Cyclomodulins and Hemolysis in E. coli as Potential Low-Cost Non-Invasive Biomarkers for Colorectal Cancer Screening
Abstract
1. Introduction
1.1. Roles of Cyclomodulins
1.1.1. Colibactin
1.1.2. Cytotoxic Necrotizing Factor
1.1.3. Cytolethal Distending Toxin
2. Materials and Methods
2.1. Patients and Clinical Material
2.2. Samples Processing
2.3. Genetic Analysis of E. coli Isolates
2.4. Statistical Analysis
3. Results
3.1. E. coli Culture Results
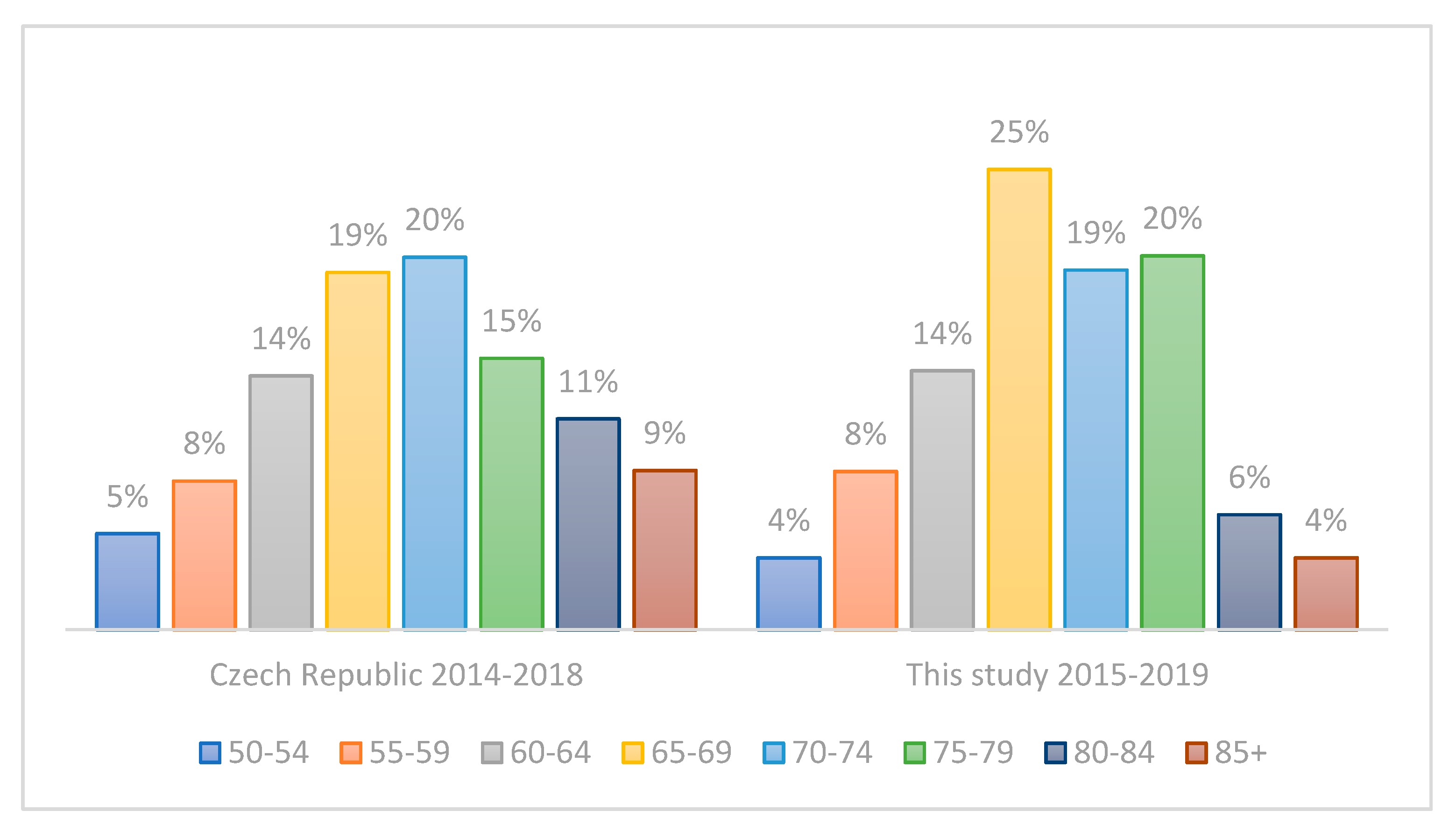
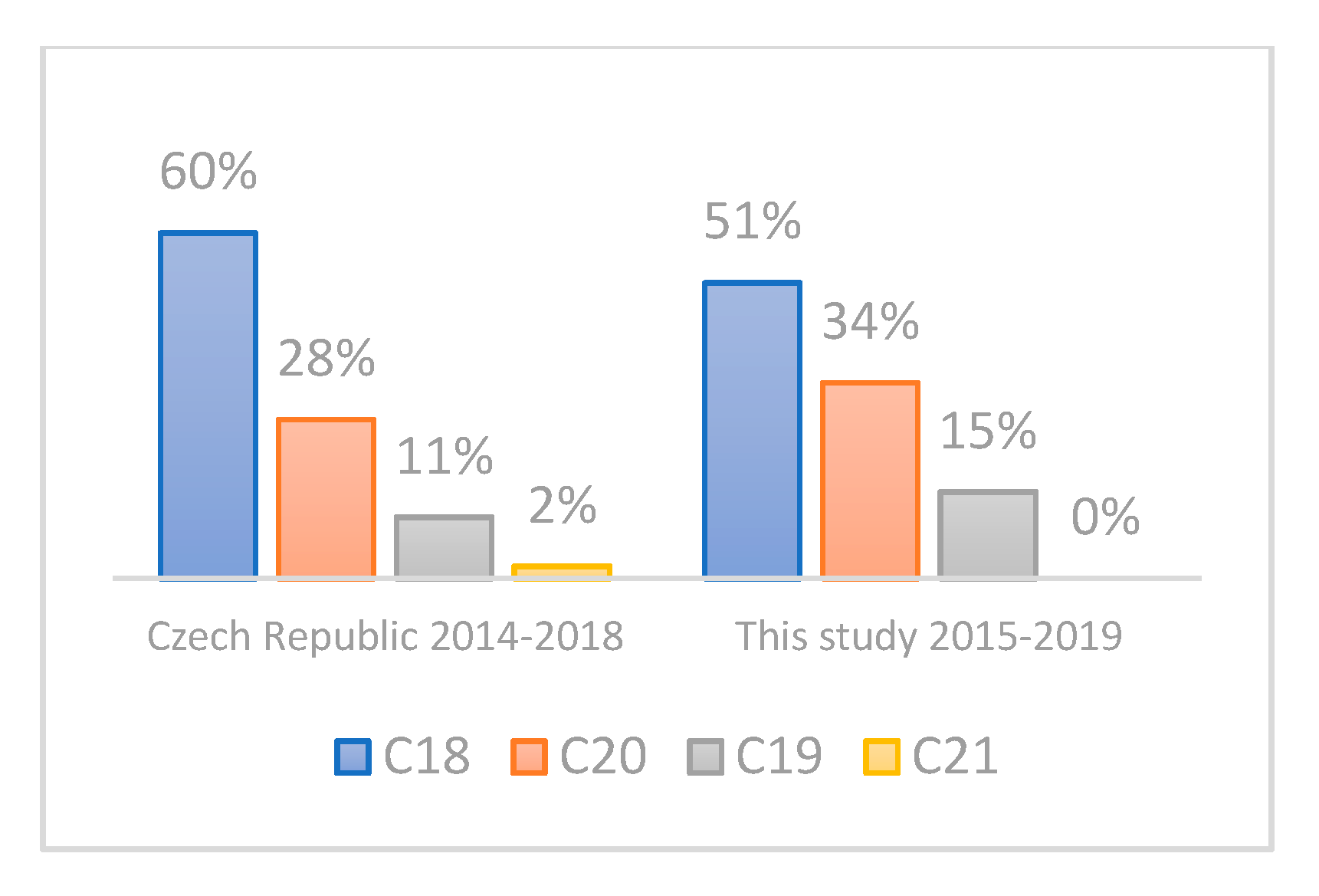
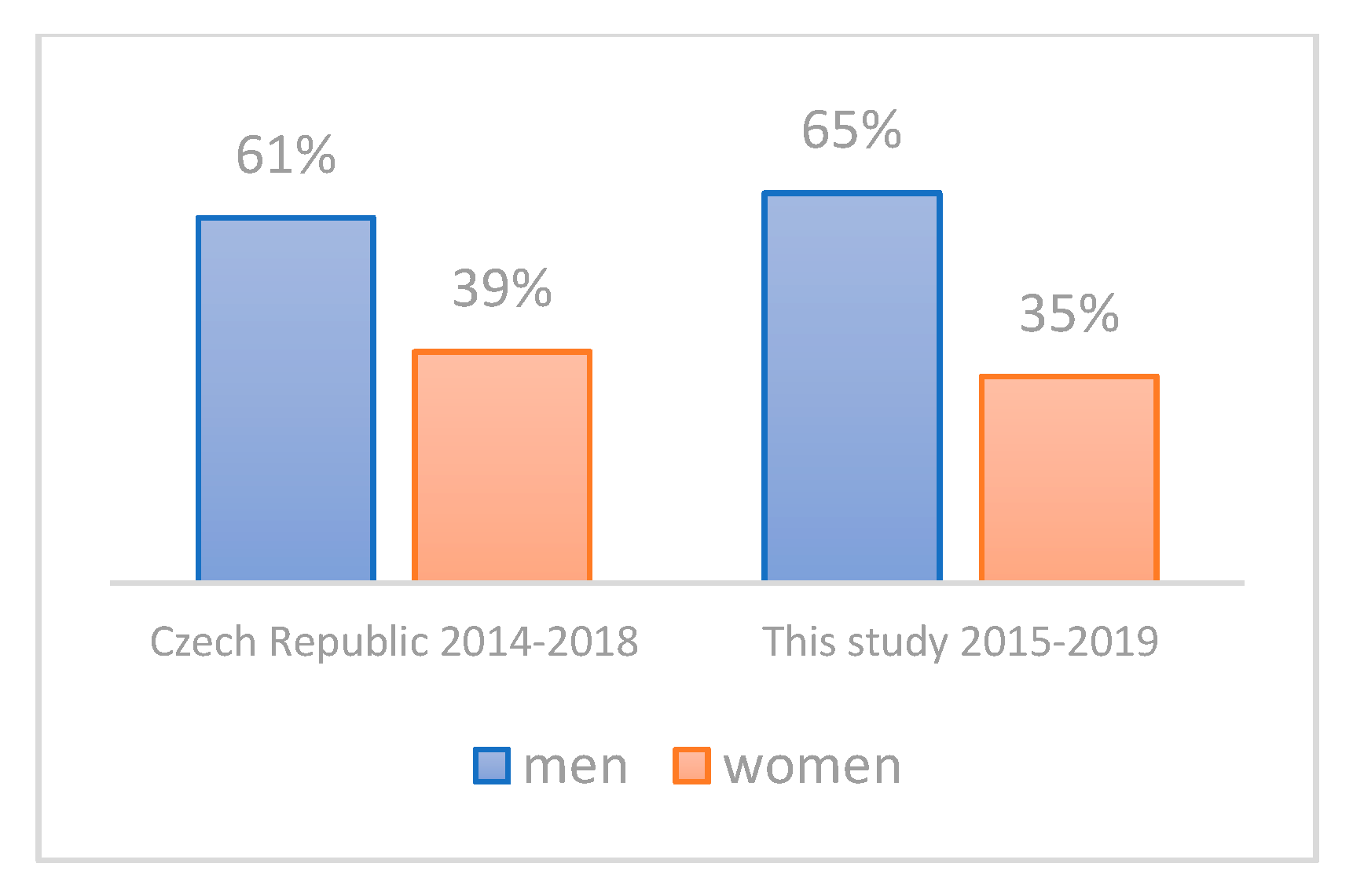
3.2. Evaluation of the Representativeness of the CRC Cohort
3.3. Prevalence of Cyclomodulin Genes and Hemolysis and Implications for Usefulness in Screening
3.4. Deciphering the Different Burden of Distinct E. coli Isolates in CRC Patient versus Controls
3.5. Evaluation of Potential Benefit of Toxin Screening in Different Age Groups, in Males versus Females, and in Different Disease Stages
4. Discussion
4.1. Colibactin Established as the Most Abundant Potential CRC Marker
4.2. Other Cyclomodulin Genes Are Detected Less Frequently and Are Mostly Linked to the Presence of Colibactin
4.3. Males Should Benefit from Toxin Detection in CRC Screening More Than Women
4.4. Cyclomodulin-Producing E. coli May Represent Harmful and Possibly Preventable Newcomers to Colon Microbiota
4.5. Open Questions and Perspectives
5. Conclusions
- Our data confirm the association of clb+, cnf+ and hemolytic E. coli strains with CRC.
- Cultures from rectal swab followed by colony PCR may represent a viable and economical alternative to non-culture detection of toxigenic E. coli in stool samples, provided that its sensitivity be successfully increased.
- It needs to be established whether the detection of cyclomodulin-producing E. coli does increase the sensitivity of current non-invasive CRC screening strategies. Clinical trials that would encompass simultaneous FIT and cyclomodulin-producing E. coli detection in the general population are urgently needed.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.L.; Freeman, D.J.; Pleasance, S.; Watson, P.; Moore, R.A.; Cochrane, K.; Allen-Vercoe, E.; Holt, R.A. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 2013, 1, 16. [Google Scholar] [CrossRef]
- Compare, D.; Nardone, G. The bacteria-hypothesis of colorectal cancer: Pathogenetic and therapeutic implications. Transl. Gastrointest. Cancer 2014, 3, 44–53. [Google Scholar] [CrossRef]
- Bonnet, M.; Buc, E.; Sauvanet, P.; Darcha, C.; Dubois, D.; Pereira, B.; Déchelotte, P.; Bonnet, R.; Pezet, D.; Darfeuille-Michaud, A. Colonization of the human gut by E. coli and colorectal cancer risk. Clin. Cancer Res. 2014, 20, 859–867. [Google Scholar] [CrossRef]
- Viljoen, K.S.; Dakshinamurthy, A.; Goldberg, P.; Blackburn, J.M. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between Fusobacterium spp., Enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS ONE 2015, 10, e0119462. [Google Scholar] [CrossRef] [PubMed]
