Susceptibility of Lactobacillaceae Strains to Aminoglycoside Antibiotics in the Light of EFSA Guidelines
Abstract
1. Introduction
2. Materials and Methods
2.1. Purpose and Scope of the Research
2.2. Strains and WGSs Used in the Study
2.3. Antimicrobial Susceptibility Testing
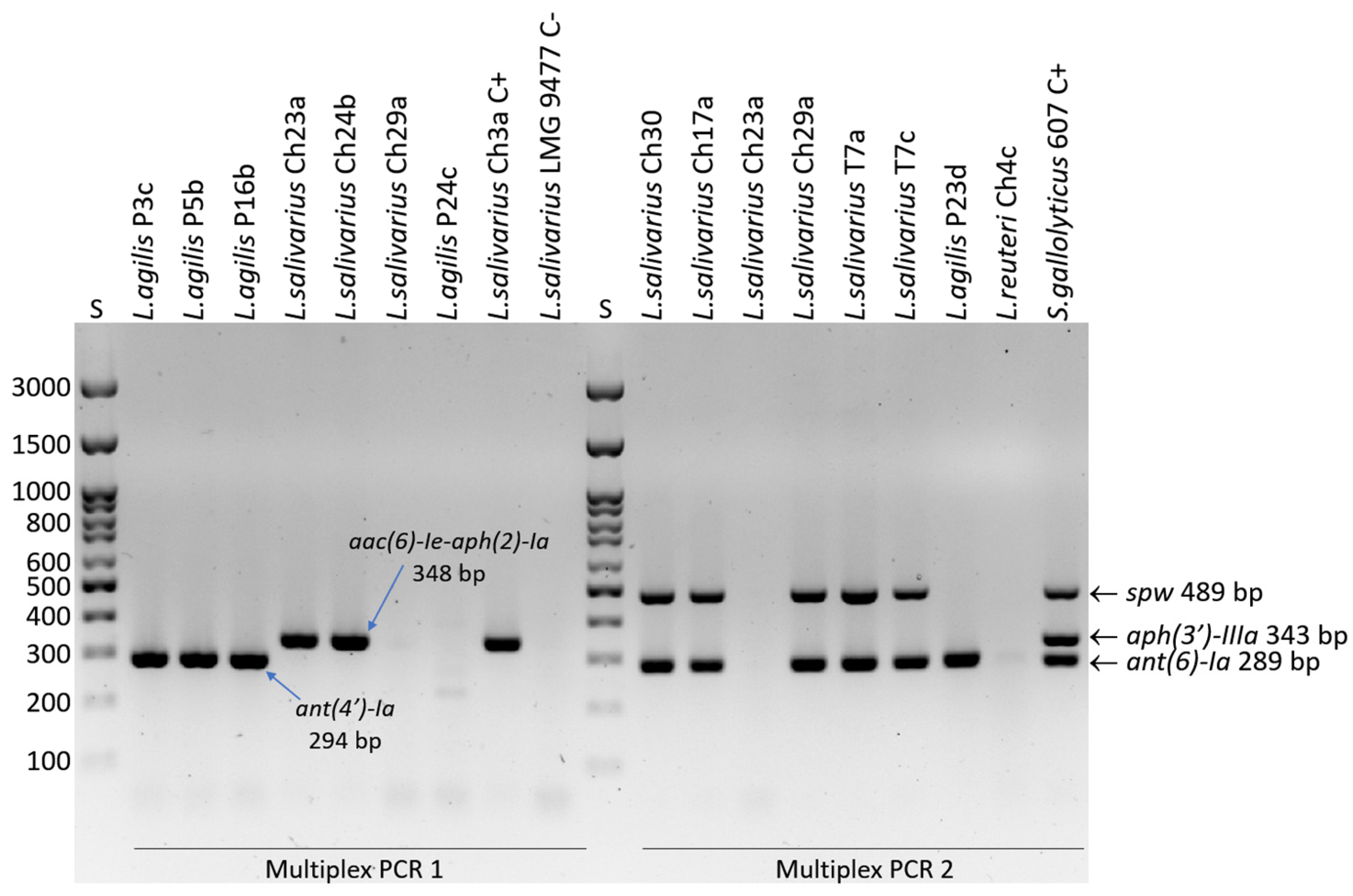
2.4. Detection of Resistance Genes
2.5. Analysis of Mutations in the rpsL Gene
3. Results
3.1. Results of AST and Resistance Gene Detection
3.2. Mutations in the rpsL Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AST | Antimicrobial susceptibility testing |
| CLSI | Clinical and Laboratory Standards Institute |
| EFSA | European Food Safety Authority |
| FEEDAP | EFSA Panel on Additives and Products or Substances used in Animal Feed |
| IDF | International Dairy Federation |
| ISO | International Organization for Standardization |
| LAB | Lactic acid bacteria |
| LSM | LAB susceptibility test medium |
| MRS | De Man-Rogosa-Sharpe |
| WGS | Whole genome sequence |
Appendix A
| Strain | Metabolic Group | Phylogenetic Group | GenBank Acc. No. | Susceptibility to STR | MIC Value | Mutations in rpsL Gene | Aminoglycoside RGs |
|---|---|---|---|---|---|---|---|
| L. gallinarum LMG 9435 | OHO | L. delbrueckii | GCA_001434975.1 | R | >1024 | Lys56 → Arg | No |
| L. gallinarum An153 | OHO | L. delbrueckii | GCA_002160635.1 | ND | ND | No | |
| L. gallinarum An101 | OHO | L. delbrueckii | GCA_002161165.1 | ND | ND | No | |
| L. gallinarum J07 | OHO | L. delbrueckii | GCA_947381795.1 | ND | ND | No | |
| L. gallinarum Chicken_20_mag_156 | OHO | L. delbrueckii | GCA_904419645.1 | ND | ND | No | |
| L. acidophilus ATCC 4356 | OHO | L. delbrueckii | GCA_034298135.1 | S | 4–16 | No | |
