Chromatin Remodeling via Retinoic Acid Action during Murine Spermatogonial Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Genotyping
2.3. WIN 18,446/RA Treatments
2.4. Cell Sorting
2.5. ATAC-Seq Preparation
2.6. Data Processing and Analysis
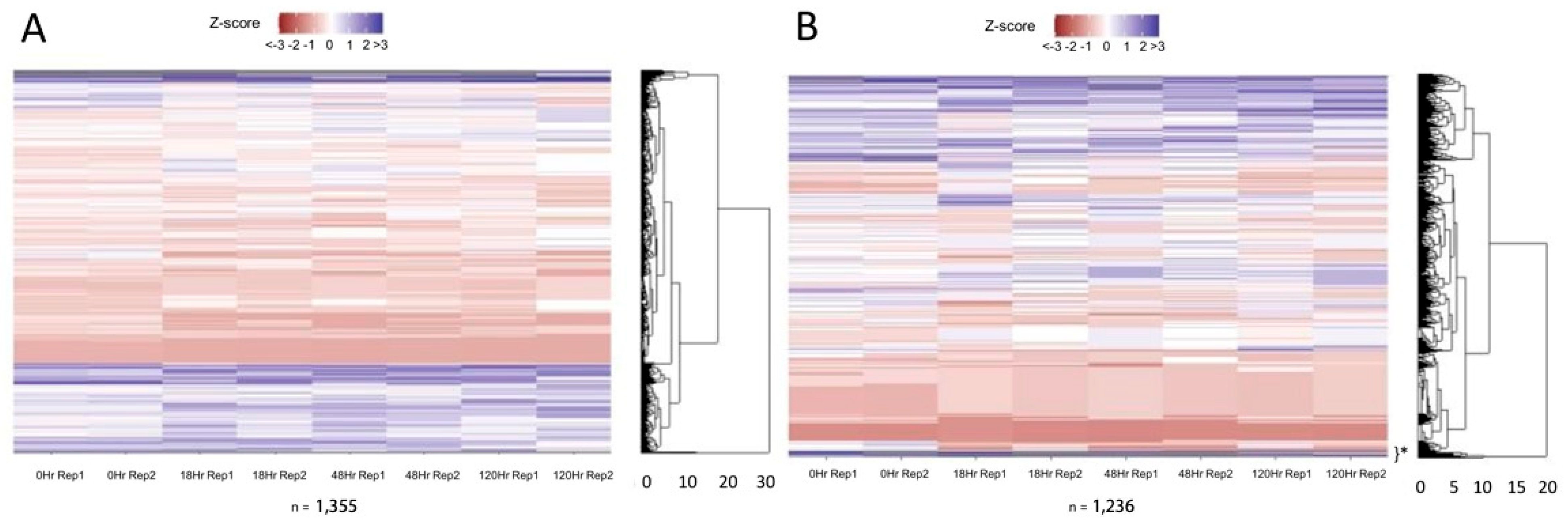
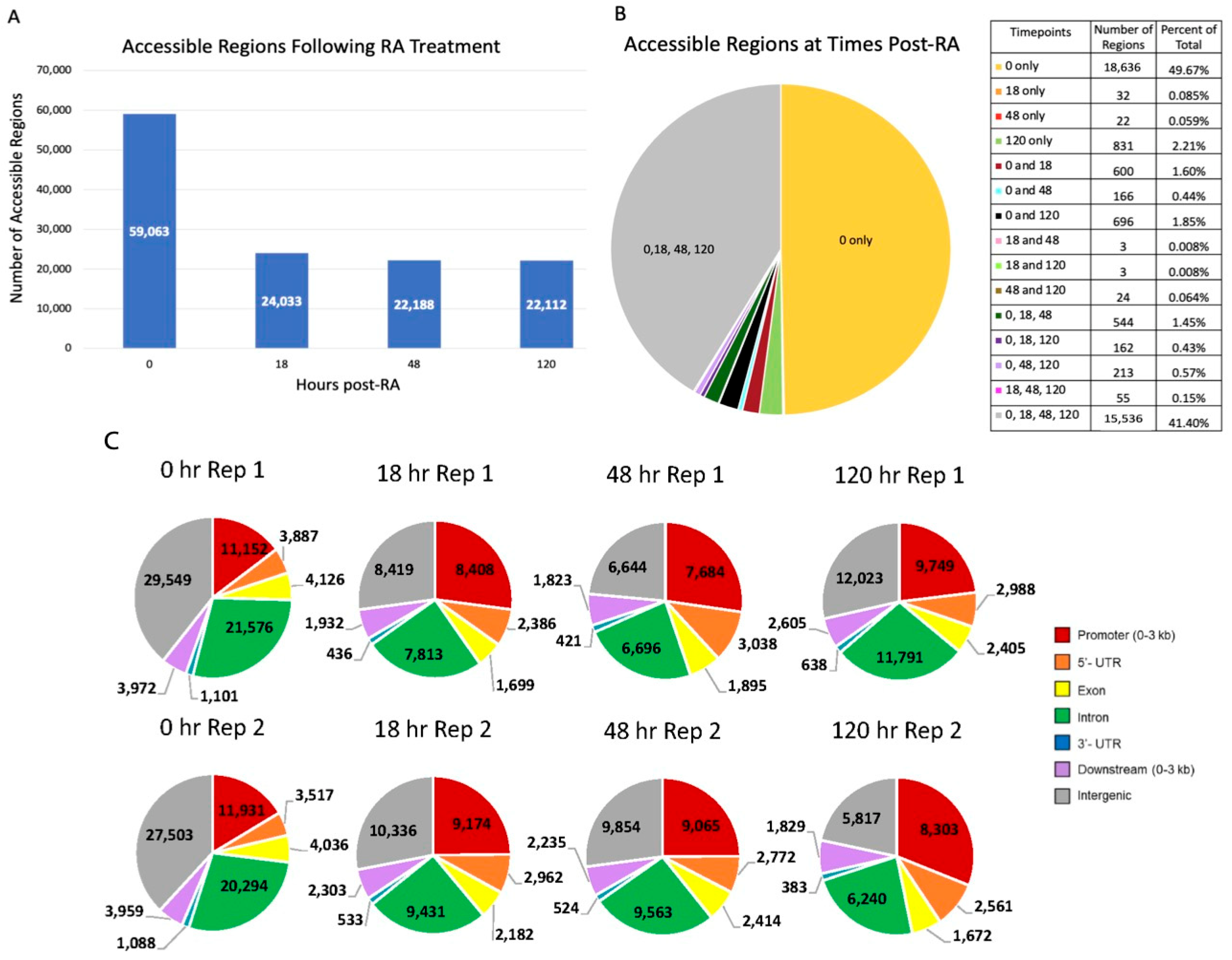
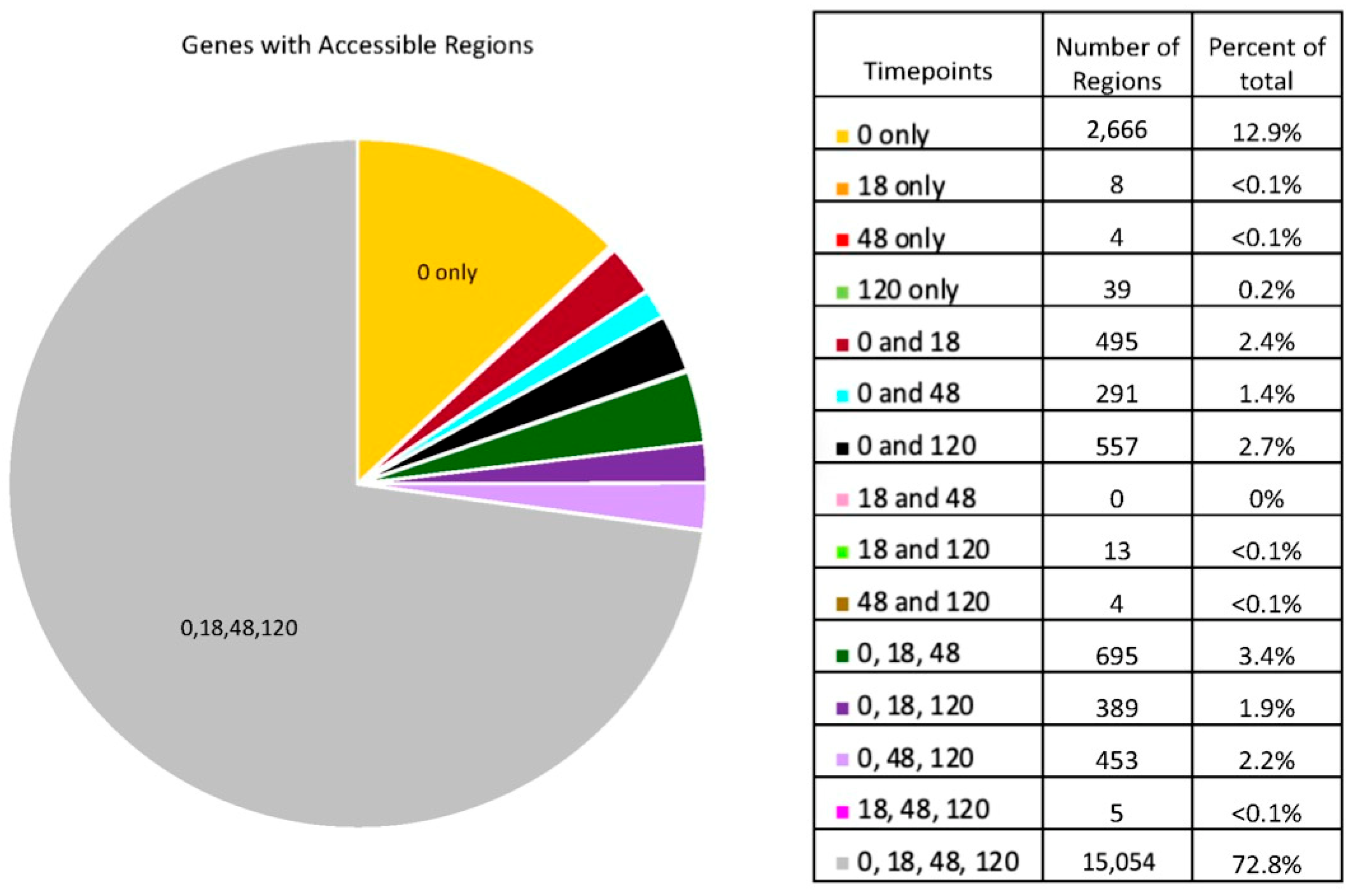
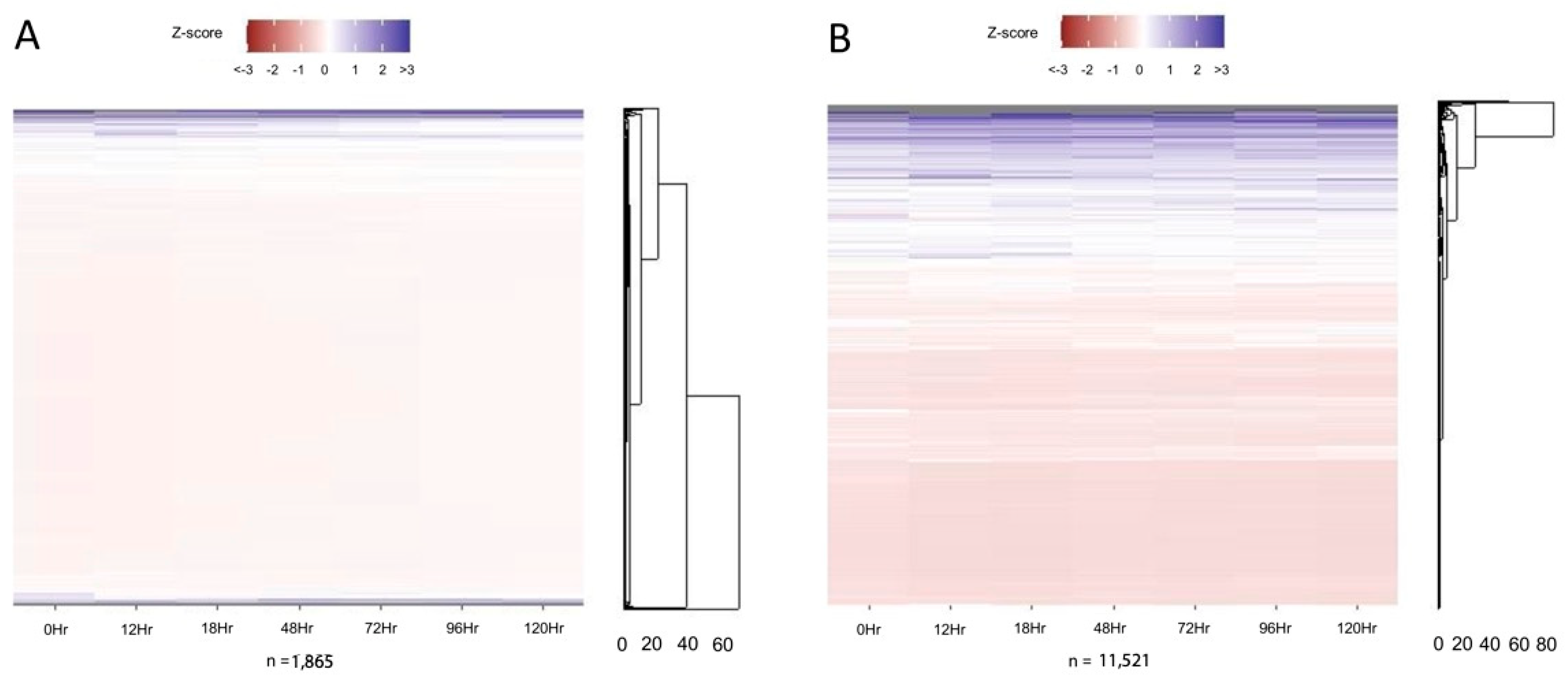
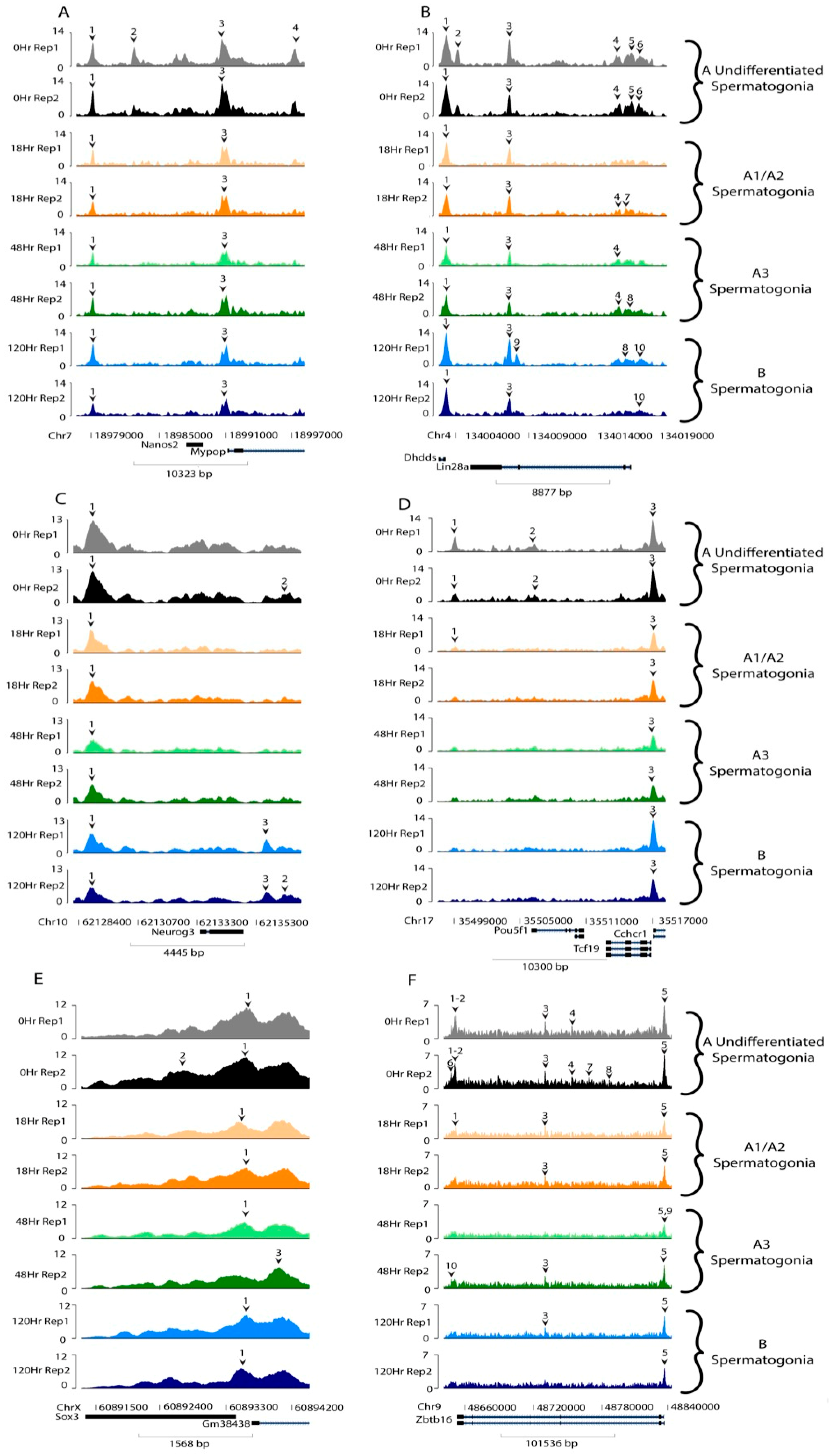
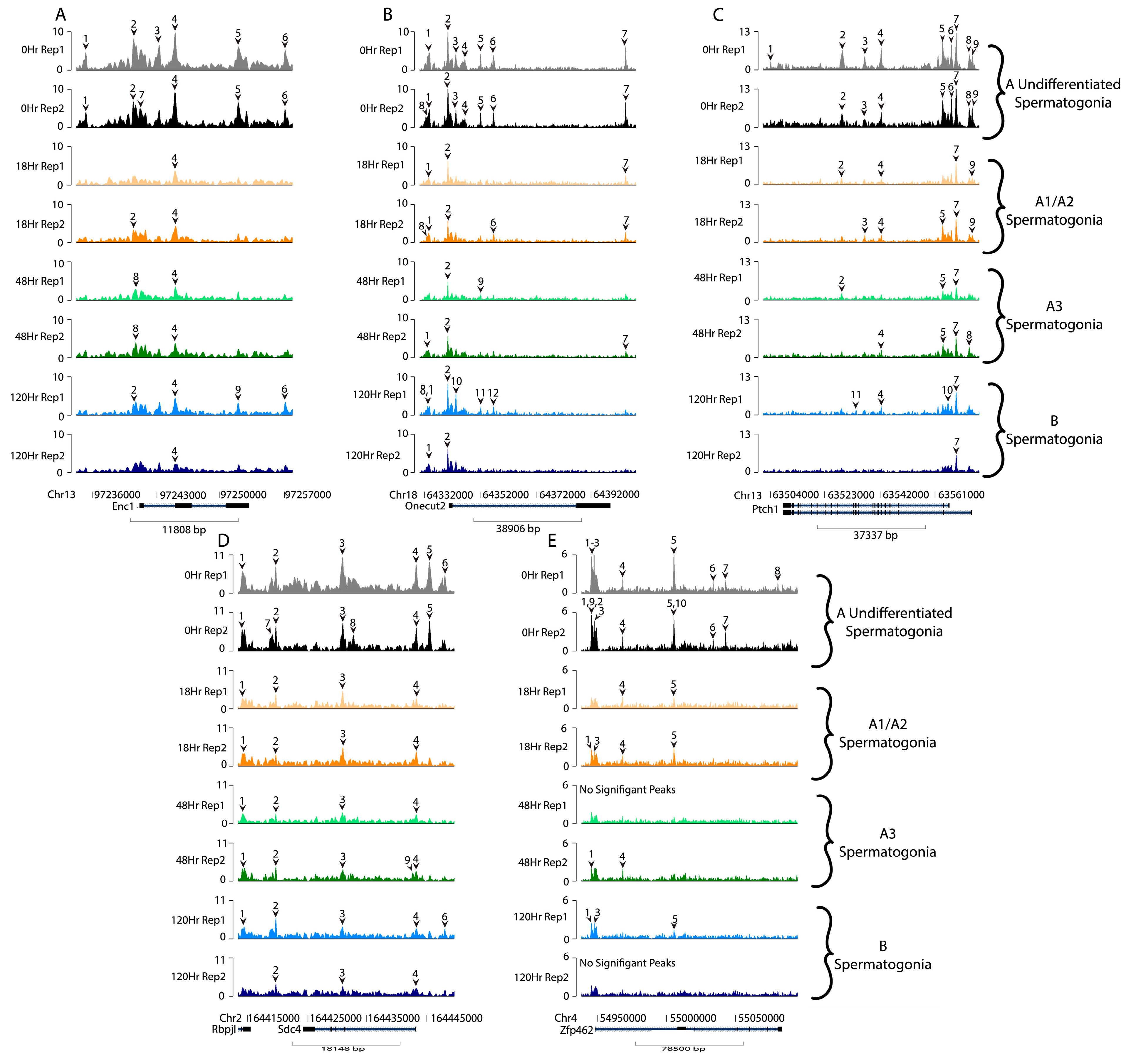
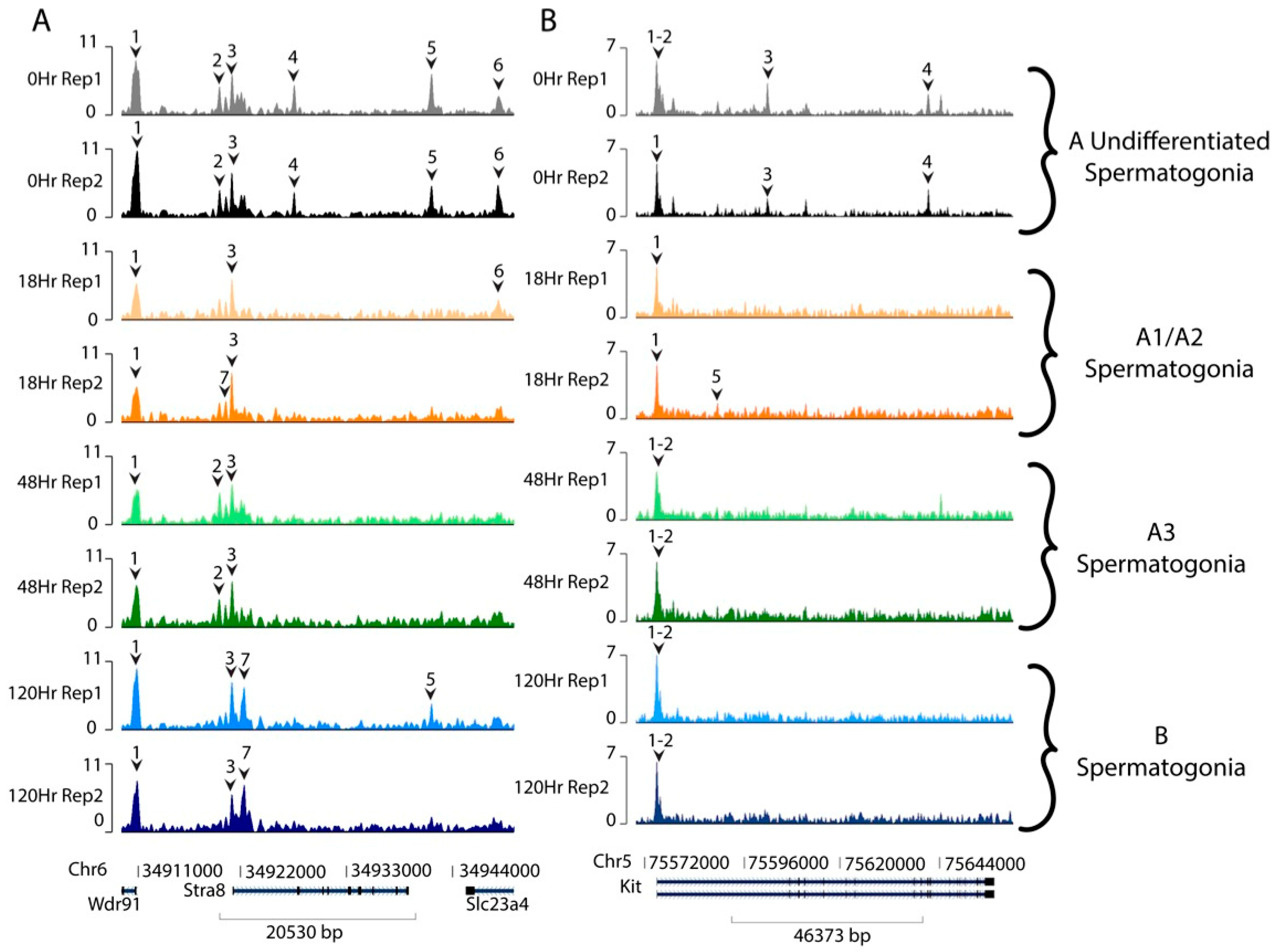
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Russell, L.D.; Ettlin, R.A.; Hikim, A.P.S.; Clegg, E.D. Histological and Histopathological Evaluation of the Testis. Int. J. Androl. 1993, 16, 83. [Google Scholar] [CrossRef]
- Leblond, C.P.; Clermont, Y. Definition of the stages of the cycle of the seminiferous epithelium in the rat. Ann. N. Y. Acad. Sci. 1952, 55, 548–573. [Google Scholar] [CrossRef] [PubMed]
- Clermont, Y. Kinetics of Spermatogenesis in Mammals: Seminiferous Epithelium Cycle and Spermatogonial Renewal. Physiol. Rev. 1972, 52, 198–236. [Google Scholar] [CrossRef] [PubMed]
- Hermo, L.; Pelletier, R.-M.; Cyr, D.G.; Smith, C.E. Surfing the Wave, Cycle, Life History, and Genes/Proteins Expressed by Testicular Germ Cells. Part 2: Changes in Spermatid Organelles Associated with Development of Spermatozoa. Microsc. Res. Tech. 2009, 73, 279–319. [Google Scholar] [CrossRef]
