Lacticaseibacillus paracasei: Occurrence in the Human Gut Microbiota and K-Mer-Based Assessment of Intraspecies Diversity
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Growth
2.2. DNA Preparation
2.3. Genome Sequencing
2.4. Genome Assembly and Annotation
2.5. Barcoding
2.6. Phylogenetic Analysis
2.7. Phylogroup-Specific Taxonomic Analysis of Human Metagenomes
3. Results
3.1. Characteristics of the L. paracasei VKM B-1144 Strain
3.2. Genome Sequencing and Assembly
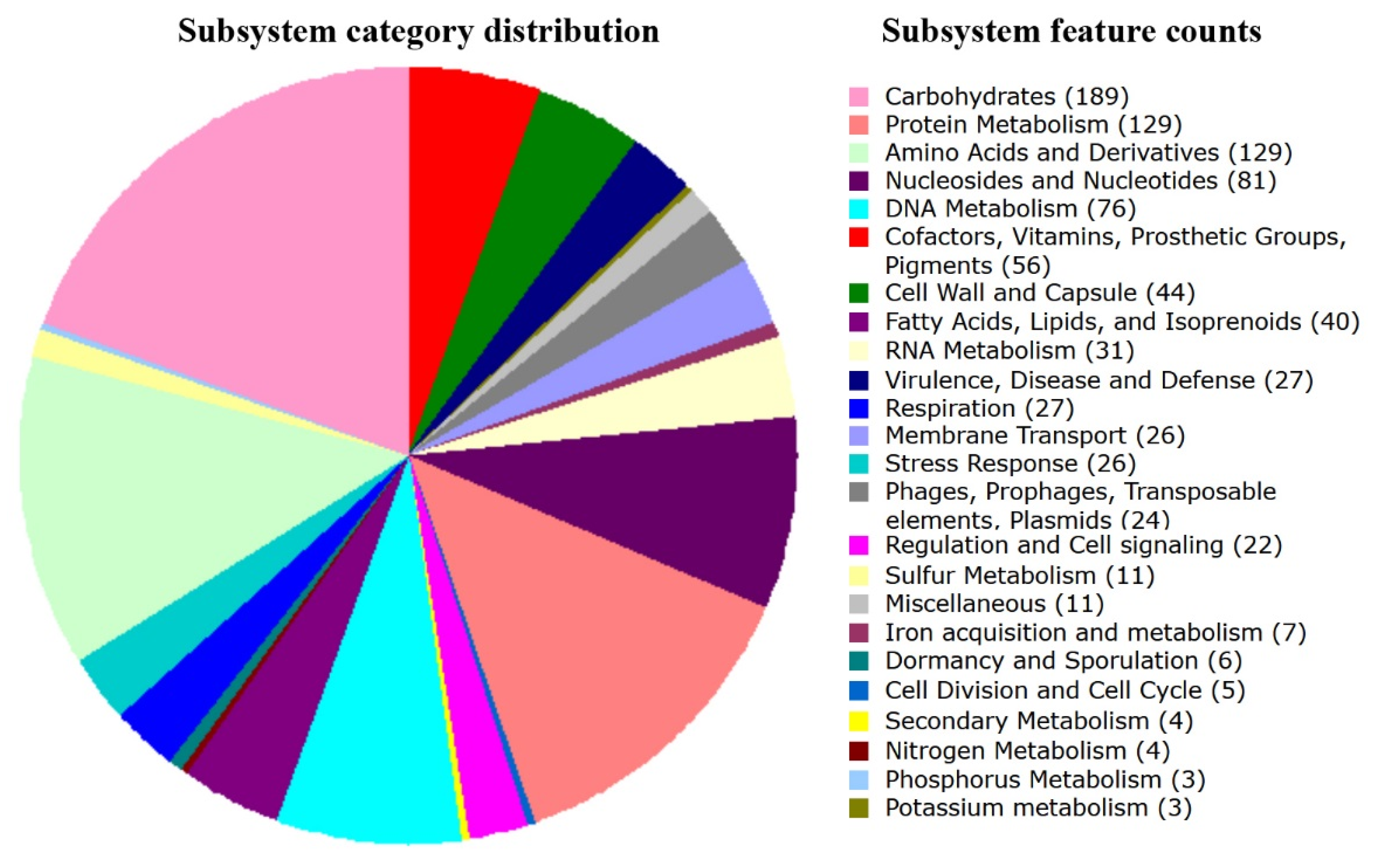
3.3. Genome Annotation and Analysis
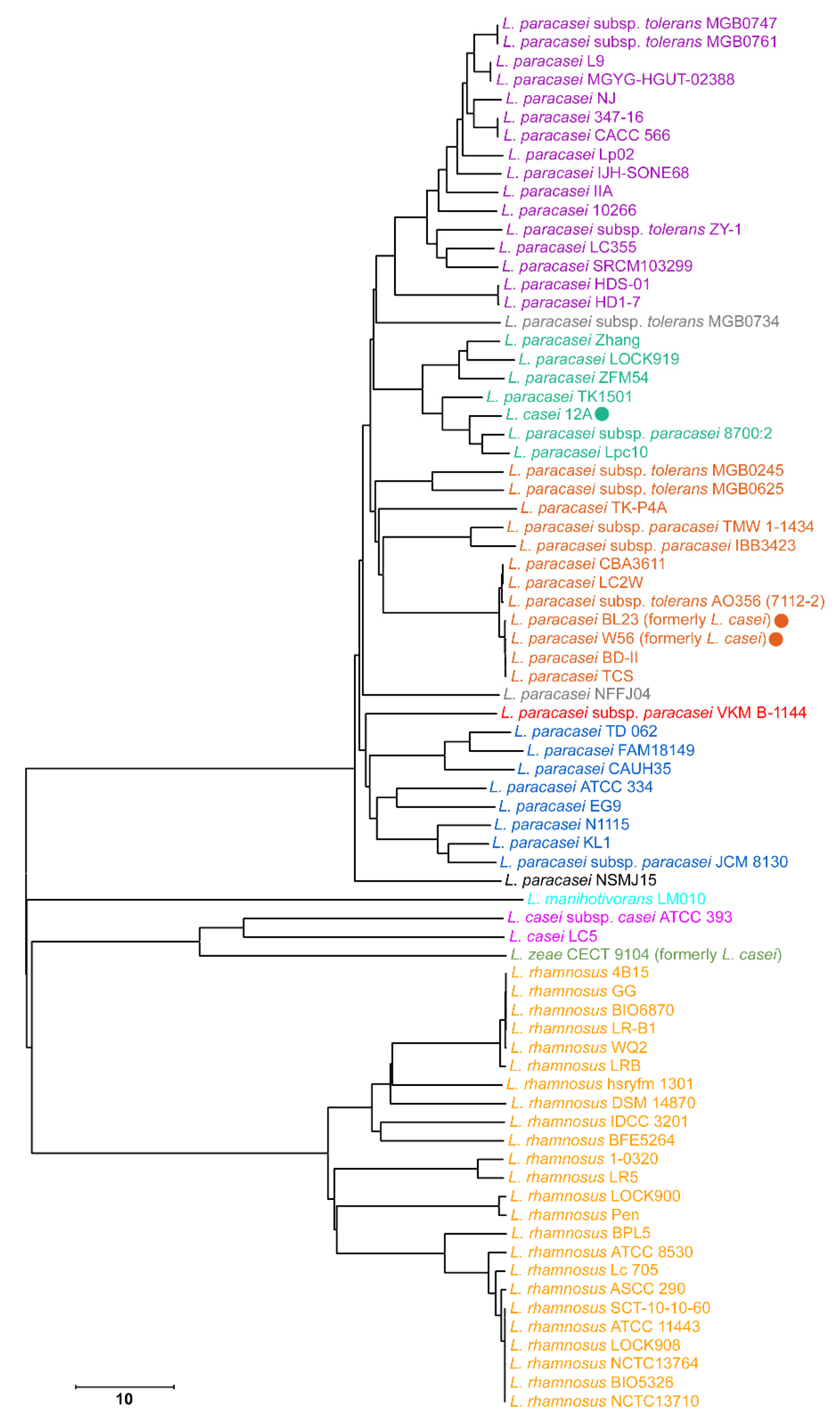
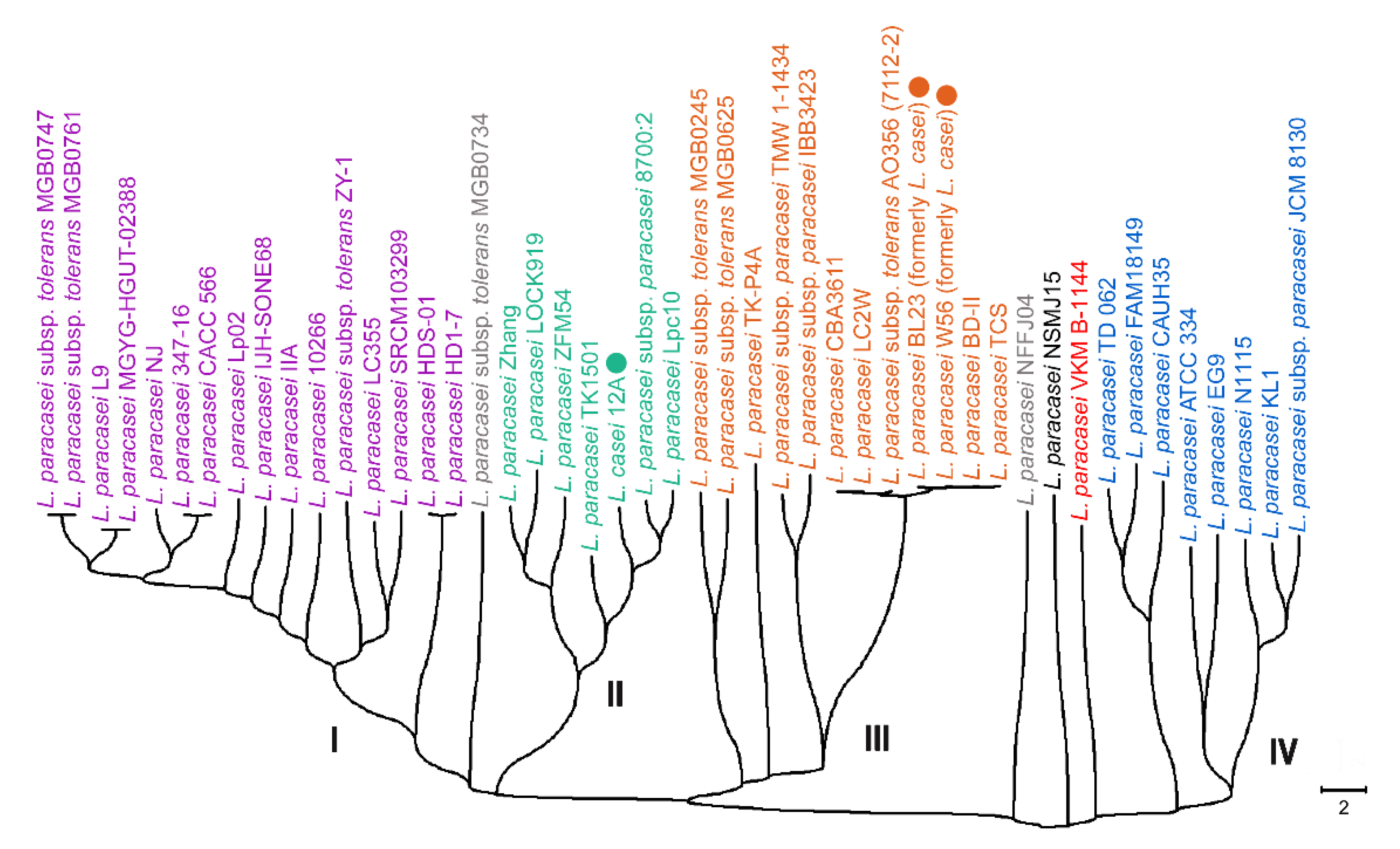
3.4. Intrageneric and Intraspecific Phylotyping of L. paracasei Genomes
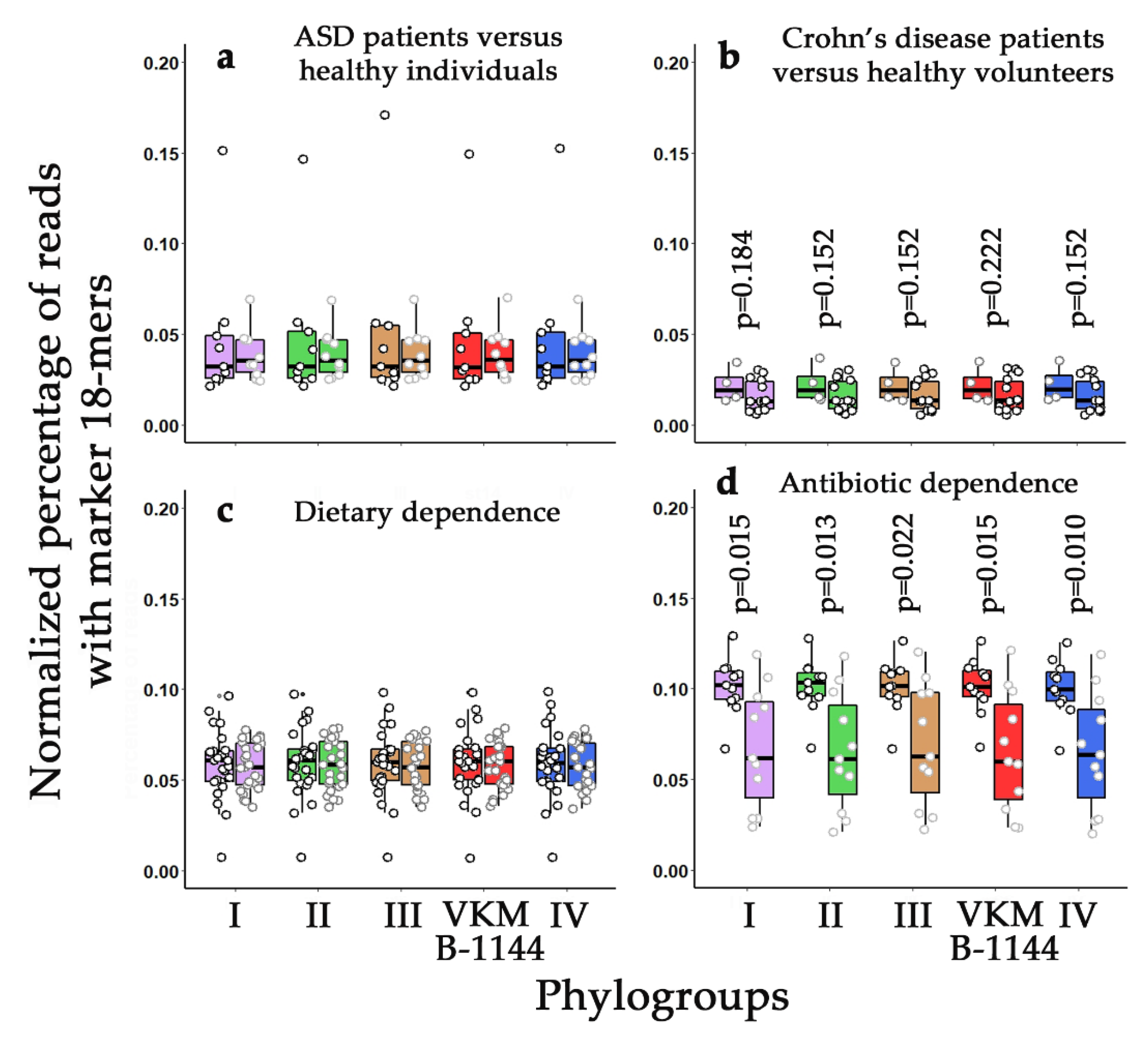
3.5. Phylogroup-Dependent Profiling of L. paracasei Presence in Human Intestinal Microbiomes
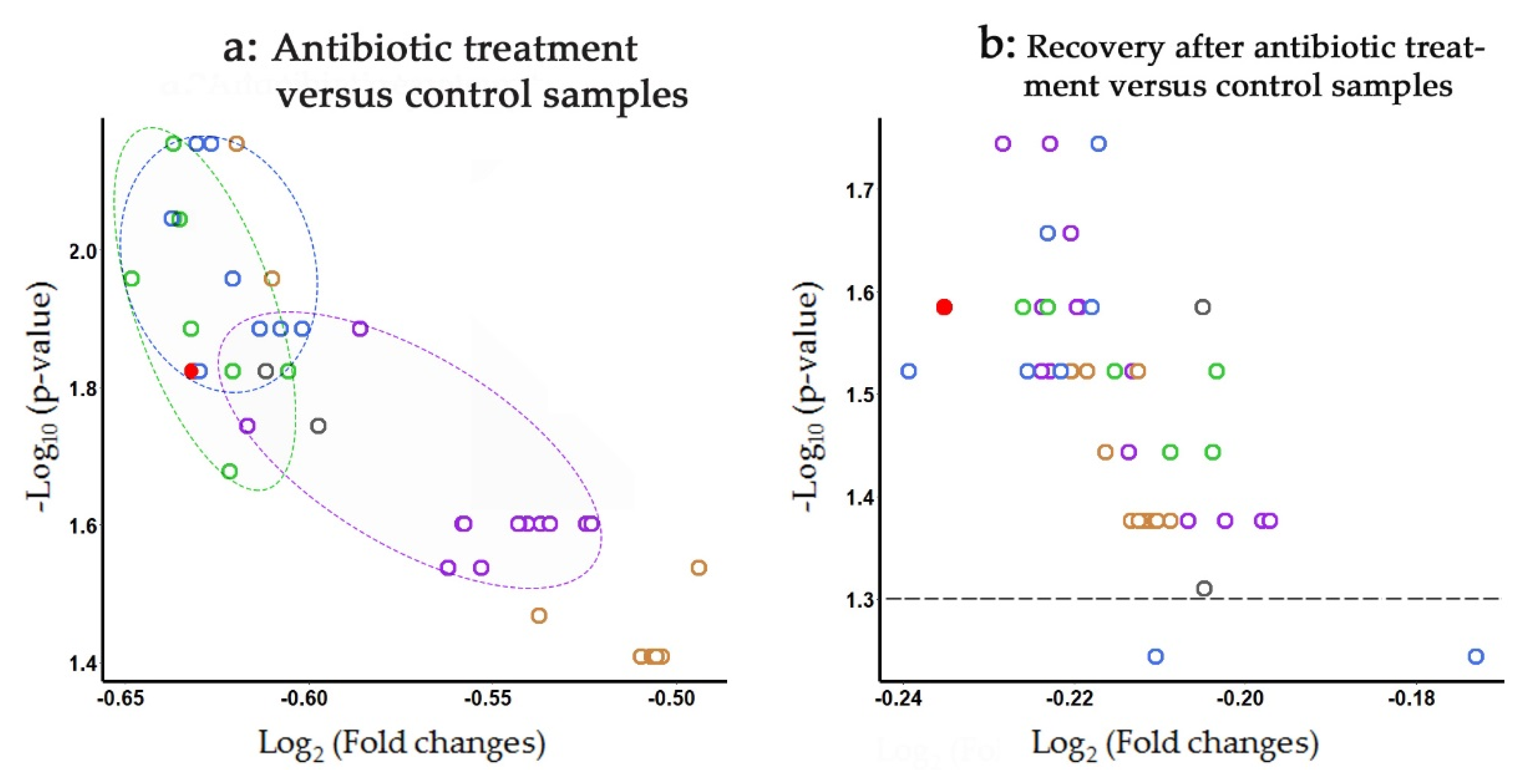
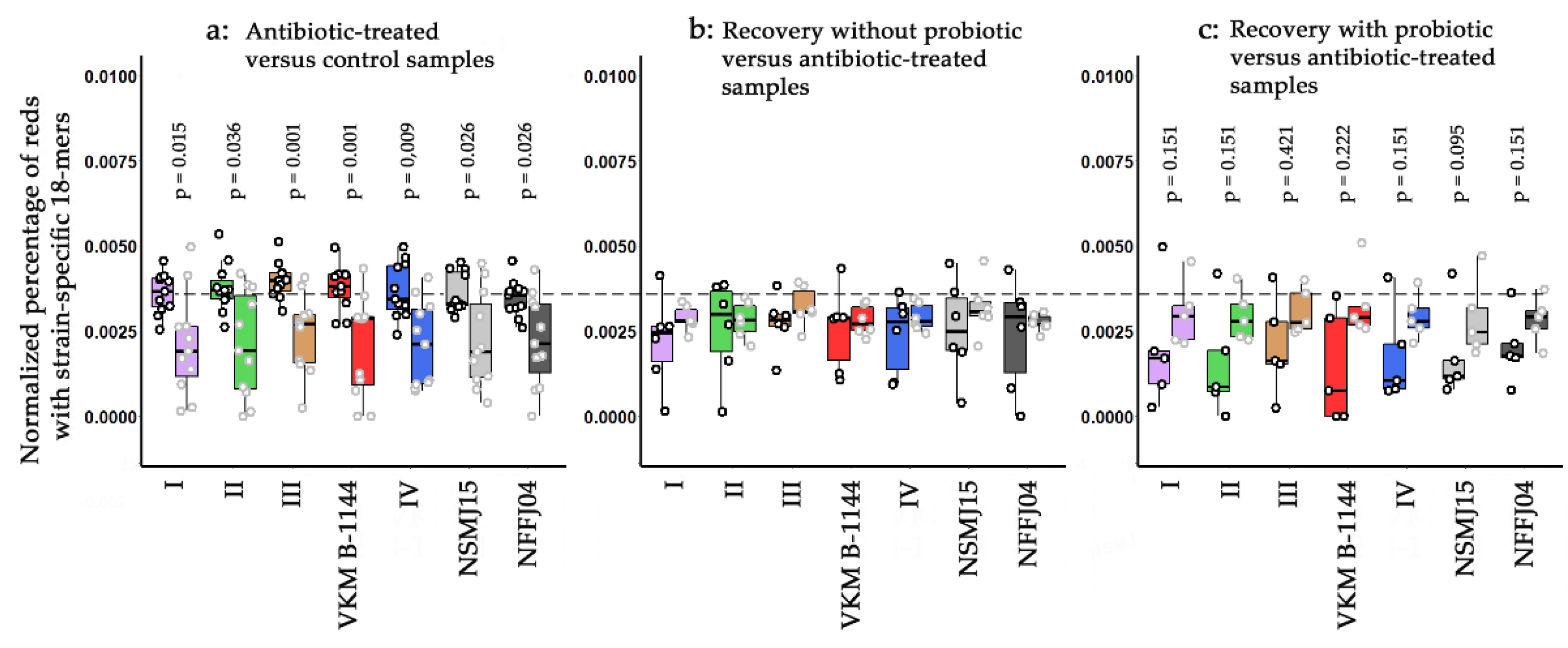
3.6. Strain-Specific Characterization of Antibiotic/Probiotic-Mediated Changes in Microbiomes
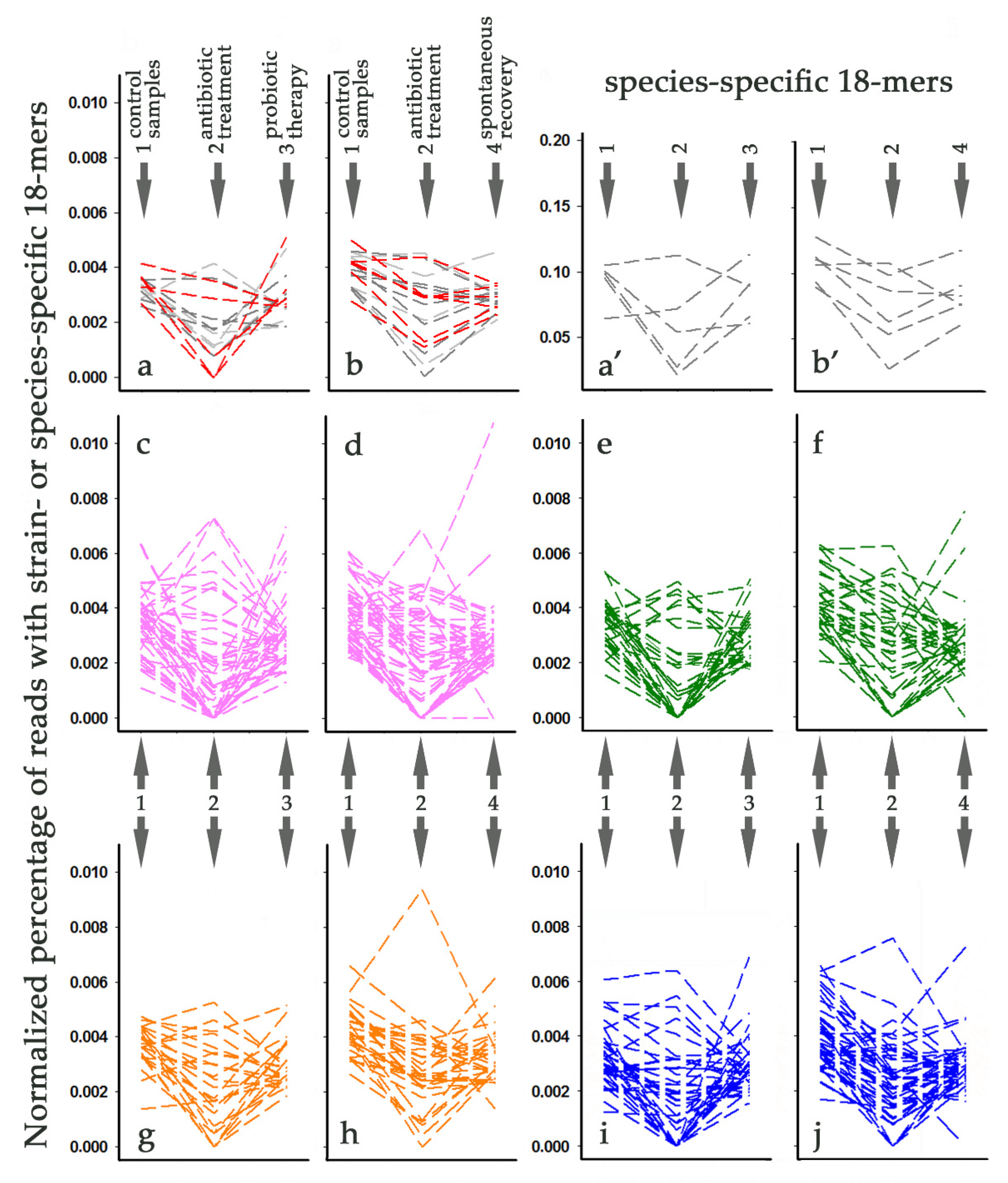
3.7. Tracking Individual Changes in the Presence of L. paracasei Strains in Response to Antibiotic Administration
3.8. Tracking Individual Changes in the Presence of L. paracasei Strains in Response to Probiotic Therapy and Spontaneous Recovery
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, Z.; Harris, H.M.; McCann, A.; Guo, C.; Argimon, S.; Zhang, W.; Yang, X.; Jeery, I.B.; Cooney, J.C.; Kagawa, T.F.; et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 2015, 6, 8322. [Google Scholar] [CrossRef]
- Dietrich, C.G.; Kottmann, T.; Alavi, M. Commercially available probiotic drinks containing Lactobacillus casei DN-114001 reduce antibiotic-associated diarrhea. World J. Gastroenterol. 2014, 20, 15837. [Google Scholar] [CrossRef]
- Collins, M.D.; Phillips, B.A.; Zanoni, P. Deoxyribonucleic acid homology studies of Lactobacillus casei, Lactobacillus paracasei sp. nov., subsp. paracasei and subsp. tolerans, and Lactobacillus rhamnosus sp. nov., comb. nov. Int. J. Syst. Bacteriol. 1989, 39, 105–108. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Panyukov, V.V.; Kiselev, S.S.; Ozoline, O.N. Unique k-mers as strain-specific barcodes for phylogenetic analysis and natural microbiome profiling. Int. J. Mol. Sci. 2020, 21, 944. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Dixit, O.V.A.; Vangchhia, B.; Condamine, B.; Dion, S.; Bridier-Nahmias, A.; Denamur, E.; Gordon, D. Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol. 2019, 21, 3107–3117. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Olier, M.; Hoede, C.; Diancourt, L.; Brisse, S.; Keroudean, M.; Glodt, J.; Picard, B.; Oswald, E.; Denamur, E. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 2011, 11, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Paramo, P.; Clermont, O.; Blanc-Potard, A.B.; Bui, H.; Le Bouguenec, C.; Denamur, E. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 2004, 21, 1085–1094. [Google Scholar] [CrossRef]
- Johnson, J.R.; Owens, K.L.; Clabots, C.R.; Weissman, S.J.; Cannon, S.B. Phylogenetic relationships among clonal groups of extraintestinal pathogenic Escherichia coli as assessed by multi-locus sequence analysis. Microbes Infect. 2006, 8, 1702–1713. [Google Scholar] [CrossRef]
- Gordon, D.M.; Clermont, O.; Tolley, H.; Denamur, E. Assigning Escherichia coli strains to phylogenetic groups: Multi-locus sequence typing versus the PCR triplex method. Environ. Microbiol. 2008, 10, 2484–2496. [Google Scholar] [CrossRef] [PubMed]
- Jaureguy, F.; Landraud, L.; Passet, V.; Diancourt, L.; Frapy, E.; Guigon, G.; Carbonnelle, E.; Lortholary, O.; Clermont, O.; Denamur, E.; et al. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genom. 2008, 9, 560. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Alba, J.M.; Baquero, F.; Canton, R.; Galan, J.C. Stratified reconstruction of ancestral Escherichia coli diversification. BMC Genom. 2019, 20, 936. [Google Scholar] [CrossRef]
- Abram, K.; Udaondo, Z.; Bleker, C.; Wanchai, V.; Wassenaar, T.M.; Robeson II, M.S.; Ussery, D.W. Mash-based analyses of Escherichia coli genomes reveal 14 distinct phylogroups. Commun. Biol. 2021, 4, 117. [Google Scholar] [CrossRef] [PubMed]
- Brulc, J.M.; Yeoman, C.J.; Wilson, M.K.; Berg Miller, M.E.; Jeraldo, P.; Jindou, S.; Goldenfeld, N.; Flint, H.J.; Lamed, R.; Borovok, I.; et al. Cellulosomics, a gene-centric approach to investigating the intraspecific diversity and adaptation of Ruminococcus flavefaciens within the rumen. PLoS ONE 2011, 6, e25329. [Google Scholar] [CrossRef]
- Rozman Grinberg, I.; Yin, G.; Borovok, Y.; Berg Miller, M.E.; Yeoman, C.J.; Dassa, B.; Yu, Z.; Mizrahi, I.; Flint, H.J.; Bayer, E.A.; et al. Functional phylotyping approach for assessing intraspecific diversity of Ruminococcus albus within the rumen microbiome. FEMS Microbiol. Lett. 2015, 362, 1–10. [Google Scholar] [CrossRef]