| L. amylovorus LMG 9496 | OHO | L. delbrueckii | GCA_002706375.1 | S/R | 8–32 | Lys101 → Thr | No |
| L. crispatus LMG 9479 | OHO | L. delbrueckii | GCA_018987235.1 | S/R | 16–32 | No | |
| L. crisptus T31e | OHO | L. delbrueckii | GCA_047782765.1 | S | 8–16 | No | |
| L. amylolyticus DSM 11664 | OHO | L. delbrueckii | GCA_004354545.1 | S [14] | UN | No | |
| L. gigeriorum DSM 23908 | OHO | L. delbrueckii | GCA_001436575.1 | R [14] | UN | Lys56 → Arg | No |
| L. delbrueckii subsp. jakobsenii DSM 26046 | OHO | L. delbrueckii | GCA_001888925.1 | R [14] | UN | No | |
| L. delbruckii subsp. delbrueckii DSM 20074 | OHO | L. delbrueckii | GCA_001908495.1 | S [14] | UN | No | |
| L. helveticus LMG 22464 | OHO | L. delbrueckii | GCA_001437535.1 | S [14] | UN | No | |
| L. taiwanensis DSM 21401 | OHO | L. delbrueckii | GCA_001436695.1 | S [14] | UN | No | |
| L. pasteurii DSM 23907 | FHE | L. delbrueckii | GCA_004354755.1 | S [14] | UN | No | |
| L. apis LMG 26964 | FHE | L. delbrueckii | GCA_002837055.1 | R [14] | UN | No |
References
- Sun, Z.; Yu, J.; Dan, T.; Zhang, W.; Zhang, H. Phylogenesis and evolution of lactic acid bacteria. In Lactic Acid Bacteria; Zhang, H., Cai, Y., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 1–101. [Google Scholar] [CrossRef]
- Gilroy, R.; Ravi, A.; Getino, M.; Pursley, I.; Horton, D.L.; Alikhan, N.F.; Baker, D.; Gharbi, K.; Hall, N.; Watson, M.; et al. Extensive microbial diversity within the chicken gut microbiome revealed by metagenomics and culture. PeerJ 2021, 9, e10941. [Google Scholar] [CrossRef]
- NCBI Database. Available online: https://www.ncbi.nlm.nih.gov/taxonomy (accessed on 27 December 2024).