- Gewiss, R.L.; Shelden, E.A.; Griswold, M.D. STRA8 Induces Transcriptional Changes in Germ Cells during Spermatogonial Development. Mol. Reprod. Dev. 2021, 88, 128–140. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, Y.; Gao, Y.; Lin, Z.; Yang, S.; Wang, T.; Wang, Q.; Xie, N.; Hua, R.; Liu, M.; et al. Single-Cell RNA-Seq Uncovers Dynamic Processes and Critical Regulators in Mouse Spermatogenesis. Cell Res. 2018, 28, 879–896. [Google Scholar] [CrossRef]
- Hermann, B.P.; Cheng, K.; Singh, A.; Roa-De La Cruz, L.; Mutoji, K.N.; Chen, I.-C.; Gildersleeve, H.; Lehle, J.D.; Mayo, M.; Westernströer, B.; et al. The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Rep. 2018, 25, 1650–1667.e8. [Google Scholar] [CrossRef]
- Green, C.D.; Ma, Q.; Manske, G.L.; Shami, A.N.; Zheng, X.; Marini, S.; Moritz, L.; Sultan, C.; Gurczynski, S.J.; Moore, B.B.; et al. A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Dev. Cell 2018, 46, 651–667.e10. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, Y.; Nie, R.; Friel, P.; Mitchell, D.; Evanoff, R.M.; Pouchnik, D.; Banasik, B.; McCarrey, J.R.; Small, C.; et al. Expression of Stimulated by Retinoic Acid Gene 8 (Stra8) and Maturation of Murine Gonocytes and Spermatogonia Induced by Retinoic Acid In Vitro1. Biol. Reprod. 2008, 78, 537–545. [Google Scholar] [CrossRef]
- Zhou, Q.; Nie, R.; Li, Y.; Friel, P.; Mitchell, D.; Hess, R.A.; Small, C.; Griswold, M.D. Expression of Stimulated by Retinoic Acid Gene 8 (Stra8) in Spermatogenic Cells Induced by Retinoic Acid: An In Vivo Study in Vitamin A-Sufficient Postnatal Murine Testes1. Biol. Reprod. 2008, 79, 35–42. [Google Scholar] [CrossRef]
