Identification of an Amylomaltase from the Halophilic Archaeon Haloquadratum walsbyi by Functional Metagenomics: Structural and Functional Insights
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Purification of eDNA
2.2. Generation and Screening of a Fosmid Library
2.3. Bioinformatic Analysis
2.4. Amplification and Cloning of the Amylomaltase Gene
2.5. Expression and Purification of Hw-A Recombinant Protein
2.6. Amylomaltase Transglycosylation and Hydrolytic Activity Assays
2.7. Thermal Shift
3. Results and Discussion
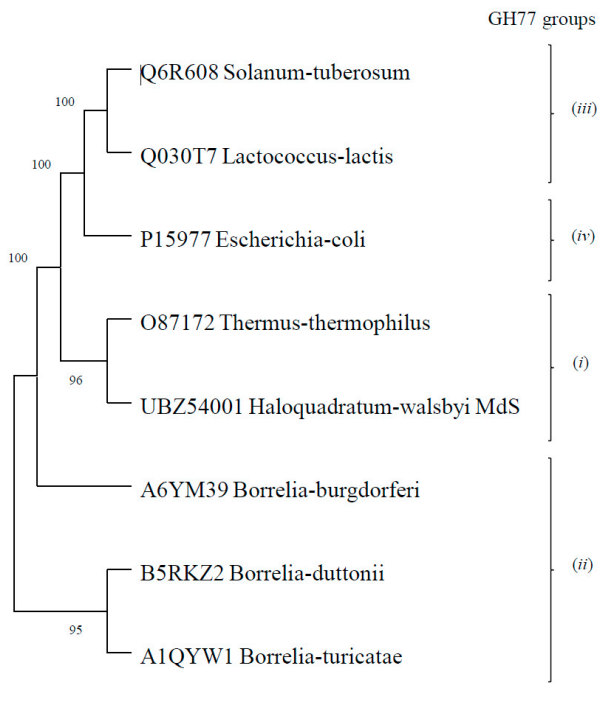
3.1. Fosmid Library Generation, Screening and Identification of an Amylomaltase Coding Sequence
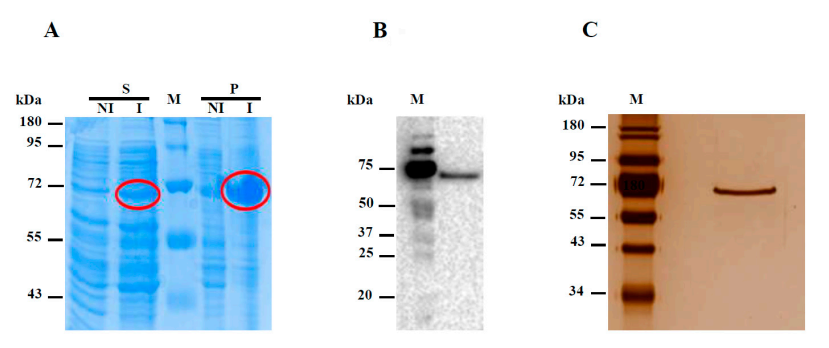
3.2. Expression of the Recombinant Hw-A Enzyme
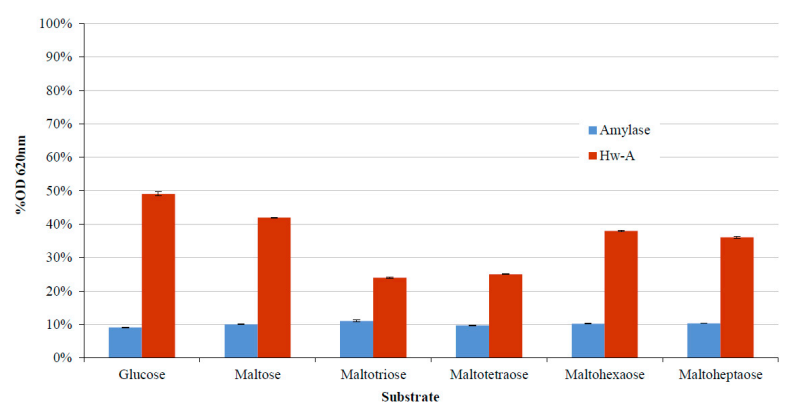
3.3. Transglycosylation Activity
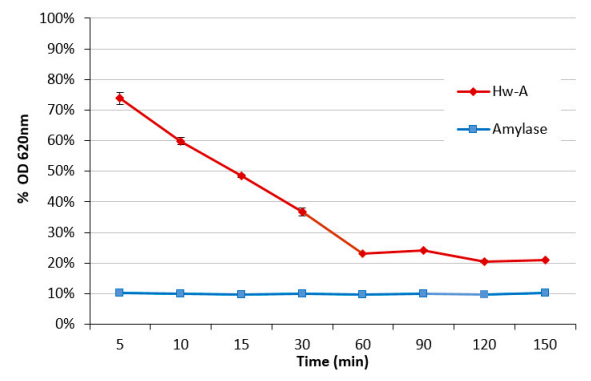
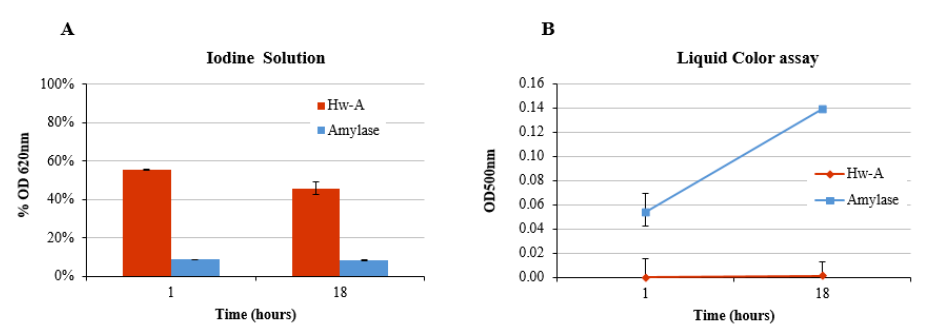
3.4. Hydrolytic Activity
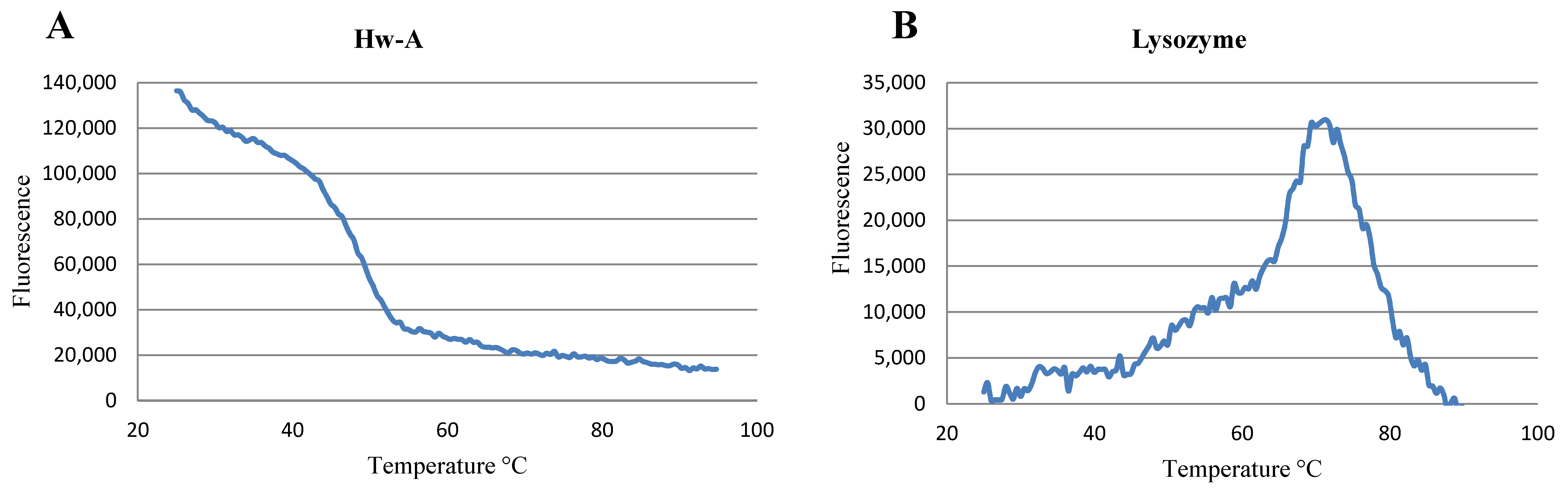
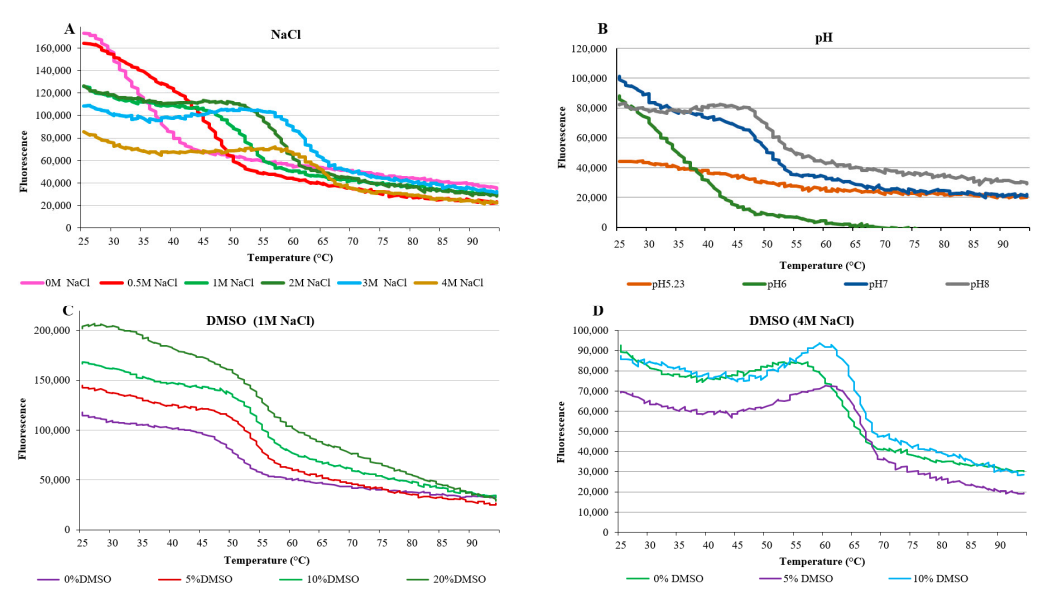
3.5. Thermal Shift Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cabrera, M.Á.; Blamey, J.M. Biotechnological Applications of Archaeal Enzymes from Extreme Environments. Biol. Res. 2018, 51, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrer, M.; Martínez-Martínez, M.; Bargiela, R.; Streit, W.R.; Golyshina, O.V.; Golyshin, P.N. Estimating the Success of Enzyme Bioprospecting through Metagenomics: Current Status and Future Trends: Enzyme Bioprospecting by Metagenomics. Microb. Biotechnol. 2016, 9, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Satyanarayana, T. Biotechnology of Cold-Active Proteases. Biology 2013, 2, 755–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piller, K.; Daniel, R.M.; Petach, H.H. Properties and Stabilization of an Extracellular Alpha-Glucosidase from the Extremely Thermophilic Archaebacteria Thermococcus Strain AN1: Enzyme Activity at 130 °C. Biochim. Biophys. Acta 1996, 1292, 197–205. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, H.; Kondo, A.; Noda, H. Biodiesel Fuel Production by Transesterification of Oils. J. Biosci. Bioeng. 2001, 92, 405–416. [Google Scholar] [CrossRef]