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Bhogoju, S.; Nahashon, S. Recent Advances in Probiotic Application in Animal Health and Nutrition: A Review. Agriculture 2022, 12, 304. [Google Scholar] [CrossRef]
- EFSA. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, 5206. [Google Scholar]
- Klare, I.; Konstabel, C.; Müller-Bertling, S.; Reissbrodt, R.; Huys, G.; Vancanneyt, M.; Swings, J.; Goossens, H.; Witte, W. Evaluation of new broth media for microdilution antibiotic susceptibility testing of Lactobacilli, Pediococci, Lactococci, and Bifidobacteria. Appl. Environ. Microbiol. 2005, 71, 8982–8986. [Google Scholar] [CrossRef]
- Egervärn, M.; Lindmark, H.; Roos, S.; Huys, G.; Lindgren, S. Effects of inoculum size and incubation time on broth microdilution susceptibility testing of lactic acid bacteria. Antimicrob. Agents Chemother. 2007, 5, 394–396. [Google Scholar] [CrossRef] [PubMed]
- Mayrhofer, S.; Zitz, U.; Birru, F.H.; Gollan, D.; Gołoś, A.K.; Kneifel, W.; Domig, K.J. Comparison of the CLSI guideline and ISO/IDF standard for antimicrobial susceptibility testing of Lactobacilli. Microb. Drug Resist. 2014, 20, 591–603. [Google Scholar] [CrossRef]
- Łepecka, A.; Szymański, P.; Rutkowska, S.; Iwanowska, K.; Kołożyn-Krajewska, D. The Influence of Environmental Conditions on the Antagonistic Activity of Lactic Acid Bacteria Isolated from Fermented Meat Products. Foods 2021, 10, 2267. [Google Scholar] [CrossRef]
- Tscherne, A.; Mantel, E.; Boskani, T.; Budniak, S.; Elschner, M.; Fasanella, A.; Feruglio, S.L.; Galante, D.; Giske, C.G.; Grunow, R.; et al. Adaptation of Brucella melitensis Antimicrobial Susceptibility Testing to the ISO 20776 Standard and Validation of the Method. Microorganisms 2022, 10, 1470. [Google Scholar] [CrossRef]
- Sabath, L.D. Chemical and physical factors influencing methicillin resistance of Staphylococcus aureus and Staphylococcus epidermidis. J. Antimicrob. Chemother. 1977, 3 (Suppl. C), 47–51. [Google Scholar] [CrossRef]
- Mendonça, A.A.; de Morais, M.A.; Cabrera, M.Z., Jr. Cysteine induces resistance of lactobacilli to erythromycin and azithromycin. Int. J. Antimicrob. Agents. 2019, 53, 352–353. [Google Scholar] [CrossRef] [PubMed]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-Wide Assessment of Antibiotic Resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2018, 85, e01738-18. [Google Scholar] [CrossRef]
- Nøhr-Meldgaard, K.; Struve, C.; Ingmer, H.; Koza, A.; Al-Nakeeb, K.; Agersø, Y. Antimicrobial susceptibility testing and tentative epidemiological cut-off values for Lactobacillaceae family species intended for ingestion. Front. Antibiot. 2023, 2, 1162636. [Google Scholar] [CrossRef] [PubMed]
- Stefańska, I.; Kwiecień, E.; Jóźwiak-Piasecka, K.; Garbowska, M.; Binek, M.; Rzewuska, M. Antimicrobial Susceptibility of Lactic Acid Bacteria Strains of Potential Use as Feed Additives—The Basic Safety and Usefulness Criterion. Front. Vet. Sci. 2021, 8, 687071. [Google Scholar] [CrossRef]