- Schrans-Stassen, B.H.G.J.; van de Kant, H.J.G.; de Rooij, D.G.; van Pelt, A.M.M. Differential Expression of C-Kit in Mouse Undifferentiated and Differentiating Type A Spermatogonia. Endocrinology 1999, 140, 5894–5900. [Google Scholar] [CrossRef]
- Yoshinaga, K.; Nishikawa, S.; Ogawa, M.; Hayashi, S.; Kunisada, T.; Fujimoto, T.; Nishikawa, S. Role of C-Kit in Mouse Spermatogenesis: Identification of Spermatogonia as a Specific Site of c-Kit Expression and Function. Development 1991, 113, 689–699. [Google Scholar] [CrossRef]
- Cheng, K.; Chen, I.-C.; Cheng, C.-H.E.; Mutoji, K.; Hale, B.J.; Hermann, B.P.; Geyer, C.B.; Oatley, J.M.; McCarrey, J.R. Unique Epigenetic Programming Distinguishes Regenerative Spermatogonial Stem Cells in the Developing Mouse Testis. iScience 2020, 23, 101596. [Google Scholar] [CrossRef]
- Maezawa, S.; Sakashita, A.; Yukawa, M.; Chen, X.; Takahashi, K.; Alavattam, K.G.; Nakata, I.; Weirauch, M.T.; Barski, A.; Namekawa, S.H. Super-Enhancer Switching Drives a Burst in Gene Expression at the Mitosis-to-Meiosis Transition. Nat. Struct. Mol. Biol. 2020, 27, 978–988. [Google Scholar] [CrossRef]
- Chiarini-Garcia, H.; Russell, L.D. High-Resolution Light Microscopic Characterization of Mouse Spermatogonia1. Biol. Reprod. 2001, 65, 1170–1178. [Google Scholar] [CrossRef]
- Agrimson, K.S.; Onken, J.; Mitchell, D.; Topping, T.B.; Chiarini-Garcia, H.; Hogarth, C.A.; Griswold, M.D. Characterizing the Spermatogonial Response to Retinoic Acid During the Onset of Spermatogenesis and Following Synchronization in the Neonatal Mouse Testis. Biol. Reprod. 2016, 95, 81. [Google Scholar] [CrossRef]
