoded by the genes ZBTB10, RP11-48B3.3, and RP11-48B3.4 (a total of 25 proteins) were studied for the locus rs440837 [A/G] ZBTB10, while the PPc of 68 TFs, 18 regulatory proteins, and 15 proteins encoded by the genes LMTK2, TECPR1, AC004967.7, ASNS, BAIAP2L1, BRI3, RP11-307C18.1, RP11-307C18.2, RP11-307C18.3, RP11-307C18.4, RP11-307C18.5, RP11-307C18.6, RP11-307C18.7, RP11-307C18.10, and RP11-307C18.11 (a total of 101 proteins) were considered for the locus rs3779195 [T/A] BAIAP2L1. Additionally, several proteins most significant for these PPcs (according to the STRING data) were included in the PPc analysis of these two UF-causal loci.
Among the many identified pathways involving PPc linked with two UF-causal loci (over 350 pathways for rs440837 [A/G] ZBTB10 (Figure 2A, Supplementary Table S10) and over 350 pathways for rs3779195 [T/A] BAIAP2L1 (Figure 2B, Supplementary Table S11)), various regulatory influences on gene transcription (including estrogen-dependent gene expression and miRNA transcription regulation, etc.), steroid hormone processes (levels, biosynthesis, androgen receptor signaling, etc.), morphogenesis (including gonad development, apoptotic process, etc.) prevail and are characterized by the TGF-beta signaling pathway (hsa04350). All of the above pathways may be important for the pathophysiology of UF.
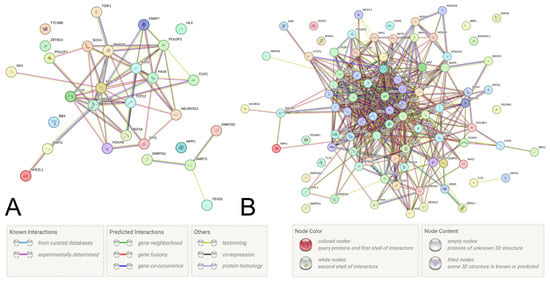
Figure 2.
Protein–protein communications determined by the UF-causal SNPs rs440837 [A/G] ZBTB10 (A) and rs3779195 [T/A] BAIAP2L15 (B) and their proxy loci (STRING program data).
3.1.2. The Characterization of the Seven UF-Associated Loci Functionality
The overwhelming majority of UF-related loci (118 out of 122 considered variants [96.72%] such as seven UF-associated polymorphisms and 115 strongly linked SNPs) had pronounced potential epigenetic influences on 13 adjoining genes (GCKR, KLRAQ1, FOXN2, NR2F2, BRI3, RP11-327J17.2, BAIAP2L1, PPP1R21, PRMT6, RP11-327J17.3, ZBTB10, JMJD1C, and RP11-48B3.4) due to their localization in such regulatorily valuable regions of these genes as (a) open chromatin (they are highly sensitive to the effects of DNase enzyme) (21.31%/26 variants); (b) evolutionarily conservative regions (4.10%/5 variants); (c) promoters (8.19%/10 variants); (d) enhancers (22.95%/28 variants); (e) regulatory motifs (90.16%/110 variants); and (f) liaison sites with regulatory proteins (13.93%/17 variants) (Supplementary Table S5).
More than 95% of UF-correlated loci (116 out of 122 variants [95.08%]) were communications to the transcription activity of 47 genes (Supplementary Tables S6 and S7). The abovementioned loci were eQTL impact promoters in several organs including both organs of the female reproductive system (uterus [RP11-460M2.1, RP11-307C18.1] and ovary [RP11-307C18.1]) and other UF-significant organism organs, such as adipose tissue [NRBP1, ATRAID, REEP3, BRI3, GTF2A1L, KRTCAP3, PPM1G, STON1-GTF2A1L, PPP1R21, MRPL35P2, PRMT6, RP11-307C18.1], the brain (pituitary) [RP11-307C18.1, FOXN2, GTF2A1L, MRPL35P2], thyroid [KRTCAP3, AC074117.10, IFT172, ATRAID, GTF2A1L, BAIAP2L1, MRPL35P2, C2orf16, PRMT6, FNDC4, GCKR, JMJD1C-AS1, LMTK2, PPM1G, ZNF512, STON1, PPP1R21, REEP3, RPL7AP50, RP11-307C18.1, TECPR1], the blood [RP11-307C18.1, KRTCAP3, PRMT6, TECPR1, NRBP1], adrenal glands [FOXN2, GTF2A1L, KRTCAP3, MRPL35P2, PRMT6, RP11-307C18.1], and also in the liver where SHBG is formed [RP11-307C18.1, RP11-307C18.2, PRMT6, GTF2A1L, RP11-327J17.2, BRI3] (Supplementary Tables S6 and S7).
A substantial proportion of the UF-related loci (34 out of 122 SNPs [27.87%]) showed significant associations with the splicing control of 13 genes (GPN1, KRTCAP3, PPP1R21, IFT172, BRI3, STON1, FNDC4, TRIM54, STON1-GTF2A1L, GTF2A1L, BAIAP2L1, GCKR, and SNX17), and they showed their sQTL influences in female reproductive system organs (the uterus [IFT17, PPP1R21], the ovaries [IFT17, FNDC4]) and other such UF-important organs as the brain (pituitary) [IFT172, FNDC4, PPP1R21], adipose tissue [GPN1, BRI3, SNX17, FNDC4, IFT172, GTF2A1L, STON1, PPP1R21, STON1-GTF2A1L], adrenal glands [GCKR, FNDC4, IFT172], the thyroid gland [IFT172, BRI3, PPP1R21, KRTCAP3], and also the liver [GCKR, FNDC4] (Supplementary Tables S8 and S9).
In the final stage of our study, we investigated PPcs involving the protein products of 52 UF candidate genes (functionally correlated with seven UF-causal loci and 115 LD SNPs, the data are presented above). The data obtained (Figure 3) indicate the hub value of PPc of hormone-related genes (such as LHCGR, FSHR, STON1, and STON1-GTF2A1L), and the involvement of PPc in the regulation of gene transcription (due to transcription factor IIA (alpha/beta subunits) [pFRD = 0.0280, SM01371]), as well as interactions with hormone ligand-binding receptors [pFRD = 0.0062,HSA-375281] and the content control of SHBG [pFRD = 3.11 × 10−6, EFO: 0004696], testosterone [pFRD = 2.83 × 10−6, EFO: 0004908], liver enzymes [pFRD = 0.0001, EFO: 0004582], fasting blood glucose [pFRD = 0.0038, EFO:0004465] [pFRD = 2.60 × 10−8, EFO: 0011008], lipids [pFRD = 2.13 × 10−5,EFO: 0004529], etc. All of the aforementioned pathways may impact the biology of UF.