- Hungate, R.E. A roll tube method for cultivation of strict anaerobes. Methods Microbiol. 1969, 3, 117–132. [Google Scholar] [CrossRef]
- Alimolaei, M.; Golchin, M. An efficient DNA extraction method for Lactobacillus casei, a difficult-to-lyse bacterium. Int. J. Enteric Pathog. 2016, 4, e32472. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Gruning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef]
- Wattam, A.R.; Brettin, T.; Davis, J.J.; Gerdes, S.; Kenyon, R.; Machi, D.; Mao, C.; Olson, R.; Overbeek, R.; Pusch, G.D.; et al. Assembly, annotation, and comparative genomics in PATRIC, the all bacterial Bioinformatics Resource Center. Methods Mol. Biol. 2018, 1704, 79–101. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [PubMed]
- Elbourne, L.D.H.; Tetu, S.; Hassan, K.; Paulsen, I.T. TransportDB 2.0: A database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 2017, 45, D320–D324. [Google Scholar] [CrossRef] [PubMed]
- Panyukov, V.V.; Kiselev, S.S.; Alikina, O.V.; Nazipova, N.N.; Ozoline, O.N. Short unique sequences in bacterial genomes as strain- and species-specific signatures. Math. Biol. Bioinf. 2017, 12, 547–558. [Google Scholar] [CrossRef]
- Sørensen, T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content. K. Dan. Vidensk. Selskab. Biol. Krifter. 1948, 4, 1–34. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Rzhetsky, A.; Nei, M. Theoretical foundation of the minimum-evolution method of phylogenetic inference. Mol. Biol. Evol. 1993, 10, 1073–1095. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wang, M.; Wan, J.; Rong, H.; He, F.; Wang, H.; Zhou, J.; Cai, C.; Wang, Y.; Xu, R.; Yin, Z.; et al. Alterations in gut glutamate metabolism associated with changes in gut microbiota composition in children with autism spectrum disorder. mSystems 2019, 4, e00321–e18. [Google Scholar] [CrossRef]
- Vaughn, B.P.; Vatanen, T.; Allegretti, J.R.; Bai, A.; Xavier, R.J.; Korzenik, J.; Gevers, D.; Ting, A.; Robson, S.C.; Moss, A.C. Increased intestinal microbial diversity following fecal microbiota transplant for active Crohn’s disease. Inflamm. Bowel Dis. 2016, 22, 2182–2190. [Google Scholar] [CrossRef]