- Poli, A.; Finore, I.; Romano, I.; Gioiello, A.; Lama, L.; Nicolaus, B. Microbial Diversity in Extreme Marine Habitats and Their Biomolecules. Microorganisms 2017, 5, 25. [Google Scholar] [CrossRef] [Green Version]
- Alcázar-Alay, S.C.; Meireles, M.A.A. Physicochemical Properties, Modifications and Applications of Starches from Different Botanical Sources. Food Sci. Technol. 2015, 35, 215–236. [Google Scholar] [CrossRef] [Green Version]
- Hashim, S.O. Starch-Modifying Enzymes. In Alkaliphiles in Biotechnology; Mamo, G., Mattiasson, B., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 172, pp. 221–244. ISBN 978-3-030-49735-4. [Google Scholar]
- Park, S.H.; Na, Y.; Kim, J.; Kang, S.D.; Park, K.-H. Properties and Applications of Starch Modifying Enzymes for Use in the Baking Industry. Food Sci. Biotechnol. 2017, 27, 299–312. [Google Scholar] [CrossRef]
- Miao, M.; Jiang, B.; Jin, Z.; BeMiller, J.N. Microbial Starch-Converting Enzymes: Recent Insights and Perspectives. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1238–1260. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, X.; Ji, H.; Zheng, D.; Jin, Z.; Bai, Y.; Svensson, B. Thermophilic 4-α-Glucanotransferase from Thermoproteus Uzoniensis Retards the Long-Term Retrogradation but Maintains the Short-Term Gelation Strength of Tapioca Starch. J. Agric. Food Chem. 2020, 68, 5658–5667. [Google Scholar] [CrossRef]
- Ahmad, N.; Mehboob, S.; Rashid, N. Starch-Processing Enzymes—Emphasis on Thermostable 4-α-Glucanotransferases. Biologia 2015, 70, 709–725. [Google Scholar] [CrossRef]
- Dou, S.; Chi, N.; Zhou, X.; Zhang, Q.; Pang, F.; Xiu, Z. Molecular Cloning, Expression, and Biochemical Characterization of a Novel Cold-Active α-Amylase from Bacillus sp. dsh19-1. Extremophiles 2018, 22, 739–749. [Google Scholar] [CrossRef]
- Amoozegar, M.A.; Safarpour, A.; Noghabi, K.A.; Bakhtiary, T.; Ventosa, A. Halophiles and Their Vast Potential in Biofuel Production. Front. Microbiol. 2019, 10, 1895. [Google Scholar] [CrossRef]