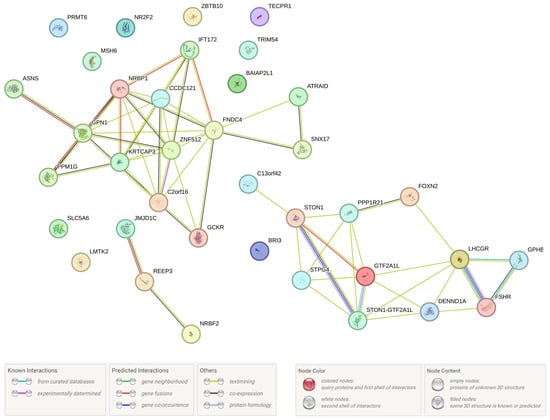
Figure 3.
Interaction of UF-correlated proteins (STRING program data).
4. Discussion
In this study, we have proven for the first time the risk (OR = 1.93)/protective effect (OR = 0.38) for UF of SHBGcon-increasing (allele G rs440837 [A/G] ZBTB10)/decreasing (allele A rs3779195 [T/A] BAIAP2L1) genetic variants of GWAS-significant for SHBGcon polymorphisms. The susceptibility to UF was also determined by the interactions of seven SHBG-related SNPs, which, while showing potential pronounced functionality, were involved in UF impact pathways of gene transcription regulation, interaction with hormone ligand-binding receptors, the content control of SHBG, testosterone, liver enzymes, fasting blood glucose, lipids, etc.
Our findings demonstrate that the AA genotype of rs3779195 [T/A] BAIAP2L1 has a protective effect against UF development (if it is present in the genotype, the risk of UF decreases by more than 60% [OR = 0.38]). It should be noted that our data on associations of rs3779195 [T/A] BAIAP2L1 with UF were obtained for the AA genotype, which is quite rare and occurs in patients (1.93%) two times less frequently than in the control (3.80%), which led to a pronounced OR index equal to 0.38. The results of two previously published GWASs showed a correlation of the allele A with low SHBGcon [18,21]. So, the SHBG-lowering genetic variant (allele A rs3779195 [T/A] BAIAP2L1) (data from the two abovementioned GWAS) was UF-protective (our results). The rs3779195 [T/A] BAIAP2L1 polymorphism and its proxy loci may have pronounced functionality—they were involved in plenty of (n = 86) “DNA-TFs/protein-regulatory” cooperations (in the BAIAP2L1/BRI3 genes region), in the regulation of the eQTL (15 genes) and sQTL (3 genes) traits in various organs of both the female reproductive system (uterus [eQTL-RP11-307C18.1], ovary [eQTL-RP11-307C18.1]), and other organs directly related to UF pathophysiology (adipose; skeletal muscles; blood; adrenal/thyroid glands;brain; and others), including in the liver (BRI3; RP11-307C18.1 [eQTL]) (our in silico data), which is the major organ of SHBG formation in the body [54].
The protein product of the BAIAP2L1 gene (brain-specific angiogenesis inhibitor 1—associated protein 2-like 1 [BAIAP2L1], also known as insulin receptor tyrosine kinase substrate [IRTKS]), is involved in a number of UF-significant processes such as morphogenesis and cell migration (involved in the protrusion of the plasma membrane and the formation of actin), proliferation, and apoptosis (due to the activation of EGFR-ERK, PI3K/AKT, and other pathways) [55,56,57]. These processes play an important role in tumorigenesis [56,57], including in malignant neoplasms of the female reproductive system (ovarian cancer) [55]. Interestingly, positive genetic correlations are observed between UF and ovarian cancer [58], and it can be assumed that one of the genes that may underlie these correlations may be the BAIAP2L1 gene. Importantly, several rs3779195 [T/A] BAIAP2L1 LD variants have also been correlated (GWAS materials) with SHBGcon and testosterone level: SHBGcon—rs112758337 (D’ = 1.00; r2 = 0.96), rs1688606 (D’ = 1.00; r2 = 0.96), and rs4268041 (D’ = 1.00; r2 = 0.99) [21]); total testosterone—rs1635612 (D’ = 1.00; r2 = 0.96) [21] and rs35903783 (D’ = 1.00; r2 = 0.41) [20]. So, the genome region in the SNP rs3779195 [T/A] BAIAP2L1, due to its assumed pronounced functional meaning in silico (epigenetic/eQTL effects) in the liver (the main organ of SHBG synthesis), is essential in SHBGcon/testosterone level regulation in the organism and, thanks to this, may affect UF risk.
The present study revealed the risk effect of the genotype GG rs440837 [A/G] ZBTB10 on UF development (its presence in the genotype increased UF risk by almost two times [OR = 1.93]). It should be noted that our data on associations of rs440837 [A/G] ZBTB10 with UF were received for the GG genotype, which occurs with a low frequency and its prevalence in patients (7.03%) is 1.6 times higher than in the control (4.31%), which led to an expressed OR index of 1.93. The allele variant G rs440837 [A/G] ZBTB10 showed associations with high SHBGcon in two previously presented GWASs [18,21]. So, the SHBGcon-boosting genetic variant (allele G rs440837 [A/G] ZBTB10, in the result of the two abovementioned GWASs) was UF-risky (the results of this study). The UF-causal locus rs440837 [A/G] ZBTB10 (jointly with LD SNPs) was potentially impactful for the “DNA-TFs engagement” (n = 22 TFs) and other epigenetic modifications (histone acetylation/methylation) at the ZBTB10/RP11-48B3.3/RP11-48B3.4 genes’ place in liver (our in silico results). It is important that the protein of the same name, encoded by the ZBTB10 gene, is an important regulator of gene transcriptional activity due to its modulating effects on the binding of RNA polymerase II to the corresponding genomic sequence [59], which may be important for the regulation of SHBG synthesis in the liver. Interestingly, a number of loci strongly associated with rs440837 [A/G] ZBTB10 also showed their GWAS relevance for SHBGcon, with such loci as rs72688090 (D’ = 0.85; r2 = 0.33) [60], rs388922 (D’ = 0.96; r2 = 0.53), rs575452 (D’ = 0.71; r2 = 0.28), and rs117921873(D’ = 1.00; r2 = 0.26) [21]. So, we would like to note the importance of the rs440837 [A/G] ZBTB10 genome region in regulating SHBGcon (due to the supposedly significant in silico epigenetic influences in the liver, in which SHBG is mainly formed) and its involvement in UF biology/risk due to this.