- Mayrhofer, S.; van Hoek, A.H.; Mair, C.; Huys, G.; Aarts, H.J.; Kneifel, W.; Domig, K.J. Antibiotic susceptibility of members of the Lactobacillus acidophilus group using broth microdilution and molecular identification of their resistance determinants. Int. J. Food Microbiol. 2010, 144, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Dec, M.; Stępień-Pyśniak, D.; Nowaczek, A.; Puchalski, A.; Urban-Chmiel, R. Phenotypic and genotypic antimicrobial resistance profiles of fecal lactobacilli from domesticated pigeons in Poland. Anaerobe. 2020, 65, 102251. [Google Scholar] [CrossRef]
- Dec, M.; Nowaczek, A.; Stępień-Pyśniak, D.; Wawrzykowski, J.; Urban-Chmiel, R. Identification and antibiotic susceptibility of lactobacilli isolated from turkeys. BMC Microbiol. 2018, 18, 168. [Google Scholar] [CrossRef]
- Dec, M.; Urban-Chmiel, R.; Stępień-Pyśniak, D.; Wernicki, A. Assessment of antibiotic susceptibility in Lactobacillus isolates from chickens. Gut Pathog. 2017, 9, 54. [Google Scholar] [CrossRef]
- Dec, M.; Urban-Chmiel, R.; Gnat, S.; Puchalski, A.; Wernicki, A. Identification of Lactobacillus strains of goose origin using MALDI-TOF mass spectrometry and 16S-23S rDNA intergenic spacer PCR analysis. Res. Microbiol. 2014, 165, 190–201. [Google Scholar] [CrossRef]
- Dec, M.; Puchalski, A.; Urban-Chmiel, R.; Wernicki, A. 16S-ARDRA and MALDI-TOF mass spectrometry as tools for identification of Lactobacillus bacteria isolated from poultry. BMC Microbiol. 2016, 16, 105. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Huang, Y.T.; Liu, P.Y.; Shih, P.W. Homopolish: A method for the removal of systematic errors in nanopore sequencing by homologous polishing. Genome Biol. 2021, 22, 95. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- ISO 10932:2010 (IDF 223:2010); Milk and Milk Products—Determination of the Minimal Inhibitory Concentration (Mic) of Antibiotics Applicable to Bifidobacteria and Non-Enterococcal Lactic Acid Bacteria (LAB). International Organization for Standardization: Geneva, Switzerland, 2010. Available online: https://www.iso.org/standard/46434.html (accessed on 13 January 2025).
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Abricate, Github. Available online: https://github.com/tseemann/abricate (accessed on 15 January 2025).
- Vakulenko, S.B.; Donabedian, S.M.; Voskresenskiy, A.M.; Zervos, M.J.; Lerner, S.A.; Chow, J.W. Multiplex PCR for detection of aminoglycoside resistance genes in enterococci. Antimicrob. Agents Chemother. 2003, 47, 1423–1426. [Google Scholar] [CrossRef] [PubMed]
- Dec, M.; Nowak, T.; Webster, J.; Wódz, K. Serotypes, Antimicrobial Susceptibility, and Potential Mechanisms of Resistance Gene Transfer in Erysipelothrix rhusiopathiae Strains from Waterfowl in Poland. Int. J. Mol. Sci. 2024, 25, 12192. [Google Scholar] [CrossRef]
- Danielsen, M.; Wind, A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 2003, 82, 1–11. [Google Scholar] [CrossRef]
- Hummel, A.S.; Hertel, C.; Holzapfel, W.H.; Franz, C.M. Antibiotic resistances of starter and probiotic strains of lactic acid bacteria. Appl. Environ. Microbiol. 2007, 73, 730–739. [Google Scholar] [CrossRef]
- CLSI Document M100-Ed34; Performance Standards for Antimicrobial Susceptibility Testing CLSI M100 Includes Updated Tables for the Clinical and Laboratory Standards Institute Antimicrobial Susceptibility Testing Standards CLSI M02, M07, and M11. A CLSI Supplement for Global Application. Clinical Lab Standards Institute: Malvern, PA, USA, 2024; p. 318.