- Meslier, V.; Laiola, M.; Roager, H.M.; De Filippis, F.; Roume, H.; Quinquis, B.; Giacco, R.; Mennella, I.; Ferracane, R.; Pons, N.; et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut 2020, 69, 1258–1268. [Google Scholar] [CrossRef]
- Montassier, E.; Valdes-Mas, R.; Batard, E.; Zmora, N.; Dori-Bachash, M.; Suez, J.; Elinav, E. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat. Microbiol. 2021, 6, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Krueger, F. Babraham Bioinformatics—Trim Galore! Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 23 September 2021).
- Briggs, M. The classification of lactobacilli by means of physiological tests. J. Gen. Microbiol. 1953, 9, 234–248. [Google Scholar] [CrossRef][Green Version]
- Sharpe, M.E. A serological classification of lactobacilli. J. Gen. Microbiol. 1955, 12, 107–122. [Google Scholar] [CrossRef]
- Sharpe, M.E. 603. Development of serologically identified lactobacilli added to cheese made without starter. J. Dairy Res. 1955, 22, 374–376. [Google Scholar] [CrossRef]
- Tilden, E.B.; Svec, M. Further studies of a differential culture technique for estimations of acidogenic bacteria in saliva. II. Species of lactobacilli isolated from saliva and their distribution in a group of children. J. Dent. Res. 1952, 31, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Simelyte, E.; Rimpilainen, M.; Lehtonen, L.; Zhang, X.; Toivanen, P. Bacterial cell wall-induced arthritis: Chemical composition and tissue distribution of four lactobacillus strains. Infect. Immun. 2000, 68, 3535–3540. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.; Mayer, R.; Benziman, M. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 1991, 55, 35–58. [Google Scholar] [CrossRef]
- Lorca, G.L.; Font de Valdez, G.; Ljungh, A. Characterization of the protein-synthesis dependent adaptive acid tolerance response in Lactobacillus acidophilus. J. Mol. Microbiol. Biotechnol. 2002, 4, 525–532. [Google Scholar]
- Jiao, Y.; Cody, G.D.; Harding, A.K.; Wilmes, P.; Schrenk, M.; Wheeler, K.E.; Banfield, J.F.; Thelen, M.P. Characterization of extracellular polymeric substances from acidophilic microbial biofilms. Appl. Environ. Microbiol. 2010, 76, 2916–2922. [Google Scholar] [CrossRef]
- Wang, L.; Wise, M.J. Glycogen with short average chain length enhances bacterial durability. Naturwissenschaften 2011, 98, 719–729. [Google Scholar] [CrossRef]