Summarizing the material obtained in the work (presented above), it can be concluded that SHBGcon-lowering genetic variant (AA genotype rs3779195 [T/A] BAIAP2L1) reduces UF risk (OR = 0.38), and SHBGcon-raising genetic variant (genotype GG rs440837 [A/G] ZBTB10) increases UF risk (OR = 1.93). There is only one paper in the literature on this topic with extremely contradictory results: after conducting an MR, the authors did not identify a link between SHBG and UF in the GWAS meta-analysis of the data from two cohorts, FinnGen and FibroGENE, and found an inverse correlation between them in one of these cohorts—FibroGENE [23]. At the same time, in the work of Misao et al., based on experimental data, a higher level of SHBG in UF cells compared to normal myometrium was found in the vast majority of the biological samples studied (21 out of 23, 91%) [61], which is completely consistent with our results and differs from the abovementioned data obtained by Wang et al. in their MR analysis [23].
It should be noted that the possible reasons for the differences between our results and the data of Wang et al. obtained by MR of UF [23], may include the following: firstly, in the work of Wang et al., an MR analysis was performed based on the combined GWAS data for UF (FinnGen and FibroGENE) and SHBG (UK Biobank [20]); the pleiotropic effects of the individual most SHBG-significant loci in both UF and UF-significant risk factors (for example, the age of menarche/menopause, etc.), through which the UF-significant effects of SHBG-significant loci may be mediated, were not considered; whereas, in our work, the independent effects of the two SHBG-significant loci in UF were found. As an example of the possibility of such “discrepancies” in the results of even one study, one can cite the work of Garitazelaia et al. [62], in which the MR analysis did not reveal reliable causal relationships between the content of sex hormones and endometriosis, but at the same time (and this was a surprise to the authors!) pleiotropic genetic associations of two loci of the FSHB gene (rs11031002, rs11031005) with endometriosis and sex hormone concentrations were found; the SNPs of the FSHB gene region (rs11031005, rs11031006) also demonstrated significant pleiotropic associations mediating endometriosis and related signs (menarche/menopause age and menstrual cycle duration) [62]. Secondly, our results may depend to a certain extent on a number of comorbid conditions common in the sample we studied. For example, more than 1/3 of the patients studied in our work (36.38%) have endometriosis (Table 1). At the same time, endometriosis is characterized by a clear association between high SHBG and an increased risk of disease [63,64]. Using the MR analysis by Qu et al. (the study analyzed GWAS data from the FinnGen cohort) showed that endometriosis is associated with an increased risk of UF, while PCOS (unlike endometriosis, low SHBG is risky for PCOS) was associated with a reduced risk of UF [65]. Thirdly, in the work of Wang et al., an inverse correlation between SHBG and UF was observed only in FibroGENE (in the MR analysis of this cohort, BMI was adjusted due to the similarity of GWAS data on SHBG and UF), and not in FinnGen, and as the authors themselves note, “care needs to be taken for bias resulting from sample overlap and collider bias caused by adjusting BMI” [23]. A critical difference that may underlie these inconsistent findings is the unequal number of instrumental variables used across the two cohorts. In the FinnGen analysis, fewer SHBG-associated SNPs were available or retained after quality control and filtering for linkage disequilibrium and pleiotropy. This reduced number of instruments likely weakened the statistical power and increased vulnerability to weak instrument bias, contributing to the null result. In contrast, the FibroGENE analysis utilized a larger set of instruments, enabling more stable estimates. However, this analysis also involved sample overlap with the SHBG GWAS, raising the possibility of bias from overfitting or collider effects, especially in the context of multivariable MR adjusting for genetically correlated traits such as BMI. These discrepancies in instrument number and dataset composition likely contributed to the divergent outcomes observed across the two cohorts. In addition, it is important to note that the information identified in the work of Wang et al., the nominally significant causal relationship between a higher level of SHBG and a lower UF risk, according to the authors themselves (!) contradicts one of the main conclusions made in their work—“ the protective effect of a higher level of total testosterone on uterine leiomyoma, as higher SHBG usually means less bioactive testosterone” [23]. Interestingly, Wang et al. also reported an inverse association between total testosterone and fibroid risk, a conclusion that aligns with our findings. However, their interpretation of the nominally inverse relationship between SHBG and fibroids as evidence for a protective effect of SHBG contradicts the well-established inverse relationship between SHBG and free testosterone. This inconsistency, combined with the methodological limitations discussed above, underscores the need for the cautious interpretation of MR findings, particularly in hormonally driven conditions with complex genetic architectures and interrelated exposures (it should be emphasized that this conclusion is made by Wang et al. and the protective role of testosterone in UF is fully consistent with the results/assumptions of our work, detailed below). Thus, there is an extremely pronounced ambiguity in the currently limited genetic data on the relationship between the genetic determinants of SHBG and UF (even in the framework of one study, for example, the Wang et al. MR analysis [23]), and further research in this area is needed.
One potential mechanism underlying the association of SHBG with UF involves the transportation/deposition of testosterone [12]. Current evidence estimates that approximately 80% of testosterone is in the SHBG-bound state, and only 1% is a free fraction and exhibits biological activity [12]. Thus, SHBGcon, by directly affecting the level of bound/free testosterone, will largely determine its effects on UF. A significant association of “SHBG–testosterone” is also evidenced by the indicators of a pronounced negative genetic correlation of SHBG with the level of free testosterone, which in women reaches a value of −0.75 [20,66]. In this case, the effects of testosterone can be realized by both its free (bioactive) and bound fractions [14]. It is assumed that testosterone bound to SHBG can enter cells by endocytosis and be released as a result of pH changes [14].
The existing literature presents limited and conflicting data regarding the association of testosterone with UF risk. For example, in the work of Wong et al., a high content of bioavailable testosterone was demonstrated to be associated with both an increased UF risk (OR = 1.33) and, concurrently, a low UF recurrence risk [13]. On the one hand, a number of studies indicate that testosterone, transformed by aromatase into estradiol in myomatous cells, promotes the development of local hyperestrogenism in the UF area, which stimulates the growth of myomatous nodes [12]. On the other hand, there is evidence of testosterone’s protective value in UF [23]. Wang et al., working with MR GWAS data from two cohorts, FinnGen and FibroGENE, revealed a reverse relationship between total testosterone and UF in each of these cohorts, as well as in their combined analysis (OR = 0.90); no correlations between free testosterone and UF were found [23]. These data disagree with the currently available unambiguous ideas about a significantly positive relationship between total and free testosterone levels: according to Ruth et al., in women the genetic correlations between these parameters are 0.65 [20]. As one of the possible reasons for the lack of a genetic link between free testosterone and UF (Wang et al.), it is suggested that in the GWAS materials Ruth et al. used in their study, data on bioavailable testosterone levels were not obtained by direct measurements, but were calculated using the Vermeulen formula (based on data on testosterone, SHBG, and albumin), and these estimates, based on model formulas regarding the binding ability of SHBG, may not correspond to the true affinity of SHBG for binding to testosterone [23].