- Zhang, F.; Gao, J.; Wang, B.; Huo, D.; Wang, Z.; Zhang, J.; Shao, Y. Whole-genome sequencing reveals the mechanisms for evolution of streptomycin resistance in Lactobacillus plantarum. J. Dairy Sci. 2018, 101, 2867–2874. [Google Scholar] [CrossRef]
- Namai, F.; Nishiyama, K.; Kitazawa, H.; Shimosato, T. Introduction of Spontaneous Mutations Using Streptomycin as a Method for Lactic Acid Bacteria Breeding. Methods Mol. Biol. 2024, 2851, 15–21. [Google Scholar] [CrossRef]
- Sirichoat, A.; Flórez, A.B.; Vázquez, L.; Buppasiri, P.; Panya, M.; Lulitanond, V.; Mayo, B. Antibiotic Susceptibility Profiles of Lactic Acid Bacteria from the Human Vagina and Genetic Basis of Acquired Resistances. Int. J. Mol. Sci. 2020, 21, 2594. [Google Scholar] [CrossRef] [PubMed]
- Dai, R.; He, J.; Zha, X.; Wang, Y.; Zhang, X.; Gao, H.; Yang, X.; Li, J.; Xin, Y.; Wang, Y.; et al. A novel mechanism of streptomycin resistance in Yersinia pestis: Mutation in the rpsL gene. PLoS Negl. Trop. Dis. 2021, 15, e0009324. [Google Scholar] [CrossRef]
- Barnard, A.M.L.; Simpson, N.J.L.; Lilley, K.S.; Salmond, G.P.C. Mutations in rpsL that confer streptomycin resistance show pleiotropic effects on virulence and the production of a carbapenem antibiotic in Erwinia carotovora. Microbiology 2010, 156, 1030–1039. [Google Scholar] [CrossRef]
- Pelchovich, G.; Schreiber, R.; Zhuravlev, A.; Gophna, U. The contribution of common rpsL mutations in Escherichia coli to sensitivity to ribosome targeting antibiotics. Int. J. Med. Microbiol. 2013, 303, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, A.D.; Etemad, N.; Hashemzadeh, M.; Khandan Dezfuli, S.; Goodarzi, H. Frequency of rrs and rpsL mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from Iranian patients. J. Glob. Antimicrob. Resist. 2017, 9, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Vera-Peña, M.Y.; Rodriguez Rodriguez, W.L. Effect of pH on the growth of three lactic acid bacteria strains isolated from sour cream. Univ. Sci. 2020, 25, 341–358. [Google Scholar] [CrossRef]
- Giraud, E.; Lelong, B.; Raimbault, M. Influence of pH and initial lactate concentration on the growth of Lactobacillus plantarum. Appl. Microbiol. Biotechnol. 1991, 36, 96–99. [Google Scholar] [CrossRef]
- Kerek, Á.; Ecsedi, B.G.; Szabó, Á.; Szimrók, Z.; Paliczné Kustán, B.; Jerzsele, Á.; Nagy, G. Stability Studies of the Dilution Series of Different Antibiotic Stock Solutions in Culture Medium Incubated at 37 °C. Antibiotics 2024, 13, 549. [Google Scholar] [CrossRef]

| Reference Strain | Other Culture Collection Numbers | WGS [GenBank Acc. No.] |
|---|---|---|
| Ligilactobacilluis salivarius LMG 9476 | DSM 20554; ATCC 11742 | GCA_002079585.1 |
| Ligilactobacillus salivarius LMG 9477 | DSM 20555; ATCC 11741; JCM 1231 | GCA_001435955.1 |
| Ligilactobacillus agilis LMG 9186 | DSM 20509; LMG 9186 | GCA_001436215.1 |
| Lactiplantibacillus plantarum ATCC 8014 | DSM 20205; JCM 1057; LMG 1284 | GCA_002631775.1 |
| Lacticaseibacillus rhamnosus ATCC 7469 | DSM 20021; JCM 1136 | GCA_001435405.1 |
| Lacticaseibacillus casei ATCC 393 | DSM 20011; JCM 1134 | GCA_000829055.1 |