- Van der Maarel, M.J.E.C.; van der Veen, B.; Uitdehaag, J.C.M.; Leemhuis, H.; Dijkhuizen, L. Properties and Applications of Starch-Converting Enzymes of the α-Amylase Family. J. Biotechnol. 2002, 94, 137–155. [Google Scholar] [CrossRef] [Green Version]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes Database (CAZy): An Expert Resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Critchley, J.H.; Zeeman, S.C.; Takaha, T.; Smith, A.M.; Smith, S.M. A Critical Role for Disproportionating Enzyme in Starch Breakdown Is Revealed by a Knock-out Mutation in Arabidopsis: Function of D-Enzyme in Starch Breakdown. Plant J. 2001, 26, 89–100. [Google Scholar] [CrossRef]
- Nguyen, D.H.D.; Park, S.-H.; Tran, P.L.; Kim, J.-W.; Le, Q.T.; Boos, W.; Park, J.-T. Characterization of the Transglycosylation Reaction of 4-a-Glucanotransferase (MalQ) and Its Role in Glycogen Breakdown in Escherichia coli. J. Microbiol. Biotechnol. 2019, 29, 357–366. [Google Scholar] [CrossRef]
- O′Neill, E.C.; Stevenson, C.E.M.; Tantanarat, K.; Latousakis, D.; Donaldson, M.I.; Rejzek, M.; Nepogodiev, S.A.; Limpaseni, T.; Field, R.A.; Lawson, D.M. Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1). J. Biol. Chem. 2015, 290, 29834–29853. [Google Scholar] [CrossRef] [Green Version]
- Imamura, K.; Matsuura, T.; Nakagawa, A.; Kitamura, S.; Kusunoki, M.; Takaha, T.; Unno, H. Structural Analysis and Reaction Mechanism of the Disproportionating Enzyme (D-enzyme) from Potato. Protein Sci. 2020, 29, 2085–2100. [Google Scholar] [CrossRef]
- Srisimarat, W.; Murakami, S.; Pongsawasdi, P.; Krusong, K. Crystallization and Preliminary X-Ray Crystallographic Analysis of the Amylomaltase from Corynebacterium Glutamicum. Acta Cryst. F Struct. Biol. Cryst. Commun. 2013, 69, 1004–1006. [Google Scholar] [CrossRef] [Green Version]
- Weiss, S.C.; Skerra, A.; Schiefner, A. Structural Basis for the Interconversion of Maltodextrins by MalQ, the Amylomaltase of Escherichia Coli. J. Biol. Chem. 2015, 290, 21352–21364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tumhom, S.; Nimpiboon, P.; Wangkanont, K.; Pongsawasdi, P. Streptococcus Agalactiae Amylomaltase Offers Insight into the Transglycosylation Mechanism and the Molecular Basis of Thermostability among Amylomaltases. Sci. Rep. 2021, 11, 6740. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Jung, T.-Y.; Seo, D.-H.; Yoon, S.-M.; Choi, H.-C.; Park, B.C.; Park, C.-S.; Woo, E.-J. Structural and Functional Analysis of Substrate Recognition by the 250s Loop in Amylomaltase from Thermus Brockianus. Proteins 2011, 79, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Leoni, C.; Gattulli, B.A.R.; Pesole, G.; Ceci, L.R.; Volpicella, M. Amylomaltases in Extremophilic Microorganisms. Biomolecules 2021, 11, 1335. [Google Scholar] [CrossRef] [PubMed]