A review article by Whitton and Baber notes the proapoptotic/antiproliferative testosterone effects on hormone-dependent organs/structures of the female reproductive system, such as the mammary gland and uterus endometrium [66], despite the fact that their cells (like myomatous cells) contain high levels of both aromatases and androgen receptors [23,67]. The proapoptotic/antiproliferative effects of testosterone may be based on various testosterone-related biological mechanisms such as the activation of the growth and maturation of primordial follicles, increased metabolic processes in oocytes (the early stages of folliculogenesis), the suppression of follicle growth, the inhibition of estrogen formation (the late stages of folliculogenesis), the stimulation of corpus luteum formation, the increased synthesis of progesterone (luteal phase) etc. [68,69].
Notably, the observed association between low testosterone concentrations (both total and free; the genetic correlations between them in women are 0.65 [20]) and increased UF risk may be mediated through established UF risk factors. These include obesity as evidenced in our cohort by positive BMI-UF association (Table 1) and cardiovascular diseases, etc. [1,2,3]. Thus, low levels of total testosterone correlate with an increased risk of metabolic syndrome, obesity/dyslipidemia, hypertension, and insulin resistance/diabetes [14].
Another potential mechanism explaining the association of SHBG-lowering/SHBG-increasing genetic variants with low/high UF risk may be the “genetic” association of SHBG with estradiol [20]. Thus, according to a large-scale study by Ruth et al., there are significant positive genetic correlations between SHBG and estradiol levels in women (0.45) [20]. Estradiol is a known driver of the growth of myomatous nodules [13]. Based on these data, it can be assumed that a low level of SHBG will be genetically correlated with a low content of estradiol [20], which will have a protective value for UF [13], and, conversely, a high level of SHBG, genetically correlated with a high concentration of estradiol, will contribute to an increase in UF risk, which is fully consistent with the results we have obtained. Also, in the work of Ruth et al., negative genetic correlations have been shown between the level of estradiol and the content of both total (−0.25) and free (−0.51) testosterone [20], which is also fully consistent with the assumption we made above (according to the results obtained in this work) about the SHBG-induced protective effect of testosterone on UF risk (which is opposite to the effect of estradiol).
So, the biomedical justification for the established UF-protective role of the SHBG-reducing genetic variant rs3779195 [T/A] BAIAP2L1 (genotype AA,OR = 0.38) may be the pronounced proapoptotic/antiproliferative effects of elevated free testosterone levels recorded at low SHBGcon, and, conversely, the value of the UF risk of the SHBG-enhancing genetic variant rs440837 [A/G] ZBTB10 (genotype GG,OR = 1.93), identified in our work, may be associated with the “weak” proapoptotic/antiproliferative effects of low levels of free testosterone, typical for a high SHBGcon.
Notably, this genetic panel (nine loci linked to SHBG levels) has been previously investigated in our studies of breast cancer [70,71,72] and endometriosis [73,74]. According to the findings from our previous work, the risk of developing breast cancer was influenced by rs10454142 PPP1R21 [71], while the likelihood of developing endometriosis was determined by rs440837 [A/G] ZBTB10 [73]. Based on these data, we can see that rs440837 [A/G] ZBTB10 was associated with both endometriosis (as reported by Ponomareva et al. [73]) and UF (according to this study). It may be a syntropic SNP/gene that influences the development of uterus-benign proliferative conditions, such as UF and endometriosis. This SNP/gene may play a significant role in determining the “common” genetic susceptibility to these conditions, as suggested by previous studies [75,76,77].
This work has a number of limitations, which include the following: (a) we obtained the results of the association of SHBGcon to significant genetic variants with UF and data on the pronounced effects of individual loci (OR = 0.38 for genotype AA rs3779195 [T/A] BAIAP2L1 and OR = 1.93 for genotype GG rs440837 [A/G] ZBTB10) in only one sample, and their replication/verification in another independent cohort is necessary; (b) UF-significant associations of polymorphisms linked with SHBGcon were obtained only in one mono-ethnic cohort of Europeans (Russians from central Russia) and confirmation of these associations in populations of other ethnic groups is needed due to the fact that the potential stratification of the population by ethnic composition can have a significant impact on the results of associative genetic studies; (c) the possible functionality of UF-significant polymorphisms (and strongly linked loci) was predicted by us only in silico, and experimental studies are needed to confirm these assumptions in silico; (d) the causal relationships between the genetic determinants of SHBGcon and UF, which we assumed in this work, need additional confirmation by Mendelian randomization; (e) there is a certain probability of non-detection of UF (for example, small-sized UF, cervical UF, etc.) during ultrasound examination of women in the control group.
It should be noted that with the existing paucity and ambiguity/inconsistency of data on the topic of “SHBG-UF” and “testosterone-UF” correlations, there is an obvious need to continue research in this area in order to accumulate experimental data on the issue of genetic links between SHBG, testosterone, and UF, their further generalization, and the establishment of unambiguous patterns existing between them. Therefore, our work is actually the first experimental genetic study on this topic.
5. Conclusions
SHBGcon-significant genetic determinants were associated with UF risk.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/life15071150/s1, Table S1: The GWAS data about associations of the studied candidate gene polymorphisms with the circulating SHBG and other sex hormone concentrations; Table S2: The regulatory potential of the studied SNPs; Table S3: The allele and genotype frequencies of the studied SNPs in the uterine leiomyoma and control groups; Table S4: Genotype combinations associated with uterine leiomyoma; Table S5: The regulatory effects of the endometriosis-associated loci and SNPs in high LD (r2 ≥ 0.80); Table S6: The eQTL effects of the endometriosis-associated SNPs in various tissues/organs; Table S7: The eQTL values of SNPs in high LD (r2 ≥ 0.80) with the endometriosis-associated polymorphisms; Table S8: The sQTL effects of the endometriosis-associated SNPs in various tissues/organs; Table S9: sQTL values of SNPs in high LD (r2 ≥ 0.80) with the endometriosis-associated polymorphisms; Table S10: Gene set enrichment analysis of biological pathways correlated with 22 TFs and 3 proteins functionally related to UF-associated rs440837 [A/G] ZBTB10 and their proxy 5 SNPs; Table S11: Gene set enrichment analysis of biological pathways correlated with 68 TFs, 18 regulatory proteins, and 15 proteins functionally related to the UF-associated rs3779195 [T/A] BAIAP2L1 and their proxy 20 SNPs.