- Zamani, S.; Taslimi, R.; Sarabi, A.; Jasemi, S.; Sechi, L.A.; Feizabadi, M.M. Enterotoxigenic Bacteroides fragilis: A possible etiological candidate for bacterially-induced colorectal precancerous and cancerous lesions. Front. Cell. Infect. Microbiol. 2020, 9, 449. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Sinha, R.; Pei, Z.; Dominianni, C.; Wu, J.; Shi, J.; Goedert, J.J.; Hayes, R.B.; Yang, L. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 1907–1911. [Google Scholar] [CrossRef]
- Eklöf, V.; Löfgren-Burström, A.; Zingmark, C.; Edin, S.; Larsson, P.; Karling, P.; Alexeyev, O.; Rutegård, J.; Wikberg, M.L.; Palmqvist, R. Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer 2017, 141, 2528–2536. [Google Scholar] [CrossRef]
- Flemer, B.; Warren, R.D.; Barrett, M.P.; Cisek, K.; Das, A.; Jeffery, I.B.; Hurley, E.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 2018, 67, 1454–1463. [Google Scholar] [CrossRef]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef]
- Malagón, M.; Ramió-Pujol, S.; Serrano, M.; Amoedo, J.; Oliver, L.; Bahí, A.; Miquel-Cusachs, J.O.; Ramirez, M.; Queralt-Moles, X.; Gilabert, P.; et al. New fecal bacterial signature for colorectal cancer screening reduces the fecal immunochemical test false-positive rate in a screening population. PLoS ONE 2020, 15, e0243158. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Jiao, N.; Zhu, R.; Zhang, Y.; Wu, D.; Wang, A.J.; Fang, S.; Tao, L.; Li, Y.; Cheng, S.; et al. Identification of microbial markers across populations in early detection of colorectal cancer. Nat. Commun. 2021, 12, 3063. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.Q.; Li, T.; Nakatsu, G.; Chen, Y.-X.; Yau, T.O.; Chu, E.; Wong, S.; Szeto, C.H.; Ng, S.C.; Chan, F.K.L.; et al. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut 2020, 69, 1248–1257. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, A.; Neefs, I.; Hoeck, S.; Peeters, M.; Van Hal, G. Towards novel non-invasive colorectal cancer screening methods: A comprehensive review. Cancers 2021, 13, 1820. [Google Scholar] [CrossRef]
- Lax, A.J.; Thomas, W. How bacteria could cause cancer: One step at a time. Trends Microbiol. 2002, 10, 293–299. [Google Scholar] [CrossRef]
- Nougayrede, J.-P. Escherichia coli Induces DNA Double-Strand Breaks in Eukaryotic Cells. Science 2006, 313, 848–851. [Google Scholar] [CrossRef]
- Cuevas-Ramos, G.; Petit, C.R.; Marcq, I.; Boury, M.; Oswald, E.; Nougayrède, J.P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11537–11542. [Google Scholar] [CrossRef]
- Putze, J.; Hennequin, C.; Nougayrède, J.-P.; Zhang, W.; Homburg, S.; Karch, H.; Bringer, M.-A.; Fayolle, C.; Carniel, E.; Rabsch, W.; et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect. Immun. 2009, 77, 4696–4703. [Google Scholar] [CrossRef] [PubMed]
- Bossuet-Greif, N.; Dubois, D.; Petit, C.; Tronnet, S.; Martin, P.; Bonnet, R.; Oswald, E.; Nougayrède, J.P. Escherichia coli ClbS is a colibactin resistance protein. Mol. Microbiol. 2016, 99, 897–908. [Google Scholar] [CrossRef]