- Kerns, J.C.; Arundel, C.; Chawla, L.S. Thiamin deficiency in people with obesity. Adv. Nutr. 2015, 6, 147–153. [Google Scholar] [CrossRef]
- Call, E.K.; Goh, Y.J.; Selle, K.; Klaenhammer, T.R.; O’Flaherty, S. Sortase-deficient lactobacilli: Effect on immunomodulation and gut retention. Microbiology 2015, 161, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Sarangi, A.N.; Mukherjee, M.; Bhowmick, S.; Tripathy, S. Reanalysis of Lactobacillus paracasei Lbs2 strain and large-scale comparative genomics places many strains into their correct taxonomic position. Microorganisms 2019, 7, 487. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C.; Sarda Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Goker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Smokvina, T.; Wels, M.; Polka, J.; Chervaux, C.; Brisse, S.; Boekhorst, J.; van Hylckama Vlieg, J.E.T.; Siezen, R.J. Lactobacillus paracasei comparative genomics: Toward species pan-genomic definition and exploitation of diversity. PLoS ONE 2013, 3, e68731. [Google Scholar] [CrossRef] [PubMed]
- Vuong, H.E.; Hsiao, E.Y. Emerging roles for the gut microbiome in autism spectrum disorder. Biol. Psychiatry 2017, 81, 411–423. [Google Scholar] [CrossRef]
- Lombardi, M.; Troisi, J. Gut reactions: How far are we from understanding and manipulating the microbiota complexity and the interaction with its host? Lessons from autism spectrum disorder studies. Nutrients 2021, 13, 3492. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tian, Y.; Cao, Y.; Li, J.; Guo, H.; Su, Y.; Tian, Y.; Wang, C.; Wang, T.; Zhang, L. Probiotic properties of Lactobacillus paracasei subsp. paracasei L1 and its growth performance-promotion in chicken by improving the intestinal microflora. Front. Physiol. 2019, 10, 937. [Google Scholar] [CrossRef] [PubMed]
- Verdu, E.F.; Bercik, P.; Verma-Gandhu, M.; Huang, X.X.; Blennerhassett, P.; Jackson, W.; Mao, Y.; Wang, L.; Rochat, F.; Collins, S.M. Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut 2006, 55, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Yamazaki, T.; Ohshio, K.; Sugamata, M.; Yoshikawa, M.; Kanauchi, O.; Morita, Y. A specific strain of lactic acid bacteria, Lactobacillus paracasei, inhibits inflammasome activation in vitro and prevents inflammation-related disorders. J. Immunol. 2020, 205, 811–821. [Google Scholar] [CrossRef]