Author Contributions
Conceptualization, M.P., V.C., and M.C. (Maria Churnosova); data curation, V.N., I.P., and A.P.; formal analysis, M.A., E.R., I.A., V.N., and D.P.; project administration, M.C. (Mikhail Churnosov); writing—original draft, M.P. and M.C. (Maria Churnosova); writing—review and editing, M.C. (Mikhail Churnosov), E.R., I.A., and I.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation, grant number 25-25-00034, https://rscf.ru/en/project/25-25-00034/, accessed on 23 April 2025.
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Local Ethical Committee of the Belgorod State University (25 April 2016, No. 4).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
The data generated in the present study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| UF | Uterine fibroids |
| SHBG | Sex hormone-binding globulin |
| SHBGcon | Sex hormone-binding globulin concentration |
| SNP | Single-nucleotide polymorphism |
| GWAS | Genome-wide association studies |
| BMI | Body mass index |
| DNA | Deoxyribonucleic acid |
| MB-MDR | Model-based multifactor dimensionality reduction |
| MDR | Multifactor dimensionality reduction |
| LD | Linkage disequilibrium |
| TFs | Transcription factors |
References
- Yang, Q.; Ciebiera, M.; Bariani, M.V.; Ali, M.; Elkafas, H.; Boyer, T.G.; Al-Hendy, A. Comprehensive Review of Uterine Fibroids: Developmental Origin, Pathogenesis, and Treatment. Endocr. Rev. 2022, 43, 678–719. [Google Scholar] [CrossRef]
- Stewart, E.A.; Nowak, R.A. Uterine Fibroids: Hiding in Plain Sight. Physiology 2022, 37, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Koltsova, A.S.; Efimova, O.A.; Pendina, A.A. A View on Uterine Leiomyoma Genesis through the Prism of Genetic, Epigenetic and Cellular Heterogeneity. Int. J. Mol. Sci. 2023, 24, 5752. [Google Scholar] [CrossRef] [PubMed]
- Cianci, S.; Gulino, F.A.; Palmara, V.; La Verde, M.; Ronsini, C.; Romeo, P.; Occhipinti, S.; Incognito, G.G.; Capozzi, V.A.; Restaino, S.; et al. Exploring Surgical Strategies for Uterine Fibroid Treatment: A Comprehensive Review of Literature on Open and Minimally Invasive Approaches. Medicina 2023, 60, 64. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Tang, H.; Xie, Z.; Deng, S. Robotic-assisted vs. laparoscopic and abdominal myomectomy for treatment of uterine fibroids: A meta-analysis. Minim. Invasive Ther. Allied Technol. 2018, 27, 249–264. [Google Scholar] [CrossRef]
- Lou, Z.; Huang, Y.; Li, S.; Luo, Z.; Li, C.; Chu, K.; Zhang, T.; Song, P.; Zhou, J. Global, regional, and national time trends in incidence, prevalence, years lived with disability for uterine fibroids, 1990–2019: An age-period-cohort analysis for the global burden of disease 2019 study. BMC Public Health 2023, 23, 916. [Google Scholar] [CrossRef]
- Shih, V.; Banks, E.; Bonine, N.G.; Harrington, A.; Stafkey-Mailey, D.; Yue, B.; Ye, J.M.; Fuldeore, R.M.; Gillard, P. Healthcare resource utilization and costs among women diagnosed with uterine fibroids compared to women without uterine fibroids. Curr. Med. Res. Opin. 2019, 35, 1925–1935. [Google Scholar] [CrossRef]
- Snieder, H.; MacGregor, A.J.; Spector, T.D. Genes control the cessation of a woman’s reproductive life: A twin study of hysterectomy and age at menopause. J. Clin. Endocrinol. Metab. 1998, 83, 1875–1880. [Google Scholar] [CrossRef]
- Luoto, R.; Kaprio, J.; Rutanen, E.M.; Taipale, P.; Perola, M.; Koskenvuo, M. Heritability and risk factors of uterine fibroids--the Finnish Twin Cohort study. Maturitas 2000, 37, 15–26. [Google Scholar] [CrossRef]
- Välimäki, N.; Kuisma, H.; Pasanen, A.; Heikinheimo, O.; Sjöberg, J.; Bützow, R.; Sarvilinna, N.; Heinonen, H.R.; Tolvanen, J.; Bramante, S.; et al. Genetic predisposition to uterine leiomyoma is determined by loci for genitourinary development and genome stability. Elife 2018, 7, e37110. [Google Scholar] [CrossRef]
- Rafnar, T.; Gunnarsson, B.; Stefansson, O.A.; Sulem, P.; Ingason, A.; Frigge, M.L.; Stefansdottir, L.; Sigurdsson, J.K.; Tragante, V.; Steinthorsdottir, V.; et al. Variants associating with uterine leiomyoma highlight genetic background shared by various cancers and hormone-related traits. Nat. Commun. 2018, 9, 3636. [Google Scholar] [CrossRef]
- Lv, M.; Yu, J.; Huang, Y.; Ma, J.; Xiang, J.; Wang, Y.; Li, L.; Zhang, Z.; Liao, H. Androgen Signaling in Uterine Diseases: New Insights and New Targets. Biomolecules 2022, 12, 1624. [Google Scholar] [CrossRef]
- Wong, J.Y.; Gold, E.B.; Johnson, W.O.; Lee, J.S. Circulating Sex Hormones and Risk of Uterine Fibroids: Study of Women’s Health Across the Nation (SWAN). J. Clin. Endocrinol. Metab. 2016, 101, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Goldman, A.L.; Bhasin, S.; Wu, F.C.W.; Krishna, M.; Matsumoto, A.M.; Jasuja, R. A Reappraisal of Testosterone’s Binding in Circulation: Physiological and Clinical Implications. Endocr. Rev. 2017, 38, 302–324. [Google Scholar] [CrossRef] [PubMed]
- Balogh, A.; Karpati, E.; Schneider, A.E.; Hetey, S.; Szilagyi, A.; Juhasz, K.; Laszlo, G.; Hupuczi, P.; Zavodszky, P.; Papp, Z.; et al. Sex hormone-binding globulin provides a novel entry pathway for estradiol and influences subsequent signaling in lymphocytes via membrane receptor. Sci. Rep. 2019, 9, 4. [Google Scholar] [CrossRef]