- Wami, H.; Wallenstein, A.; Sauer, D.; Stoll, M.; von Bünau, R.; Oswald, E.; Müller, R.; Dobrindt, U. Diversity and prevalence of colibactin- and yersiniabactin encoding mobile genetic elements in enterobacterial populations: Insights into evolution and co-existence of two bacterial secondary metabolite determinants. bioRxiv 2021, 49. [Google Scholar] [CrossRef]
- Xue, M.; Kim, C.S.; Healy, A.R.; Wernke, K.M.; Wang, Z.; Frischling, M.C.; Shine, E.E.; Wang, W.; Herzon, S.B.; Crawford, J.M. Structure elucidation of colibactin and its DNA cross-links. Science 2019, 365, 6457. [Google Scholar] [CrossRef]
- Li, Z.-R.; Li, J.; Cai, W.; Lai, J.Y.H.; McKinnie, S.M.K.; Zhang, W.-P.; Moore, B.S.; Zhang, W.; Qian, P.-Y. Macrocyclic colibactin induces DNA double-strand breaks via copper-mediated oxidative cleavage. Nat. Chem. 2019, 11, 880–889. [Google Scholar] [CrossRef]
- Wilson, M.R.; Jiang, Y.; Villalta, P.W.; Stornetta, A.; Boudreau, P.D.; Carrá, A.; Brennan, C.A.; Chun, E.; Ngo, L.; Samson, L.D.; et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science 2019, 363, eaar7785. [Google Scholar] [CrossRef]
- Xue, M.; Wernke, K.M.; Herzon, S.B. Depurination of colibactin-derived interstrand cross-links. Biochemistry 2020, 59, 892–900. [Google Scholar] [CrossRef]
- Dalmasso, G.; Cougnoux, A.; Delmas, J.; Darfeuille-Michaud, A.; Bonnet, R. The bacterial genotoxin colibactin promotes colon tumor growth by modifying the tumor microenvironment. Gut Microbes 2014, 5, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Kusibab, P.J.D.; Berger, H.; Battistini, F.; Bouwman, B.A.M.; Iftekhar, A.; Katainen, R.; Crosetto, N.; Orozco, M.; Aaltonen, L.A. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat Med. 2020, 26, 1063–1069. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Rosendahl Huber, A.; van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 2020, 580, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Muzny, D.M.; Bainbridge, M.N.; Chang, K.; Dinh, H.H.; Drummond, J.A.; Fowler, G.; Kovar, C.L.; Lewis, L.R.; Morgan, M.B.; Newsham, I.F.; et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Iftekhar, A.; Berger, H.; Bouznad, N.; Heuberger, J.; Boccellato, F.; Dobrindt, U.; Hermeking, H.; Sigal, M.; Meyer, T.F. Genomic aberrations after short-term exposure to colibactin-producing E. coli transform primary colon epithelial cells. Nat. Commun. 2021, 12, 1003. [Google Scholar] [CrossRef]
- Aghabozorgi, A.S.; Ebrahimi, R.; Bahiraee, A.; Tehrani, S.S.; Nabizadeh, F.; Setayesh, L.; Jafarzadeh-Esfehani, R.; Ferns, G.A.; Avan, A.; Rashidi, Z. The genetic factors associated with Wnt signaling pathway in colorectal cancer. Life Sci. 2020, 256, 118006. [Google Scholar] [CrossRef]
- Iyadorai, T.; Mariappan, V.; Vellasamy, K.M.; Wanyiri, J.W.; Roslani, A.C.; Lee, G.K.; Sears, C.; Vadivelu, J. Prevalence and association of pks+ Escherichia coli with colorectal cancer in patients at the University Malaya Medical Centre, Malaysia. PLoS ONE 2020, 15, e0228217. [Google Scholar] [CrossRef]