- Chondrou, P.; Karapetsas, A.; Kiousi, D.E.; Vasileiadis, S.; Ypsilantis, P.; Botaitis, S.; Alexopoulos, A.; Plessas, S.; Bezirtzoglou, E.; Galanis, A. Assessment of the immunomodulatory properties of the probiotic strain Lactobacillus paracasei K5 in vitro and in vivo. Microorganisms 2020, 8, 709. [Google Scholar] [CrossRef] [PubMed]
- Buchholz, U.; Bernard, H.; Werber, D.; Bohmer, M.M.; Remschmidt, C.; Wilking, H.; Delere, Y.; an der Heiden, M.; Adlhoch, C.; Dreesman, J.; et al. German outbreak of Escherichia coli O104:H4 associated with sprouts. N. Engl. J. Med. 2011, 365, 1763–1770. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.-Z.; et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 2018, 174, 1388–1405. [Google Scholar] [CrossRef]
| N | Name of Strain | Phylo- Group | Number of Unique 18-Mers Absent in the Genomes of Other Genera and Plasmids | N | Name of Strain | Phylo- Group | Number of Unique 18-Mers Absent in the Genomes of Other Genera and Plasmids |
|---|---|---|---|---|---|---|---|
| L. paracasei | 40 | EG9 | IV | 1,243,018 | |||
| 1 | MGB0747 | I | 1,342,841 | 41 | N1115 | IV | 1,235,840 |
| 2 | MGB0761 | I | 1,342,893 | 42 | KL1 | IV | 1,234,653 |
| 3 | L9 | I | 1,326,474 | 43 | JCM 8130 | IV | 1,286,351 |
| 4 | MGYG-HGUT-02388 | I | 1,326,474 | 44 | MGB0734 | - | 1,327,753 |
| 5 | NJ | I | 1,335,035 | 45 | NFFJ04 | - | 1,287,217 |
| 6 | 347–16 | I | 1,354,652 | 46 | VKM B-1144 | - | 1,274,372 |
| 7 | CACC 566 | I | 1,354,718 | 47 | NSMJ15 | - | 1,208,720 |
| 8 | Lp02 | I | 1,307,893 | L. casei | |||
| 9 | IJH-SONE68 | I | 1,316,002 | 48 | ATCC 393 | 1,216,652 | |
| 10 | IIA | I | 1,334,702 | 49 | LC5 | 1,398,284 | |
| 11 | 10266 | I | 1,306,215 | L. zeae | |||
| 12 | ZY-1 | I | 1,354,745 | 50 | CECT 9104 (formerly L. casei) | 1,326,946 | |
| 13 | LC355 | I | 1,311,398 | L. manihotivorans | |||
| 14 | SRCM103299 | I | 1,333,804 | 51 | LM010 | 1,381,360 | |
| 15 | HDS-01 | I | 1,310,186 | L. rhamnosus | |||
| 16 | HD1–7 | I | 1,310,389 | 52 | 4B15 | 1,357,067 | |
| 17 | Zhang | II | 1,267,650 | 53 | GG | 1,357,107 | |
| 18 | LOCK919 | II | 1,373,093 | 54 | BIO6870 | 1,356,905 | |
| 19 | ZFM54 | II | 1,310,426 | 55 | LR-B1 | 1,357,098 | |
| 20 | TK1501 | II | 1,289,097 | 56 | WQ2 | 1,351,500 | |
| 21 | 12A (annotated as L. casei) | II | 1,301,297 | 57 | LRB | 1,333,164 | |
| 22 | 8700:2 | II | 1,319,012 | 58 | hsryfm 1301 | 1,352,189 | |