- Ohlsson, C.; Wallaschofski, H.; Lunetta, K.L.; Stolk, L.; Perry, J.R.; Koster, A.; Petersen, A.K.; Eriksson, J.; Lehtimäki, T.; Huhtaniemi, I.T.; et al. Genetic determinants of serum testosterone concentrations in men. PLoS Genet. 2011, 7, e1002313. [Google Scholar] [CrossRef] [PubMed]
- Prescott, J.; Thompson, D.J.; Kraft, P.; Chanock, S.J.; Audley, T.; Brown, J.; Leyland, J.; Folkerd, E.; Doody, D.; Hankinson, S.E.; et al. Genome-wide association study of circulating estradiol, testosterone, and sex hormone-binding globulin in postmenopausal women. PLoS ONE 2012, 7, e37815. [Google Scholar] [CrossRef]
- Coviello, A.D.; Haring, R.; Wellons, M.; Vaidya, D.; Lehtimäki, T.; Keildson, S.; Lunetta, K.L.; He, C.; Fornage, M.; Lagou, V.; et al. A genome-wide association meta-analysis of circulating sex hormone-binding globulin reveals multiple Loci implicated in sex steroid hormone regulation. PLOS Genet. 2012, 8, e1002805. [Google Scholar] [CrossRef]
- Ruth, K.S.; Campbell, P.J.; Chew, S.; Lim, E.M.; Hadlow, N.; Stuckey, B.G.; Brown, S.J.; Feenstra, B.; Joseph, J.; Surdulescu, G.L.; et al. Genome-wide association study with 1000 genomes imputation identifies signals for nine sex hormone-related phenotypes. Eur. J. Hum. Genet. 2016, 24, 284–290. [Google Scholar] [CrossRef]
- Ruth, K.S.; Day, F.R.; Tyrrell, J.; Thompson, D.J.; Wood, A.R.; Mahajan, A.; Beaumont, R.N.; Wittemans, L.; Martin, S.; Busch, A.S.; et al. Using human genetics to understand the disease impacts of testosterone in men and women. Nat. Med. 2020, 26, 252–258. [Google Scholar] [CrossRef]
- Harrison, S.; Davies, N.M.; Howe, L.D.; Hughes, A. Testosterone and socioeconomic position: Mendelian randomization in 306,248 men and women in UK Biobank. Sci. Adv. 2021, 7, eabf8257. [Google Scholar] [CrossRef]
- Haas, C.B.; Hsu, L.; Lampe, J.W.; Wernli, K.J.; Lindström, S. Cross-ancestry Genome-wide Association Studies of Sex Hormone Concentrations in Pre- and Postmenopausal Women. Endocrinology 2022, 163, bqac020, Erratum in Endocrinology 2022, 164, bqac207. [Google Scholar] [CrossRef]
- Wang, H.; Li, C.; Chen, L.; Zhang, M.; Ren, T.; Zhang, S. Causal relationship between female reproductive factors, sex hormones and uterine leiomyoma: A Mendelian randomization study. Reprod. Biomed. Online 2024, 48, 103584. [Google Scholar] [CrossRef]
- Abramova, M.; Churnosova, M.; Efremova, O.; Aristova, I.; Reshetnikov, E.; Polonikov, A.; Churnosov, M.; Ponomarenko, I. Effects of pre-pregnancy over-weight/obesity on the pattern of association of hypertension susceptibility genes with preeclampsia. Life 2022, 12, 2018. [Google Scholar] [CrossRef] [PubMed]
- Churnosov, M.; Abramova, M.; Reshetnikov, E.; Lyashenko, I.V.; Efremova, O.; Churnosova, M.; Ponomarenko, I. Polymorphisms of hypertension susceptibility genes as a risk factors of preeclampsia in the Caucasian population of central Russia. Placenta 2022, 129, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Sergeeva, K.N.; Sorokina, I.N. Assessment of the relationship between marriage and migration characteristics of the population of the Belgorod region in dynamics over 130 years. Res. Results Biomed. 2024, 10, 374–388. (In Russian) [Google Scholar] [CrossRef]
- Ponomarenko, I.; Reshetnikov, E.; Polonikov, A.; Verzilina, I.; Sorokina, I.; Yermachenko, A.; Dvornyk, V.; Churnosov, M. Candidate genes for age at menarche are associated with uterine leiomyoma. Front. Genet. 2021, 11, 512940. [Google Scholar] [CrossRef]
- Reshetnikov, E.; Ponomarenko, I.; Golovchenko, O.; Sorokina, I.; Batlutskaya, I.; Yakunchenko, T.; Dvornyk, V.; Polonikov, A.; Churnosov, M. The VNTR polymorphism of the endothelial nitric oxide synthase gene and blood pressure in women at the end of pregnancy. Taiwan. J. Obstet. Gynecol. 2019, 58, 390–395. [Google Scholar] [CrossRef]
- Novakov, V.; Novakova, O.; Churnosova, M.; Sorokina, I.; Aristova, I.; Polonikov, A.; Reshetnikov, E.; Churnosov, M. Intergenic Interactions of SBNO1, NFAT5 and GLT8D1 Determine the Susceptibility to Knee Osteoarthritis among Europeans of Russia. Life 2023, 13, 405. [Google Scholar] [CrossRef]
- Minyaylo, O.; Ponomarenko, I.; Reshetnikov, E.; Dvornyk, V.; Churnosov, M. Functionally significant polymorphisms of the MMP-9 gene are associated with peptic ulcer disease in the Caucasian population of Central Russia. Sci. Rep. 2021, 11, 13515. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef]
- Eliseeva, N.; Ponomarenko, I.; Reshetnikov, E.; Dvornyk, V.; Churnosov, M. LOXL1 gene polymorphism candidates for exfoliation glaucoma are also associated with a risk for primary open-angle glaucoma in a Caucasian population from central Russia. Mol. Vis. 2021, 27, 262–269. [Google Scholar]
- Pavlova, N.; Demin, S.; Churnosov, M.; Reshetnikov, E.; Aristova, I.; Churnosova, M.; Ponomarenko, I. The Modifying Effect of Obesity on the Association of Matrix Metalloproteinase Gene Polymorphisms with Breast Cancer Risk. Biomedicines 2022, 10, 2617. [Google Scholar] [CrossRef]
- Polonikov, A.; Kharchenko, A.; Bykanova, M.; Sirotina, S.; Ponomarenko, I.; Bocharova, A.; Vagaytseva, K.; Stepanov, V.; Bushueva, O.; Churnosov, M.; et al. Polymorphisms of CYP2C8, CYP2C9 and CYP2C19 and risk of coronary heart disease in Russian population. Gene 2017, 627, 451–459. [Google Scholar] [CrossRef]