- Oswald, E.; De Rycke, J.; Lintermans, P.; Van Muylem, K.; Mainil, J.; Daube, G.; Pohl, P. Virulence factors associated with cytotoxic necrotizing factor type two in bovine diarrheic and septicemic strains of Escherichia coli. J. Clin. Microbiol. 1991, 29, 2522–2527. [Google Scholar] [CrossRef]
- Bouzari, S.; Oloomi, M.; Oswald, E. Detection of the cytolethal distending toxin locus cdtB among diarrheagenic Escherichia coli isolates from humans in Iran. Res. Microbiol. 2005, 156, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Hilali, F.; Ruimy, R.; Saulnier, P.; Barnabé, C.; Lebouguénec, C.; Tibayrenc, M.; Andremont, A. Prevalence of virulence genes and clonality in Escherichia coli strains that cause bacteremia in cancer patients. Infect. Immun. 2000, 68, 3983–3989. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bisicchia, R.; Ciammarughi, R.; Caprioli, A.; Falbo, V.; Ruggeri, F.M. Toxin production and haemagglutination in strains of Escherichia coli from diarrhoea in Brescia, Italy. J. Hyg. 1985, 95, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Alonso, P.; Blanco, J.; Blanco, M.; Gonzalez, E.A. Frequent production of toxins by Escherichia coli strains isolated from human urinary tract infections: Relation with haemagglutination. FEMS Microbiol. Lett. 1987, 48, 391–396. [Google Scholar] [CrossRef]
- Falzano, L.; Fiorentini, P.; Boquet, P.; Donelli, G. Interaction of Escherichia coli cytotoxic necrotizing factor type 1 (cnf1) with cultured cells. J. Chem. Inf. Model. 1993, 53, 1689–1699. [Google Scholar] [CrossRef]
- Caprioli, A.; Falbo, V.; Roda, L.G.; Ruggeri, F.M.; Zona, C. Partial purification and characterization of an Escherichia coli toxic factor that induces morphological cell alterations. Infect. Immun. 1983, 39, 1300–1306. [Google Scholar] [CrossRef]
- De Rycke, J.; Phan-Thanh, L.; Bernard, S. Immunochemical identification and biological characterization of cytotoxic necrotizing factor from Escherichia coli. J. Clin. Microbiol. 1989, 27, 983–988. [Google Scholar] [CrossRef]
- Fiorentini, C.; Fabbri, A.; Flatau, G.; Donelli, G.; Matarrese, P.; Lemichez, E.; Falzano, L.; Boquet, P. Escherichia coli cytotoxic necrotizing factor 1 (CNF1), a toxin that activates the Rho GTPase. J. Biol. Chem. 1997, 272, 19532–19537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Aung, K.M.; Uhlin, B.E.; Wai, S.N. Reversible senescence of human colon cancer cells after blockage of mitosis/cytokinesis caused by the CNF1 cyclomodulin from Escherichia coli. Sci. Rep. 2018, 8, 17780. [Google Scholar] [CrossRef]
- Mehdipour, S.; Doosti, A.; Ghasemi Dehkordi, P. Detection of cytolethal distending toxin (cdt) and cytotoxic necrotizing factor (cnf) genes among Escherichia coli isolates from Iranian sheep carcasses. Comp. Clin. Path. 2012, 21, 1683–1688. [Google Scholar] [CrossRef]