| 23 | Lpc10 | II | 1,345,201 | 59 | DSM 14870 | 1,346,181 | |
| 24 | MGB0245 | III | 1,331,017 | 60 | IDCC 3201 | 1,323,375 | |
| 25 | MGB0625 | III | 1,391,072 | 61 | BFE5264 | 1,338,512 | |
| 26 | TK-P4A | III | 1,352,170 | 62 | 1–0320 | 1,350,218 | |
| 27 | TMW 1–1434 | III | 1,323,010 | 63 | LR5 | 1,370,793 | |
| 28 | IBB3423 | III | 1,400,224 | 64 | LOCK900 | 1,332,726 | |
| 29 | CBA3611 | III | 1,316,237 | 65 | Pen | 1,329,920 | |
| 30 | LC2W | III | 1,316,047 | 66 | BPL5 | 1,392,367 | |
| 31 | AO356 (7112–2) | III | 1,310,307 | 67 | ATCC 8530 | 1,362,386 | |
| 32 | BL23 (formerly L. casei) | III | 1,337,551 | 68 | Lc 705 | 1,377,678 | |
| 33 | W56 (formerly L. casei) | III | 1,337,353 | 69 | ASCC 290 | 1,357,004 | |
| 34 | BD-II | III | 1,337,137 | 70 | SCT-10–10-60 | 1,377,318 | |
| 35 | TCS | III | 1,337,717 | 71 | ATCC 11443 | 1,377,473 | |
| 36 | TD 062 | IV | 1,129,425 | 72 | LOCK908 | 1,377,399 | |
| 37 | FAM18149 | IV | 1,162,779 | 73 | NCTC13764 | 1,377,359 | |
| 38 | CAUH35 | IV | 1,187,210 | 74 | BIO5326 | 1,377,176 | |
| 39 | ATCC 334 | IV | 1,229,640 | 75 | NCTC13710 | 1,377,380 | |
| N | Strain 1 | PG 2 | Number of | N | Strain | Number of | |||
|---|---|---|---|---|---|---|---|---|---|
| Species- Specific 18-Mers | Strain-Specific 18-Mers | PG | Species- Specific 18-Mers | Strain-Specific 18-Mers | |||||
| 1 | MGB0747 | I | 1,201,260 | 83 | 25 | MGB0245 | III | 1,184,705 | 50,602 |
| 2 | MGB0761 | I | 1,201,306 | 81 | 26 | MGB0625 | III | 1,228,586 | 48,092 |
| 3 | L9 | I | 1,186,502 | 0 | 27 | TK-P4A | III | 1,212,483 | 86,180 |
| 4 | MGYG-HGUT | I | 1,186,506 | 0 | 28 | TMW 1.1434 | III | 1,199,252 | 25,090 |
| 5 | NJ | I | 1,188,566 | 17,470 | 29 | IBB3423 | III | 1,250,212 | 38,367 |
| 6 | 347–16 | I | 1,207,017 | 130 | 30 | CBA3611 | III | 1,188,329 | 89 |
| 7 | CACC 566 | I | 1,207,087 | 216 | 31 | LC2W | III | 1,188,152 | 38 |
| 8 | Lp02 | I | 1,177,634 | 11,840 | 32 | subsp. 7112–2 | III | 1,182,816 | 121 |
| 9 | IJH-SONE68 | I | 1,183,896 | 26,679 | 33 | BL23 | III | 1,199,366 | 427 |
| 10 | IIA | I | 1,198,699 | 16,607 | 34 | W56 | III | 1,199,237 | 571 |
| 11 | subsp 10266 | I | 1,172,488 | 25,784 | 35 | BD-II | III | 1,199,063 | 144 |
| 12 | ZY-1 | I | 1,216,121 | 46,665 | 36 | TCS | III | 1,199,506 | 244 |
| 13 | LC355 | I | 1,179,984 | 17,903 | 37 | NFFJ04 | - | 1,155,507 | 71,319 |