- Aravin, A.A.; Sachidanandam, R.; Bourc’his, D.; Schaefer, C.; Pezic, D.; Toth, K.F.; Bestor, T.; Hannon, G.J. A PiRNA Pathway Primed by Individual Transposons Is Linked to De Novo DNA Methylation in Mice. Mol. Cell 2008, 31, 785–799. [Google Scholar] [CrossRef]
- Lee, J.H.; Engel, W.; Nayernia, K. Stem Cell Protein Piwil2 Modulates Expression of Murine Spermatogonial Stem Cell Expressed Genes. Mol. Reprod. Dev. 2006, 73, 173–179. [Google Scholar] [CrossRef]
- Hogarth, C.A.; Evanoff, R.; Mitchell, D.; Kent, T.; Small, C.; Amory, J.K.; Griswold, M.D. Turning a Spermatogenic Wave into a Tsunami: Synchronizing Murine Spermatogenesis Using WIN 18,4461. Biol. Reprod. 2013, 88. 40, 1–9. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of Native Chromatin for Fast and Sensitive Epigenomic Profiling of Open Chromatin, DNA-Binding Proteins and Nucleosome Position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Corces, M.R.; Trevino, A.E.; Hamilton, E.G.; Greenside, P.G.; Sinnott-Armstrong, N.A.; Vesuna, S.; Satpathy, A.T.; Rubin, A.J.; Montine, K.S.; Wu, B.; et al. An Improved ATAC-Seq Protocol Reduces Background and Enables Interrogation of Frozen Tissues. Nat. Methods 2017, 14, 959–962. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Lerdrup, M.; Johansen, J.V.; Agrawal-Singh, S.; Hansen, K. An Interactive Environment for Agile Analysis and Visualization of ChIP-Sequencing Data. Nat. Struct. Mol. Biol. 2016, 23, 349–357. [Google Scholar] [CrossRef] [PubMed]