| Lacticaseibacillus zeae LMG 17315 | DSM 20178; JCM 11302; ATCC 15820 | GCA_001433745.1 |
| Limosilactobacillus reuteri subsp. reuteri LMG 9213 | ATCC 23272; JCM 1112; DSM 20016 | GCA_000010005.1 |
| Limosilactobacillus reuteri LMG 18238 | ATCC 55148 | ERR3330657 |
| Limosilactobacillus ingluviei LMG 20380 | JCM 12531; DSM 15946; CCUG 45722 | GCA_001435775.1 |
| Limosilactibacillus ingluviei LMG 22056 | DSM 14792; JCM 11425 | GCA_001437235.1 |
| Limosilactibacillus oris LMG 9848 | DSM 4864; ATCC 49062 | GCA_001434465.1 |
| Lactobacillus paragasseri LMG 13134 | LMG 11444; JCM 5344; ATCC 9857 | GCA_003307295.1 |
| Lactobacillus acidophilus ATCC 4356 | DSM 20079; JCM 1132; NCIB 8690 | GCA_034298135.1 |
| Lactobacillus gallinarum LMG 9435 | JCM 2011; ATCC 33199; DSM 10532 | GCA_001434975.1 |
| Lactobacillus kitasatonis LMG 23133 | LMG 22685; JCM 1039, DSM 16761 | GCA_000615285.1 |
| Lactobacillus amylovorus LMG 9496 | JCM 1126; ATCC 33620; DSM 20531 | GCA_002706375.1 |
| Lactobacillus johnsonii LMG 9436 | ATCC 33200; JCM 2012; DSM 10533 | GCA_001433975.1 |
| Lactobacillus crispatus LMG 9479 | ATCC 33820; DSM 20584; JCM 1185 | GCA_018987235.1 |
| Wild-type Strains | ||
| Ligilactobacillus salivarius (n = 44) Ligilactobacillus agilis (n = 20) Lactiplantibacillus plantarum (n = 17) Limosilactobacillus ingluviei (n = 20) Limosilactobacillus reuteri (n = 16) Lactobacillus crispatus (n = 4) |
| Antibiotic→ | Kanamycin | Streptomycin | Spectinomycin | Gentamicin | Neomycin | Resistance Genes Detected | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| EFSA Cut-Offs→ | R > 64 µg/mL | R > 64 µg/mL or R > 32 µg/mL A or R > 16 µg/mL B | NA | R > 16 µg/mL or R > 8 µg/mL C or R > 32 µg/mL D | NA | |||||
| Strain | 24 h | 48 h | 24 h | 48 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| L. salivarius LMG 9476 | 64–128 | 128–512 | 32 | 32–64 | 32 | 4–8 | 8 | 8–16 | 8–16 | |
| L. salivarius LMG 9477 | 64–128 | 128–256 | 32 | 32–64 | 64 | 4–8 | 4–8 | 8–16 | 8–16 | |
| L. agilis LMG 9186 | 512 | 512 | 128 | 128–256 | 256 | 16–32 | 16–32 | 16–32 | 32–64 | |
| L. reuteri subsp. reuteri LMG 9213 | 16–32 | 16–32 | 8–16 | 8–32 | 128 | 0.5–1 | 0.5–2 | 1–4 | 2–4 | |
| L. reuteri LMG 18238 | 64–256 | 128–256 | 16–64 | 32–64 | 128 | 4 | 4–8 | 16 | 16 | |
| L. oris LMG 9848 | 16–32 | 32–64 | 8–16 | 16–64 | 128 | 0.5–1 | 1 | 2–4 | 2–4 | |
| L. ingluviei LMG 20380 | 32 | 64 | 64–128 | 64–128 | 256 | 2–4 | 2–4 | 8 | 8 | tetW, tetM, tetL, ermB |
| L. ingluviei LMG 22056 | 64–128 | 128–256 | 32 | 32–64 | 256 | 2 | 4–8 | 16 | 16 | tetW, lnuC |
| L. plantarum ATCC 8014 | 4–8 | 16 | 4–8 | 4–16 | 128 | 0.25 | 0.25 | 1–2 | 1–4 | |
| L. rhamnosus ATCC 7469 | 16 | 32–64 | 4 | 8–16 | 64 | 1–2 | 2–4 | 2–8 | 4–16 | |
| L. casei ATCC 393 | 16 | 32–64 | 8 | 16 | 32 | 1–2 | 2 | 4 | 8 | |
| L. zeae LMG 17315 | 32–64 | 64–128 | 16–32 | 32 | 64 | 4 | 4 | 8–16 | 8–32 | |
| L. johnsonii LMG 9436 | 32–64 | 64 | 8–8 | 8–16 | 32 | 4 | 4–8 | 8–32 | 8–32 | |
| L. paragasseri LMG 13134 | 64 | 64 | 2–4 | 4–8 | 8 | 2–4 | 2–4 | 16–32 | 32–64 | |
| L. acidophilus ATCC 4356 | 4 | 4 | 8 | 4–16 | 16 | 1–2 | 1–2 | 1–4 | 4 | |
| L. gallinarum LMG 9435 | 16 | 16 | >1024 | >1024 | 16 | 0.25 | 0.25–0.5 | 4 | 8 | tetW, rpsLArg56 |