| 14 | SRCM 103299 | I | 1,194,448 | 23,021 | 38 | NSMJ15 | - | 1,060,651 | 77,519 |
| 15 | HD1.7 | I | 1,201,260 | 83 | 39 | VKM B-1144 | - | 1,100,024 | 73,388 |
| 16 | HDS-01 | I | 1,201,306 | 81 | 40 | TD 062 | IV | 1,000,877 | 22,825 |
| 17 | MGB0734 | - | 1,178,026 | 65,524 | 41 | FAM18149 | IV | 1,028,072 | 28,405 |
| 18 | Zhang | II | 1,143,566 | 8885 | 42 | CAUH35 | IV | 1,052,575 | 38,375 |
| 19 | LOCK919 | II | 1,230,630 | 28,394 | 43 | ATCC 334 | IV | 1,143,681 | 38,932 |
| 20 | ZFM54 | II | 1,169,160 | 22,145 | 44 | EG9 | IV | 1,093,799 | 37,831 |
| 21 | TK1501 | II | 1,154,098 | 19,871 | 45 | N1115 | IV | 1,105,817 | 25,748 |
| 22 | 12A | II | 1,103,714 | 28,753 | 46 | KL1 | IV | 1,165,966 | 20,265 |
| 23 | subsp. 8700:2 | II | 1,184,388 | 23,564 | 47 | JCM 8130 | IV | 1,134,441 | 43,983 |
| 24 | Lpc10 | II | 1,179,984 | 39,486 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frolova, M.; Yudin, S.; Makarov, V.; Glazunova, O.; Alikina, O.; Markelova, N.; Kolzhetsov, N.; Dzhelyadin, T.; Shcherbakova, V.; Trubitsyn, V.; et al. Lacticaseibacillus paracasei: Occurrence in the Human Gut Microbiota and K-Mer-Based Assessment of Intraspecies Diversity. Life 2021, 11, 1246. https://doi.org/10.3390/life11111246
Frolova M, Yudin S, Makarov V, Glazunova O, Alikina O, Markelova N, Kolzhetsov N, Dzhelyadin T, Shcherbakova V, Trubitsyn V, et al. Lacticaseibacillus paracasei: Occurrence in the Human Gut Microbiota and K-Mer-Based Assessment of Intraspecies Diversity. Life. 2021; 11(11):1246. https://doi.org/10.3390/life11111246
Chicago/Turabian StyleFrolova, Maria, Sergey Yudin, Valentin Makarov, Olga Glazunova, Olga Alikina, Natalia Markelova, Nikolay Kolzhetsov, Timur Dzhelyadin, Viktoria Shcherbakova, Vladimir Trubitsyn, and et al. 2021. "Lacticaseibacillus paracasei: Occurrence in the Human Gut Microbiota and K-Mer-Based Assessment of Intraspecies Diversity" Life 11, no. 11: 1246. https://doi.org/10.3390/life11111246
APA StyleFrolova, M., Yudin, S., Makarov, V., Glazunova, O., Alikina, O., Markelova, N., Kolzhetsov, N., Dzhelyadin, T., Shcherbakova, V., Trubitsyn, V., Panyukov, V., Zaitsev, A., Kiselev, S., Shavkunov, K., & Ozoline, O. (2021). Lacticaseibacillus paracasei: Occurrence in the Human Gut Microbiota and K-Mer-Based Assessment of Intraspecies Diversity. Life, 11(11), 1246. https://doi.org/10.3390/life11111246