- Przylas, I.; Tomoo, K.; Terada, Y.; Takaha, T.; Fujii, K.; Saenger, W.; Sträter, N. Crystal Structure of Amylomaltase from Thermus Aquaticus, a Glycosyltransferase Catalysing the Production of Large Cyclic Glucans. J. Mol. Biol. 2000, 296, 873–886. [Google Scholar] [CrossRef]
- Kuchtová, A.; Janeček, Š. In Silico Analysis of Family GH77 with Focus on Amylomaltases from Borreliae and Disproportionating Enzymes DPE2 from Plants and Bacteria. Biochim. Et Biophys. Acta Proteins Proteom. 2015, 1854, 1260–1268. [Google Scholar] [CrossRef]
- Steichen, J.M.; Petty, R.V.; Sharkey, T.D. Domain Characterization of a 4-α-Glucanotransferase Essential for Maltose Metabolism in Photosynthetic Leaves. J. Biol. Chem. 2008, 283, 20797–20804. [Google Scholar] [CrossRef] [Green Version]
- Mareček, F.; Møller, M.S.; Svensson, B.; Janeček, Š. A Putative Novel Starch-Binding Domain Revealed by in Silico Analysis of the N-Terminal Domain in Bacterial Amylomaltases from the Family GH77. 3 Biotech. 2021, 11, 229. [Google Scholar] [CrossRef]
- Leoni, C.; Volpicella, M.; Fosso, B.; Manzari, C.; Piancone, E.; Dileo, M.C.G.; Arcadi, E.; Yakimov, M.; Pesole, G.; Ceci, L.R. A Differential Metabarcoding Approach to Describe Taxonomy Profiles of Bacteria and Archaea in the Saltern of Margherita Di Savoia (Italy). Microorganisms 2020, 8, 936. [Google Scholar] [CrossRef]
- Placido, A.; Hai, T.; Ferrer, M.; Chernikova, T.N.; Distaso, M.; Armstrong, D.; Yakunin, A.F.; Toshchakov, S.V.; Yakimov, M.M.; Kublanov, I.V.; et al. Diversity of Hydrolases from Hydrothermal Vent Sediments of the Levante Bay, Vulcano Island (Aeolian Archipelago) Identified by Activity-Based Metagenomics and Biochemical Characterization of New Esterases and an Arabinopyranosidase. Appl. Microbiol. Biotechnol. 2015, 99, 10031–10046. [Google Scholar] [CrossRef] [Green Version]