| L. kitasatonis LMG 23133 | 8–16 | 8–16 | 1–2 | 2–8 | 16 | 2 | 2–4 | 32 | 32–64 | |
| L. amylovorus LMG 9496 | 8–16 | 16 | 8–16 | 8–32 | 4 | 1 | 2 | 8 | 16 | rpsLThr101 |
| L. crispatus LMG 9479 | 128 | 128 | 16 | 16–32 | 32 | 4–8 | 8 | 16–32 | 32 | |
| Total resistant | 6 [31.6%] | 7 [36.8%] | 3 [15.8%] | 5 [26.3%] | NA | 1 [5.3%] | 1 [5.3%] | NA | NA | |
| MIC Value→ [µg/mL] | ≤0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | ≥1024 | No. [%] of Resistant |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kanamycin | ||||||||||||||
| L. salivarius (n = 44) | 3 | 20 | 19 | 2 bif | 41 [93.2] | |||||||||
| L. agilis (n = 20) | 3 | 10 | 4 | 2 ant(4)Ia | 1 | 17 [85.0] | ||||||||
| L. plantarum (n = 17) | 3 | 2 | 2 | 8 | 2 | 10 [58.8] | ||||||||
| L. ingluviei (n = 18) | 1 | 2 | 9 | 5 | 1 | 6 [33.3] | ||||||||
| L. reuteri (n = 15) | 1 | 1 | 5 | 4 | 4 | 4 [26.7] | ||||||||
| L. crispatus (n = 4) | 1 | 3 | 0 | |||||||||||
| Streptomycin | ||||||||||||||
| L. salivarius (n = 44) | 3 | 13 | 19 | 3 | 2 spw aadE | 4 spw(3) aadE(3) | 9 [20.4] | |||||||
| L. agilis (n = 20) | 1 | 3 | 5 | 8 | 2 | 1 aadE | 3 [15.0] | |||||||
| L. plantarum (n = 17) | 1 | 2 | 7 | 6 | 1 | NA | ||||||||
| L. ingluviei (n = 20) | 2 | 8 | 10 | 0 | ||||||||||
| L. reuteri (n = 16) | 3 | 6 | 2 | 4 | 1 | 0 | ||||||||
| L. crispatus (n = 4) | 2 | 1 | 1 | 0 | ||||||||||
| Gentamicin | ||||||||||||||
| L. salivarius (n = 44) | 1 | 5 | 14 | 17 | 5 | 1 bif | 1 bif | 2 [4.5] | ||||||
| L. agilis (n = 20) | 2 | 3 | 9 | 4 | 1 | 1 | 1 [5.0] | |||||||
| L. plantarum (n = 17) | 2 | 2 | 6 | 1 | 6 | 0 | ||||||||
| L. ingluviei (n = 20) | 1 | 2 | 12 | 3 | 1 | 1 | 0 | |||||||
| L. reuteri (n = 16) | 3 | 5 | 3 | 3 | 2 | 0 | ||||||||
| L. crispatus (n = 4) | 2 | 2 | 0 | |||||||||||
| Neomycin | ||||||||||||||
| L. salivarius (n = 41) | 4 | 11 | 20 | 5 | 1 | NA | ||||||||
| L. agilis (n = 20) | 1 | 5 | 3 | 6 | 1 | 3 ant(4)Ia(2) | 1 ant(4)Ia | NA | ||||||
| L. plantarum (n = 11) | 1 | 3 | 1 | 3 | 2 | 1 | NA | |||||||
| L. ingluviei (n = 20) | 1 | 7 | 8 | 3 | 1 | NA | ||||||||
| L. reuteri (n = 13) | 2 | 2 | 4 | 3 | 1 | 1 | NA | |||||||
| L. crispatus (n = 4) | 1 | 2 | 1 | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dec, M.; Herman-Ostrzyżek, K.; Zomer, A.; Urban-Chmiel, R. Susceptibility of Lactobacillaceae Strains to Aminoglycoside Antibiotics in the Light of EFSA Guidelines. Life 2025, 15, 732. https://doi.org/10.3390/life15050732
Dec M, Herman-Ostrzyżek K, Zomer A, Urban-Chmiel R. Susceptibility of Lactobacillaceae Strains to Aminoglycoside Antibiotics in the Light of EFSA Guidelines. Life. 2025; 15(5):732. https://doi.org/10.3390/life15050732
Chicago/Turabian StyleDec, Marta, Klaudia Herman-Ostrzyżek, Aldert Zomer, and Renata Urban-Chmiel. 2025. "Susceptibility of Lactobacillaceae Strains to Aminoglycoside Antibiotics in the Light of EFSA Guidelines" Life 15, no. 5: 732. https://doi.org/10.3390/life15050732
APA StyleDec, M., Herman-Ostrzyżek, K., Zomer, A., & Urban-Chmiel, R. (2025). Susceptibility of Lactobacillaceae Strains to Aminoglycoside Antibiotics in the Light of EFSA Guidelines. Life, 15(5), 732. https://doi.org/10.3390/life15050732