- Polonikov, A.; Rymarova, L.; Klyosova, E.; Volkova, A.; Azarova, I.; Bushueva, O.; Bykanova, M.; Bocharova, I.; Zhabin, S.; Churnosov, M.; et al. Matrix metalloproteinases as target genes for gene regulatory networks driving molecular and cellular pathways related to a multistep pathogenesis of cerebrovascular disease. J. Cell. Biochem. 2019, 120, 16467–16482. [Google Scholar] [CrossRef]
- Golovchenko, I.; Aizikovich, B.; Golovchenko, O.; Reshetnikov, E.; Churnosova, M.; Aristova, I.; Ponomarenko, I.; Churnosov, M. Sex Hormone Candidate Gene Polymorphisms Are Associated with Endometriosis. Int. J. Mol. Sci. 2022, 23, 13691. [Google Scholar] [CrossRef]
- Polonikov, A.; Bykanova, M.; Ponomarenko, I.; Sirotina, S.; Bocharova, A.; Vagaytseva, K.; Stepanov, V.; Churnosov, M.; Bushueva, O.; Solodilova, M.; et al. The contribution of CYP2C gene subfamily involved in epoxygenase pathway of arachidonic acids metabolism to hypertension susceptibility in Russian population. Clin. Exp. Hypertens. 2017, 39, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, T.; Churnosova, M.; Abramova, M.; Ponomarenko, I.; Reshetnikov, E.; Aristova, I.; Sorokina, I.; Churnosov, M. Risk Effects of rs1799945 Polymorphism of the HFE Gene and Intergenic Interactions of GWAS-Significant Loci for Arterial Hypertension in the Caucasian Population of Central Russia. Int. J. Mol. Sci. 2023, 24, 8309. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Che, R.; Jack, J.R.; Motsinger-Reif, A.A.; Brown, C.C. An adaptive permutation approach for genome-wide association study: Evaluation and recommendations for use. BioData Min. 2014, 7, 9. [Google Scholar] [CrossRef]
- Gauderman, W.; Morrison, J. QUANTO 1.1: A Computer Program for Power and Sample Size Calculations Genetic–Epidemiology Studies. 2006. Available online: http://hydra.usc.edu/gxe (accessed on 18 November 2024).
- Ivanova, T.; Churnosova, M.; Abramova, M.; Plotnikov, D.; Ponomarenko, I.; Reshetnikov, E.; Aristova, I.; Sorokina, I.; Churnosov, M. Sex-Specific Features of the Correlation between GWAS-Noticeable Polymorphisms and Hypertension in Europeans of Russia. Int. J. Mol. Sci. 2023, 24, 7799. [Google Scholar] [CrossRef]
- Calle, M.L.; Urrea, V.; Malats, N.; Van Steen, K. Mbmdr: An R package for exploring gene-gene interactions associated with binary or quantitative traits. Bioinformatics 2010, 26, 2198–2199. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lou, X.Y.; Chen, G.B.; Yan, L.; Ma, J.Z.; Zhu, J.; Elston, R.C.; Li, M.D. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am. J. Hum. Genet. 2007, 80, 1125–1137. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.B.; Xu, Y.; Xu, H.M.; Li, M.D.; Zhu, J.; Lou, X.Y. Practical and theoretical considerations in study design for detecting gene-gene interactions using MDR and GMDR approaches. PLoS ONE 2011, 6, e16981. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.H.; Gilbert, J.C.; Tsai, C.T.; Chiang, F.T.; Holden, T.; Barney, N.; White, B.C. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J. Theor. Biol. 2006, 241, 252–261. [Google Scholar] [CrossRef]
- Ponomarenko, I.V. Using the method of Multifactor Dimensionality Reduction (MDR) and its modifications for analysis of gene-gene and gene-environment interactions in genetic-epidemiological studies (review). Res. Results Biomed. 2019, 5, 4–21. [Google Scholar] [CrossRef]
- Pavlova, N.; Demin, S.; Churnosov, M.; Reshetnikov, E.; Aristova, I.; Churnosova, M.; Ponomarenko, I. Matrix Metalloproteinase Gene Polymorphisms Are Associated with Breast Cancer in the Caucasian Women of Russia. Int. J. Mol. Sci. 2022, 23, 12638. [Google Scholar] [CrossRef]
- Reshetnikova, Y.; Churnosova, M.; Stepanov, V.; Bocharova, A.; Serebrova, V.; Trifonova, E.; Ponomarenko, I.; Sorokina, I.; Efremova, O.; Orlova, V.; et al. Maternal Age at Menarche Gene Polymorphisms Are Associated with Offspring Birth Weight. Life 2023, 13, 1525. [Google Scholar] [CrossRef]
- Novakov, V.; Novakova, O.; Churnosova, M.; Aristova, I.; Ponomarenko, M.; Reshetnikova, Y.; Churnosov, V.; Sorokina, I.; Ponomarenko, I.; Efremova, O.; et al. Polymorphism rs143384 GDF5 reduces the risk of knee osteoarthritis development in obese individuals and increases the disease risk in non-obese population. Arthroplasty 2024, 6, 12. [Google Scholar] [CrossRef]
- Reshetnikov, E.; Churnosova, M.; Reshetnikova, Y.; Stepanov, V.; Bocharova, A.; Serebrova, V.; Trifonova, E.; Ponomarenko, I.; Sorokina, I.; Efremova, O.; et al. Maternal Age at Menarche Genes Determines Fetal Growth Restriction Risk. Int. J. Mol. Sci. 2024, 25, 2647. [Google Scholar] [CrossRef]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Hammond, G.L. Plasma steroid-binding proteins: Primary gatekeepers of steroid hormone action. J. Endocrinol. 2016, 230, R13–R25. [Google Scholar] [CrossRef] [PubMed]
- Chao, A.; Tsai, C.L.; Jung, S.M.; Chuang, W.C.; Kao, C.; Hsu, A.; Chen, S.H.; Lin, C.Y.; Lee, Y.C.; Lee, Y.S.; et al. BAI1-Associated Protein 2-Like 1 (BAIAP2L1) Is a Potential Biomarker in Ovarian Cancer. PLoS ONE 2015, 10, e0133081. [Google Scholar] [CrossRef]
- Song, Y.; Zhuang, G.; Li, J.; Zhang, M. BAIAP2L2 facilitates the malignancy of prostate cancer (PCa) via VEGF and apoptosis signaling pathways. Genes Genom. 2021, 43, 421–432. [Google Scholar] [CrossRef]
- Lu, Y.; Zhou, X.Y.; Zhou, C.L.; Liu, J.; Yong, T.; Fan, Y.; Wang, C. Insulin receptor tyrosine kinase substrate (IRTKS) promotes the tumorigenesis of pancreatic cancer via PI3K/AKT signaling. Hum. Cell 2022, 35, 1885–1899. [Google Scholar] [CrossRef]