- Chiara, M.; Placido, A.; Picardi, E.; Ceci, L.R.; Horner, D.S.; Pesole, G. A-GAME: Improving the Assembly of Pooled Functional Metagenomics Sequence Data. BMC Genom. 2018, 19, 44. [Google Scholar] [CrossRef] [Green Version]
- Goecks, J.; Nekrutenko, A.; Taylor, J.; Galaxy Team, T. Galaxy: A Comprehensive Approach for Supporting Accessible, Reproducible, and Transparent Computational Research in the Life Sciences. Genome Biol. 2010, 11, R86. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Magoc, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Namiki, T.; Hachiya, T.; Tanaka, H.; Sakakibara, Y. MetaVelvet: An Extension of Velvet Assembler to de Novo Metagenome Assembly from Short Sequence Reads. Nucleic Acids Res. 2012, 40, e155. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Lomsadze, A.; Borodovsky, M. Ab Initio Gene Identification in Metagenomic Sequences. Nucleic Acids Res. 2010, 38, e132. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The Protein Families Database. Nucl. Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehboob, S.; Ahmad, N.; Rashid, N.; Imanaka, T.; Akhtar, M. Pcal_0768, a Hyperactive 4-α-Glucanotransferase from Pyrobacculum Calidifontis. Extremophiles 2016, 20, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.G.; Camakaris, H.M.; Janssen, P.H.; Dyall-Smith, M.L. Cultivation of Walsby’s Square Haloarchaeon. FEMS Microbiol. Lett. 2004, 238, 469–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolhuis, H.; Poele, E.M.T.; Rodriguez-Valera, F. Isolation and Cultivation of Walsby’s Square Archaeon. Environ. Microbiol. 2004, 6, 1287–1291. [Google Scholar] [CrossRef]
- Podell, S.; Ugalde, J.A.; Narasingarao, P.; Banfield, J.F.; Heidelberg, K.B.; Allen, E.E. Assembly-Driven Community Genomics of a Hypersaline Microbial Ecosystem. PLoS ONE 2013, 8, e61692. [Google Scholar] [CrossRef] [Green Version]
- Bolhuis, H.; Martín-Cuadrado, A.B.; Rosselli, R.; Pašić, L.; Rodriguez-Valera, F. Transcriptome Analysis of Haloquadratum walsbyi: Vanity Is but the Surface. BMC Genom. 2017, 18, 510. [Google Scholar] [CrossRef]
- Reed, C.J.; Lewis, H.; Trejo, E.; Winston, V.; Evilia, C. Protein Adaptations in Archaeal Extremophiles. Archaea 2013, 2013, 373275. [Google Scholar] [CrossRef] [Green Version]