- Pickett, C.L.; Lee, R.B.; Eyigor, A.; Elitzur, B.; Fox, E.M.; Strockbine, N.A. Patterns of Variations in Escherichia coli Strains that Produce Cytolethal Distending Toxin. Infect. Immun. 2004, 72, 684–690. [Google Scholar] [CrossRef]
- Kurnick, S.A.; Mannion, A.J.; Feng, Y.; Madden, C.M.; Chamberlain, P.; Fox, J.G. Genotoxic Escherichia coli Strains Encoding Colibactin, Cytolethal Distending Toxin, and Cytotoxic Necrotizing Factor in Laboratory Rats. Comp. Med. 2019, 69, 103–113. [Google Scholar] [CrossRef]
- Elwell, C.A.; Dreyfus, L.A. DNase I homologous residues in CdtB are critical for cytolethal distending toxin-mediated cell cycle arrest. Mol. Microbiol. 2000, 37, 952–963. [Google Scholar] [CrossRef]
- Fedor, Y.; Vignard, J.; Nicolau-Travers, M.L.; Boutet-Robinet, E.; Watrin, C.; Salles, B.; Mirey, G. From single-strand breaks to double-strand breaks during S-phase: A new mode of action of the Escherichia coli Cytolethal Distending Toxin. Cell. Microbiol. 2013, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, C.A.; Balbo, P.B.; Pesci, E.C.; Cottle, D.L.; Mirabito, P.M.; Pickett, C.L. Campylobacter jejuni cytolethal distending toxin causes a G2-phase cell cycle block. Infect. Immun. 1998, 66, 1934–1940. [Google Scholar] [CrossRef]
- Pérès, S.Y.; Marchès, O.; Daigle, F.; Nougayrède, J.P.; Hérault, F.; Tasca, C.; De Rycke, J.; Oswald, E. A new cytolethal distending toxin (CDT) from Escherichia coli producing CNF2 blocks HeLa cell division in G2/M phase. Mol. Microbiol. 1997, 24, 1095–1107. [Google Scholar] [CrossRef]
- Jinadasa, R.N.; Bloom, S.E.; Weiss, R.S.; Duhamel, G.E. Cytolethal distending toxin: A conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology 2011, 157, 1851–1875. [Google Scholar] [CrossRef] [PubMed]
- Fahrer, J.; Huelsenbeck, J.; Jaurich, H.; Dörsam, B.; Frisan, T.; Eich, M.; Roos, W.P.; Kaina, B.; Fritz, G. Cytolethal distending toxin (CDT) is a radiomimetic agent and induces persistent levels of DNA double-strand breaks in human fibroblasts. DNA Repair 2014, 18, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Shiloh, Y. ATM and ATR: Networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 2001, 11, 71–77. [Google Scholar] [CrossRef]
- Li, L.Q.; Sharipo, A.; Chaves-Olarte, E.; Masucci, M.G.; Levitsky, V.; Thelestam, M.; Frisan, T. The Haemophilus ducreyi cytolethal distending toxin activates sensors of DNA damage and repair complexes in proliferating and non-proliferating cells. Cell. Microbiol. 2002, 4, 87–99. [Google Scholar] [CrossRef]
- He, Z.; Gharaibeh, R.Z.; Newsome, R.C.; Pope, J.L.; Dougherty, M.W.; Tomkovich, S.; Pons, B.; Mirey, G.; Vignard, J.; Hendrixson, D.R.; et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 2019, 68, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Guidi, R.; Guerra, L.; Levi, L.; Stenerlöw, B.; Fox, J.G.; Josenhans, C.; Masucci, M.G.; Frisan, T. Chronic exposure to the cytolethal distending toxins of Gram-negative bacteria promotes genomic instability and altered DNA damage response. Cell Microbiol. 2013, 15, 98–113. [Google Scholar] [CrossRef]