- R Core Team R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org (accessed on 16 December 2021).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer: New York, NY, USA, 2016; pp. 1–182. ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 16 December 2021).
- de Vries, A.; Ripley, B. Ggdendro: Create Dendrograms and Tree Diagrams Using “Ggplot2”. Available online: http://andrie.github.io/ggdendro/ (accessed on 16 December 2021).
- Wickham, H.; Vaughan, D.; Girlich, M. Tidyr: Tidy Messy Data. Available online: https://tidyr.tidyverse.org/ (accessed on 16 December 2021).
- Hogarth, C.A.; Arnold, S.; Kent, T.; Mitchell, D.; Isoherranen, N.; Griswold, M.D. Processive Pulses of Retinoic Acid Propel Asynchronous and Continuous Murine Sperm Production1. Biol. Reprod. 2015, 92, 37. [Google Scholar] [CrossRef]
- Evans, E.; Hogarth, C.; Mitchell, D.; Griswold, M. Riding the Spermatogenic Wave: Profiling Gene Expression Within Neonatal Germ and Sertoli Cells During a Synchronized Initial Wave of Spermatogenesis in Mice1. Biol. Reprod. 2014, 90, 108. [Google Scholar] [CrossRef]
- Buaas, F.W.; Kirsh, A.L.; Sharma, M.; McLean, D.J.; Morris, J.L.; Griswold, M.D.; de Rooij, D.G.; Braun, R.E. Plzf Is Required in Adult Male Germ Cells for Stem Cell Self-Renewal. Nat. Genet. 2004, 36, 647–652. [Google Scholar] [CrossRef]
- Costoya, J.A.; Hobbs, R.M.; Barna, M.; Cattoretti, G.; Manova, K.; Sukhwani, M.; Orwig, K.E.; Wolgemuth, D.J.; Pandolfi, P.P. Essential Role of Plzf in Maintenance of Spermatogonial Stem Cells. Nat. Genet. 2004, 36, 653–659. [Google Scholar] [CrossRef]