- Terada, Y.; Fujii, K.; Takaha, T.; Okada, S. Thermus Aquaticus ATCC 33923 Amylomaltase Gene Cloning and Expression and Enzyme Characterization: Production of Cycloamylose. Appl. Environ. Microbiol. 1999, 65, 910–915. [Google Scholar] [CrossRef] [Green Version]
- Palmer, T.N.; Ryman, B.E.; Whelan, W.J. The Action Pattern of Amylomaltase from Escherichia Coli. Eur. J. Biochem. 1976, 69, 105–115. [Google Scholar] [CrossRef]
- Lavinder, J.J.; Hari, S.B.; Sullivan, B.J.; Magliery, T.J. High-Throughput Thermal Scanning: A General, Rapid Dye-Binding Thermal Shift Screen for Protein Engineering. J. Am. Chem. Soc. 2009, 131, 3794–3795. [Google Scholar] [CrossRef] [Green Version]
- Kohlstaedt, M.; von der Hocht, I.; Hilbers, F.; Thielmann, Y.; Michel, H. Development of a Thermofluor assay for stability determination of membrane proteins using the Na+/H+ antiporter NhaA and cytochrome c oxidase. Acta Crystallogr. D Biol. Crystallogr. 2015, 71 Pt 5, 1112–1122. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leoni, C.; Manzari, C.; Tran, H.; Golyshin, P.N.; Pesole, G.; Volpicella, M.; Ceci, L.R. Identification of an Amylomaltase from the Halophilic Archaeon Haloquadratum walsbyi by Functional Metagenomics: Structural and Functional Insights. Life 2022, 12, 85. https://doi.org/10.3390/life12010085
Leoni C, Manzari C, Tran H, Golyshin PN, Pesole G, Volpicella M, Ceci LR. Identification of an Amylomaltase from the Halophilic Archaeon Haloquadratum walsbyi by Functional Metagenomics: Structural and Functional Insights. Life. 2022; 12(1):85. https://doi.org/10.3390/life12010085
Chicago/Turabian StyleLeoni, Claudia, Caterina Manzari, Hai Tran, Peter N. Golyshin, Graziano Pesole, Mariateresa Volpicella, and Luigi R. Ceci. 2022. "Identification of an Amylomaltase from the Halophilic Archaeon Haloquadratum walsbyi by Functional Metagenomics: Structural and Functional Insights" Life 12, no. 1: 85. https://doi.org/10.3390/life12010085
APA StyleLeoni, C., Manzari, C., Tran, H., Golyshin, P. N., Pesole, G., Volpicella, M., & Ceci, L. R. (2022). Identification of an Amylomaltase from the Halophilic Archaeon Haloquadratum walsbyi by Functional Metagenomics: Structural and Functional Insights. Life, 12(1), 85. https://doi.org/10.3390/life12010085