- Graillot, V.; Dormoy, I.; Dupuy, J.; Shay, J.W.; Huc, L.; Mirey, G.; Vignard, J. Genotoxicity of Cytolethal Distending Toxin (CDT) on isogenic human colorectal cell lines: Potential promoting effects for colorectal carcinogenesis. Front. Cell. Infect. Microbiol. 2016, 6, 34. [Google Scholar] [CrossRef]
- Johnson, J.R.; Johnston, B.; Kuskowski, M.A.; Nougayrede, J.P.; Oswald, E. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J. Clin. Microbiol. 2008, 46, 3906–3911. [Google Scholar] [CrossRef] [PubMed]
- Pass, M.A.; Odedra, R.; Batt, R.M. Multiplex PCRs for identification of Escherichia coli virulence genes. J. Clin. Microbiol. 2000, 38, 2001–2004. [Google Scholar] [CrossRef]
- Tóth, I.; Hérault, F.; Beutin, L.; Oswald, E. Production of cytolethal distending toxins by pathogenic Escherichia coli strains isolated from human and animal sources: Establishment of the existence of a new cdt variant (type IV). J. Clin. Microbiol. 2003, 41, 4285–4291. [Google Scholar] [CrossRef]
- Morgan, R.N.; Saleh, S.E.; Farrag, H.A.; Aboulwafa, M.M. Prevalence and pathologic effects of colibactin and cytotoxic necrotizing factor-1 (Cnf 1) in Escherichia coli: Experimental and bioinformatics analyses. Gut Pathog. 2019, 11, 22. [Google Scholar] [CrossRef]
- Ballén, V.; Gabasa, Y.; Ratia, C.; Ortega, R.; Tejero, M.; Soto, S. Antibiotic resistance and virulence profiles of Klebsiella pneumoniae strains isolated from different clinical sources. Front. Cell. Infect. Microbiol. 2021, 11, 738223. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Buc, E.; Dubois, D.; Sauvanet, P.; Raisch, J.; Delmas, J.; Darfeuille-Michaud, A.; Pezet, D.; Bonnet, R. High Prevalence of Mucosa-Associated, E. coli Producing Cyclomodulin and Genotoxin in Colon Cancer. PLoS ONE 2013, 8, e56964. [Google Scholar] [CrossRef] [PubMed]
- Lidin-Janson, G.; Kaijser, B.; Lincoln, K.; Olling, S.; Wedel, H. The homogeneity of the faecal coliform flora of normal school-girls, characterized by serological and biochemical properties. Med. Microbiol. Immunol. 1978, 164, 247–253. [Google Scholar] [CrossRef]
- Raisch, J.; Buc, E.; Bonnet, M.; Sauvanet, P.; Vazeille, E.; de Vallée, A.; Déchelotte, P.; Darcha, C.; Pezet, D.; Bonnet, R.; et al. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J. Gastroenterol. 2014, 20, 6560–6572. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, Y.; Tsunematsu, Y.; Matsuzaki, N.; Hirayama, Y.; Higashiguchi, F.; Sato, M.; Iwashita, Y.; Miyoshi, N.; Mutoh, M.; Ishikawa, H.; et al. Characterization of colibactin-producing Escherichia coli isolated from japanese patients with colorectal cancer. Jpn. J. Infect. Dis. 2020, 73, 437–442. [Google Scholar] [CrossRef]
- Iida, Y.; Kawai, K.; Tsuno, N.H.; Ishihara, S.; Yamaguchi, H.; Sunami, E.; Kitayama, J.; Watanabe, T. Proximal shift of colorectal cancer along with aging. Clin. Colorectal Cancer 2014, 13, 213–218. [Google Scholar] [CrossRef]