- Raverot, G.; Weiss, J.; Park, S.Y.; Hurley, L.; Jameson, J.L. Sox3 Expression in Undifferentiated Spermatogonia Is Required for the Progression of Spermatogenesis. Dev. Biol. 2005, 283, 215–225. [Google Scholar] [CrossRef]
- Sada, A.; Suzuki, A.; Suzuki, H.; Saga, Y. The RNA-Binding Protein NANOS2 Is Required to Maintain Murine Spermatogonial Stem Cells. Science 2009, 325, 1394–1398. [Google Scholar] [CrossRef]
- Suzuki, A.; Tsuda, M.; Saga, Y. Functional Redundancy among Nanos Proteins and a Distinct Role of Nanos2 during Male Germ Cell Development. Development 2007, 134, 77–83. [Google Scholar] [CrossRef]
- Yoshida, S.; Takakura, A.; Ohbo, K.; Abe, K.; Wakabayashi, J.; Yamamoto, M.; Suda, T.; Nabeshima, Y. Neurogenin3 Delineates the Earliest Stages of Spermatogenesis in the Mouse Testis. Dev. Biol. 2004, 269, 447–458. [Google Scholar] [CrossRef]
- Zheng, K.; Wu, X.; Kaestner, K.H.; Wang, P.J. The Pluripotency Factor LIN28 Marks Undifferentiated Spermatogonia in Mouse. BMC Dev. Biol. 2009, 9, 38. [Google Scholar] [CrossRef]
- Anderson, E.L.; Baltus, A.E.; Roepers-Gajadien, H.L.; Hassold, T.J.; de Rooij, D.G.; van Pelt, A.M.M.; Page, D.C. Stra8 and Its Inducer, Retinoic Acid, Regulate Meiotic Initiation in Both Spermatogenesis and Oogenesis in Mice. Proc. Natl. Acad. Sci. USA 2008, 105, 14976–14980. [Google Scholar] [CrossRef]