- Zhao, C.; Shang, A.; Wu, H.; Li, Q.; Peng, L.; Yue, C. Causal relationship between genetically predicted uterine leiomyoma and cancer risk: A two-sample Mendelian randomization. Front. Endocrinol. 2024, 15, 1429165. [Google Scholar] [CrossRef]
- Giordano, C.; Accattatis, F.M.; Gelsomino, L.; Del Console, P.; Győrffy, B.; Giuliano, M.; Veneziani, B.M.; Arpino, G.; De Angelis, C.; De Placido, P.; et al. miRNAs in the Box: Potential Diagnostic Role for Extracellular Vesicle-Packaged miRNA-27a and miRNA-128 in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 15695. [Google Scholar] [CrossRef]
- Sinnott-Armstrong, N.; Tanigawa, Y.; Amar, D.; Mars, N.; Benner, C.; Aguirre, M.; Venkataraman, G.R.; Wainberg, M.; Ollila, H.M.; Kiiskinen, T.; et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 2021, 53, 185–194. [Google Scholar] [CrossRef]
- Misao, R.; Nakanishi, Y.; Fujimoto, J.; Tamaya, T. Expression of sex hormone-binding globulin mRNA in uterine leiomyoma, myometrium and endometrium of human subjects. Gynecol. Endocrinol. 1995, 9, 317–323, Erratum in Nat. Genet. 2021, 53, 1622. [Google Scholar] [CrossRef]
- Garitazelaia, A.; Rueda-Martínez, A.; Arauzo, R.; de Miguel, J.; Cilleros-Portet, A.; Marí, S.; Bilbao, J.R.; Fernandez-Jimenez, N.; García-Santisteban, I.A. Systematic Two-Sample Mendelian Randomization Analysis Identifies Shared Genetic Origin of Endometriosis and Associated Phenotypes. Life 2021, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Dinsdale, N.L.; Crespi, B.J. Endometriosis and polycystic ovary syndrome are diametric disorders. Evol. Appl. 2021, 14, 1693–1715. [Google Scholar] [CrossRef]
- Misao, R.; Hori, M.; Ichigo, S.; Fujimoto, J.; Tamaya, T. Levels of sex hormone-binding globulin (SHBG) and corticosteroid-binding globulin (CBG) messenger ribonucleic acid (mRNAs) in ovarian endometriosis. Reprod. Nutr. Dev. 1995, 35, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Chen, L.; Guo, S.; Liu, Y.; Wu, H. Genetic liability to multiple factors and uterine leiomyoma risk: A Mendelian randomization study. Front. Endocrinol. 2023, 14, 1133260. [Google Scholar] [CrossRef] [PubMed]
- Sinnott-Armstrong, N.; Naqvi, S.; Rivas, M.; Pritchard, J.K. GWAS of three molecular traits highlights core genes and pathways alongside a highly polygenic background. Elife 2021, 10, e58615. [Google Scholar] [CrossRef]
- Whitton, K.; Baber, R. Androgen-based therapies in women. Best Pract. Res. Clin. Endocrinol. Metab. 2024, 38, 101783. [Google Scholar] [CrossRef]
- Løssl, K.; Freiesleben, N.C.; Wissing, M.L.; Birch Petersen, K.; Holt, M.D.; Mamsen, L.S.; Anderson, R.A.; Andersen, C.Y. Biological and Clinical Rationale for Androgen Priming in Ovarian Stimulation. Front. Endocrinol. 2020, 11, 627. [Google Scholar] [CrossRef]
- Lissaman, A.C.; Girling, J.E.; Cree, L.M.; Campbell, R.E.; Ponnampalam, A.P. Androgen signalling in the ovaries and endometrium. Mol. Hum. Reprod. 2023, 29, gaad017. [Google Scholar] [CrossRef]
- Pasenov, K.N. Features of associations of SHBG-related genes with breast cancer in women, depending on the presence of hereditary burden and mutations in the BRCA1/CHEK2 genes. Res. Results Biomed. 2024, 10, 69–88. (In Russian) [Google Scholar] [CrossRef]
- Ponomarenko, I.; Pasenov, K.; Churnosova, M.; Sorokina, I.; Aristova, I.; Churnosov, V.; Ponomarenko, M.; Reshetnikov, E.; Churnosov, M. Sex-Hormone-Binding Globulin Gene Polymorphisms and Breast Cancer Risk in Caucasian Women of Russia. Int. J. Mol. Sci. 2024, 25, 2182. [Google Scholar] [CrossRef]
- Ponomarenko, I.; Pasenov, K.; Churnosova, M.; Sorokina, I.; Aristova, I.; Churnosov, V.; Ponomarenko, M.; Reshetnikova, Y.; Reshetnikov, E.; Churnosov, M. Obesity-Dependent Association of the rs10454142 PPP1R21 with Breast Cancer. Biomedicines 2024, 12, 818. [Google Scholar] [CrossRef]
- Ponomareva, T.; Altukhova, O.; Churnosova, M.; Aristova, I.; Reshetnikov, E.; Churnosov, M.; Ponomarenko, I. Gene polymorphisms determining sex hormone-binding globulin levels and endometriosis risk. Int. J. Mol. Sci. 2025, 23, 13691. [Google Scholar]
- Ponomareva, T.A. Genetic variants of sex hormone-binding globulin and hormonal profile in patients with genital endometriosis. Res. Results Biomed. 2025, 11, 75–90. (In Russian) [Google Scholar] [CrossRef]
- Gallagher, C.S.; Mäkinen, N.; Harris, H.R.; Rahmioglu, N.; Uimari, O.; Cook, J.P.; Shigesi, N.; Ferreira, T.; Velez-Edwards, D.R.; Edwards, T.L.; et al. Genome-wide association and epidemiological analyses reveal common genetic origins between uterine leiomyomata and endometriosis. Nat. Commun. 2019, 10, 4857, Erratum in Nat. Commun. 2022, 13, 5543. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, M.S.; Reshetnikov, E.A.; Churnosova, M.M.; Reshetnikova, Y.N.; Churnosov, V.I.; Ponomarenko, I.V. Comorbidity and syntropy of benign proliferative diseases of the female reproductive system: Non-genetic, genetic, and epigenetic factors (review). Res. Results Biomed. 2023, 9, 544–556. [Google Scholar] [CrossRef]
- McGrath, I.M.; Montgomery, G.W.; Mortlock, S. Insights from Mendelian randomization and genetic correlation analyses into the relationship between endometriosis and its comorbidities. Hum. Reprod. Update 2023, 29, 655–674. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).