- Watanabe, D.; Murakami, H.; Ohno, H.; Tanisawa, K.; Konishi, K.; Tsunematsu, Y.; Sato, M.; Miyoshi, N.; Wakabayashi, K.; Watanabe, K.; et al. Association between dietary intake and the prevalence of tumourigenic bacteria in the gut microbiota of middle-aged Japanese adults. Sci. Rep. 2020, 10, 15221. [Google Scholar] [CrossRef]
- Wassenaar, T.M. E.coli and colorectal cancer: A complex relationship that deserves a critical mindset. Crit. Rev. Microbiol. 2018, 44, 619–632. [Google Scholar] [CrossRef]
- Fabian, N.J.; Mannion, A.J.; Feng, Y.; Madden, C.M.; Fox, J.G. Intestinal colonization of genotoxic Escherichia coli strains encoding colibactin and cytotoxic necrotizing factor in small mammal pets. Vet. Microbiol. 2020, 240, 108506. [Google Scholar] [CrossRef]
- Suchanek, S.; Majek, O.; Vojtechova, G.; Minarikova, P.; Rotnaglova, B.; Seifert, B.; Minarik, M.; Kozeny, P.; Dusek, L.; Zavoral, M. Colorectal cancer prevention in the Czech Republic: Time trends in performance indicators and current situation after 10 years of screening. Eur. J. Cancer Prev. 2014, 23, 18–26. [Google Scholar] [CrossRef] [PubMed]

| Target Gene | Primer Name | Primer Sequence | 1L (bp) | 2Ta (°C) | Reference |
|---|---|---|---|---|---|
| clbB | clbB—F | GAT TTG GAT ACT GGC GAT AAC CG | 579 | 57 | [57] |
| clbB—R | CCA TTT CCC GTT TGA GCA CAC | ||||
| clbN | clbN—F | GTT TTG CTC GCC AGA TAG TCA TTC | |||
| clb N—R | CAG TTC GGG TAT GTG TGG AAG G | ||||
| cnf1 | cnf 1—F | GGCGACAAATGCAGTATTGCTTGG | 552 | 63 | [58] |
| cnf 1—R | GACGTTGGTTGCGGTAATTTTGGG | ||||
| cdtB | CDTs1—F | GAAAGTAAATGGAATATAAATGTCCG | 466 | 56 | [59] |
| CDTas1—R | AAATCACCAAGAATCATCCAGTTA | ||||
| CDTs2—F | GAAAATAAATGGAACACACATGTCCG | ||||
| CDTas2—R | AAATCTCCTGCAATCATCCAGTTA | ||||
| chuA | chuA.1F | GACGAACCA ACGGTCAGGAT | 279 | 60 | [60] |
| chuA.2R | TGCCGCCAGTACC AAAGACA | ||||
| yjaA | yjaA.1F | TGAAGTGTCAGGAGACGCT G | 211 | ||
| yjaA.2R | ATGGAGAATGCGTTCCTCAAC | ||||
| TspE4.C2 | TspE4C2.1F | GAGTAATGTCGGGGCATTCA | 152 | ||
| TspE4C2.2R | CGCGCCAACAAAGTATTACG |
| n | % | |
|---|---|---|
| Hernia | 47 | 42 |
| Gallstones | 33 | 30 |
| Non-CRC malignancy | 13 | 12 |
| Hemorrhoids | 10 | 9 |
| Other | 8 | 7 |
| All Participants | |||
|---|---|---|---|
| All | CRC+ | Controls | |
| n | 263 | 140 | 123 |
| Males | 155 | 93 | 62 |
| Females | 108 | 47 | 61 |
| Mean age | 69.1 | 69.4 | 68.6 |
| SD | 8.49 | 8.15 | 8.86 |
| E. coli-positive participants | |||
| n | 241 | 130 | 111 |
| isolates recovered | 360 | 208 | 152 |
| Males | 141 | 86 | 55 |
| isolates recovered | 225 | 147 | 78 |
| Females | 100 | 44 | 56 |
| isolates recovered | 135 | 61 | 74 |
| Mean age | 69.2 | 69.5 | 68.9 |
| SD | 8.43 | 8.07 | 8.85 |
| CRC | Controls | p | Detection Rate (%) | |
|---|---|---|---|---|
| E. coli-positive participants | 130 | 111 | ||
| clb+ | 59 | 34 | 0.019 | 45.4 |
| cnf+ | 39 | 19 | 0.020 | 30.3 |
| cdt+ | 10 | 8 | 0.886 | - |