- Endo, T.; Romer, K.A.; Anderson, E.L.; Baltus, A.E.; de Rooij, D.G.; Page, D.C. Periodic Retinoic Acid–STRA8 Signaling Intersects with Periodic Germ-Cell Competencies to Regulate Spermatogenesis. Proc. Natl. Acad. Sci. USA 2015, 112, E2347–E2356. [Google Scholar] [CrossRef]
- Koubova, J.; Menke, D.B.; Zhou, Q.; Capel, B.; Griswold, M.D.; Page, D.C. Retinoic Acid Regulates Sex-Specific Timing of Meiotic Initiation in Mice. Proc. Natl. Acad. Sci. USA 2006, 103, 2474–2479. [Google Scholar] [CrossRef]
- Kojima, M.L.; de Rooij, D.G.; Page, D.C. Amplification of a Broad Transcriptional Program by a Common Factor Triggers the Meiotic Cell Cycle in Mice. eLife 2019, 8, e43738. [Google Scholar] [CrossRef]
- Gewiss, R.; Schleif, M.C.; Griswold, M. The Role of Retinoic Acid in the Commitment to Meiosis. Asian J. Androl. 2021, 23, 549. [Google Scholar] [CrossRef]
- Shirakawa, T.; Yaman-Deveci, R.; Tomizawa, S.-i.; Kamizato, Y.; Nakajima, K.; Sone, H.; Sato, Y.; Sharif, J.; Yamashita, A.; Takada-Horisawa, Y.; et al. An Epigenetic Switch Is Crucial for Spermatogonia to Exit the Undifferentiated State toward a Kit-Positive Identity. Development 2013, 140, 3565–3576. [Google Scholar] [CrossRef]
- Gan, H.; Wen, L.; Liao, S.; Lin, X.; Ma, T.; Liu, J.; Song, C.; Wang, M.; He, C.; Han, C.; et al. Dynamics of 5-Hydroxymethylcytosine during Mouse Spermatogenesis. Nat. Commun. 2013, 4, 1995. [Google Scholar] [CrossRef]
- Oakes, C.C.; La Salle, S.; Smiraglia, D.J.; Robaire, B.; Trasler, J.M. Developmental Acquisition of Genome-Wide DNA Methylation Occurs Prior to Meiosis in Male Germ Cells. Dev. Biol. 2007, 307, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Margolin, G.; Khil, P.P.; Kim, J.; Bellani, M.A.; Camerini-Otero, R.D. Integrated Transcriptome Analysis of Mouse Spermatogenesis. BMC Genomics 2014, 15, 39. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Starmer, J.; Fedoriw, A.M.; Yee, D.; Magnuson, T. Repression of the Soma-Specific Transcriptome by Polycomb-Repressive Complex 2 Promotes Male Germ Cell Development. Genes Dev. 2014, 28, 2056–2069. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, S.S.; Low, D.H.P.; Yi, C.; Lee, C.L.; Oatley, J.M.; Payne, C.J.; Carrell, D.T.; Guccione, E.; Cairns, B.R. Transcription and Imprinting Dynamics in Developing Postnatal Male Germline Stem Cells. Genes Dev. 2015, 29, 2312–2324. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schleif, C.; Gewiss, R.; Griswold, M. Chromatin Remodeling via Retinoic Acid Action during Murine Spermatogonial Development. Life 2023, 13, 690. https://doi.org/10.3390/life13030690
Schleif C, Gewiss R, Griswold M. Chromatin Remodeling via Retinoic Acid Action during Murine Spermatogonial Development. Life. 2023; 13(3):690. https://doi.org/10.3390/life13030690
Chicago/Turabian StyleSchleif, Christine, Rachel Gewiss, and Michael Griswold. 2023. "Chromatin Remodeling via Retinoic Acid Action during Murine Spermatogonial Development" Life 13, no. 3: 690. https://doi.org/10.3390/life13030690
APA StyleSchleif, C., Gewiss, R., & Griswold, M. (2023). Chromatin Remodeling via Retinoic Acid Action during Murine Spermatogonial Development. Life, 13(3), 690. https://doi.org/10.3390/life13030690