| Hemolytic | 45 | 23 | 0.017 | 34.6 |
| Positive for at least one of the following | ||||
| clb+, cnf+ | 61 | 35 | 0.015 | 46.9 |
| clb+, hemolytic | 64 | 37 | 0.013 | 49.2 |
| clb+, cnf+, hemolytic | 64 | 37 | 0.013 | 49.2 |
| CRC | Detection Rate (%) | |
|---|---|---|
| E. coli-positive participants aged 50–74 | 91 | |
| of those | ||
| clb+ | 40 | 44.0 |
| Positive for at least one of the following | ||
| clb+, cnf+, hemolytic | 44 | 48.3 |
| E. coli-positive participants aged 75–90 | 39 | |
| of those | ||
| clb+ | 19 | 48.7 |
| Positive for at least one of the following | ||
| clb+, cnf+, hemolytic | 20 | 51.3 |
| CRC | Detection Rate (%) | |
|---|---|---|
| E. coli-positive males aged 50–90 | 85 | |
| of those | ||
| clb+ | 42 | 49.4 |
| Positive for at least one of the following | ||
| clb+, cnf+, hemolytic | 44 | 51.8 |
| E. coli-positive males aged 50–74 | 59 | |
| of those | ||
| clb+ | 26 | 44.1 |
| Positive for at least one of the following | ||
| clb+, cnf+, hemolytic | 28 | 47.5 |
| E. coli-positive males aged 75–90 | 26 | |
| of those | ||
| clb+ | 16 | 61.5 |
| Positive for at least one of the following | ||
| clb+, cnf+, hemolytic | 16 | 61.5 |
| CRC | Detection Rate (%) | |
|---|---|---|
| E. coli-positive females aged 50–90 | 45 | |
| of those | ||
| clb+ | 17 | 37.8 |
| Positive for at least one of the following | ||
| clb+, cnf+, hemolytic | 20 | 44.4 |
| E. coli-positive females aged 50–74 | 32 | |
| of those | ||
| clb+ | 14 | 43.8 |
| Positive for at least one of the following | ||
| clb+, cnf+, hemolytic | 16 | 50.0 |
| E. coli-positive females aged 75–90 | 13 | |
| of those | ||
| clb+ | 3 | 23.1 |
| Positive for at least one of the following | ||
| clb+, cnf+, hemolytic | 4 | 30.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mezerová, K.; Starý, L.; Zbořil, P.; Klementa, I.; Stašek, M.; Špička, P.; Skalický, P.; Raclavský, V. Cyclomodulins and Hemolysis in E. coli as Potential Low-Cost Non-Invasive Biomarkers for Colorectal Cancer Screening. Life 2021, 11, 1165. https://doi.org/10.3390/life11111165
Mezerová K, Starý L, Zbořil P, Klementa I, Stašek M, Špička P, Skalický P, Raclavský V. Cyclomodulins and Hemolysis in E. coli as Potential Low-Cost Non-Invasive Biomarkers for Colorectal Cancer Screening. Life. 2021; 11(11):1165. https://doi.org/10.3390/life11111165
Chicago/Turabian StyleMezerová, Kristýna, Lubomír Starý, Pavel Zbořil, Ivo Klementa, Martin Stašek, Petr Špička, Pavel Skalický, and Vladislav Raclavský. 2021. "Cyclomodulins and Hemolysis in E. coli as Potential Low-Cost Non-Invasive Biomarkers for Colorectal Cancer Screening" Life 11, no. 11: 1165. https://doi.org/10.3390/life11111165
APA StyleMezerová, K., Starý, L., Zbořil, P., Klementa, I., Stašek, M., Špička, P., Skalický, P., & Raclavský, V. (2021). Cyclomodulins and Hemolysis in E. coli as Potential Low-Cost Non-Invasive Biomarkers for Colorectal Cancer Screening. Life, 11(11), 1165. https://doi.org/10.3390/life11111165







