Over Expression of the Cyanobacterial Pgr5-Homologue Leads to Pseudoreversion in a Gene Coding for a Putative Esterase in Synechocystis 6803
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth Conditions
2.2. Strain Construction
2.3. Spectroscopy and Microscopy
2.4. Chlorophyll Extraction
2.5. Biochemical Assays
2.6. DNA Extraction, Library Preparation and Sequencing
2.7. Bioinformatics Analysis
2.8. RNA Extraction
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nugent, J.H.A. Photosynthetic electron transport in plants and bacteria. Trends Biochem. Sci. 1984, 9, 354–357. [Google Scholar] [CrossRef]
- Sonoike, K.; Hihara, Y.; Ikeuchi, M. Physiological Significance of the Regulation of Photosystem Stoichiometry upon High Light Acclimation of Synechocystis sp. PCC 6803. Plant Cell Physiol. 2001, 42, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Murakami, A.; Aizawa, K.; Ohki, K. Short-term and Long-term Adaptation of the Photosynthetic Apparatus: Homeostatic Properties of Thylakoids. In The Molecular Biology of Cyanobacteria; Springer: Dordrecht, The Netherlands, 1994; pp. 677–692. [Google Scholar]
- El Bissati, K.; Delphin, E.; Murata, N.; Etienne, A.-L.; Kirilovsky, D. Photosystem II fluorescence quenching in the cyanobacterium Synechocystis PCC 6803: Involvement of two different mechanisms. Biochim. Biophys. Acta Bioenerg. 2000, 1457, 229–242. [Google Scholar] [CrossRef]
- Horton, P.; Ruban, A.V. Regulation of Photosystem II. Photosynth. Res. 1992, 34, 375–385. [Google Scholar] [CrossRef]
- Höhner, R.; Aboukila, A.; Kunz, H.-H.; Venema, K. Proton Gradients and Proton-Dependent Transport Processes in the Chloroplast. Front. Plant Sci. 2016, 7, 218. [Google Scholar] [CrossRef]
- Kramer, D.M.; Evans, J.R. The Importance of Energy Balance in Improving Photosynthetic Productivity. Plant Physiol. 2011, 155, 70–78. [Google Scholar] [CrossRef]
- Mullineaux, C.W. Electron transport and light-harvesting switches in cyanobacteria. Front. Plant Sci. 2014, 5, 7. [Google Scholar] [CrossRef]
- Johnson, G.N. Physiology of PSI cyclic electron transport in higher plants. Biochim. Biophys. Acta-Bioenerg. 2011, 1807, 384–389. [Google Scholar] [CrossRef]
- Guedeney, G.; Corneille, S.; Cuiné, S.; Peltier, G. Evidence for an association of ndh B, ndh J gene products and ferredoxin-NADP-reductase as components of a chloroplastic NAD(P)H dehydrogenase complex. FEBS Lett. 1996, 378, 277–280. [Google Scholar] [CrossRef]
- Yamamoto, H.; Peng, L.; Fukao, Y.; Shikanai, T. An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 2011, 23, 1480–1493. [Google Scholar] [CrossRef]
- Schuller, J.M.; Birrell, J.A.; Tanaka, H.; Konuma, T.; Wulfhorst, H.; Cox, N.; Schuller, S.K.; Thiemann, J.; Lubitz, W.; Sétif, P.; et al. Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer. Science 2019, 363, 257–260. [Google Scholar] [CrossRef] [PubMed]
- DalCorso, G.; Pesaresi, P.; Masiero, S.; Aseeva, E.; Schünemann, D.; Finazzi, G.; Joliot, P.; Barbato, R.; Leister, D. A Complex Containing PGRL1 and PGR5 Is Involved in the Switch between Linear and Cyclic Electron Flow in Arabidopsis. Cell 2008, 132, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Munekage, Y.; Hojo, M.; Meurer, J.; Endo, T.; Tasaka, M.; Shikanai, T. PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 2002, 110, 361–371. [Google Scholar] [CrossRef]
- Cleland, R.E.; Bendall, D.S. Photosystem I cyclic electron transport: Measurement of ferredoxin-plastoquinone reductase activity. Photosynth. Res. 1992, 34, 409–418. [Google Scholar] [CrossRef]
- Arnon, D.I.; Allen, M.B.; Whatley, F.R. Photosynthesis by isolated chloroplasts. Nature 1954, 174, 394–396. [Google Scholar] [CrossRef]
- Suorsa, M.; Järvi, S.; Grieco, M.; Nurmi, M.; Pietrzykowska, M.; Rantala, M.; Kangasjärvi, S.; Paakkarinen, V.; Tikkanen, M.; Jansson, S.; et al. PROTON GRADIENT REGULATION5 is essential for proper acclimation of Arabidopsis photosystem I to naturally and artificially fluctuating light conditions. Plant Cell 2012, 24, 2934–2948. [Google Scholar] [CrossRef]
- Johnson, X.; Steinbeck, J.; Dent, R.M.; Takahashi, H.; Richaud, P.; Ozawa, S.I.; Houille-Vernes, L.; Petroutsos, D.; Rappaport, F.; Grossman, A.R.; et al. Proton gradient regulation 5-mediated cyclic electron flow under ATP- or redox-limited conditions: A study of ΔATPase pgr5 and ΔrbcL pgr5 mutants in the green alga Chlamydomonas reinhardtii. Plant Physiol. 2014, 165, 438–452. [Google Scholar] [CrossRef]
- Nandha, B.; Finazzi, G.; Joliot, P.; Hald, S.; Johnson, G.N. The role of PGR5 in the redox poising of photosynthetic electron transport. Biochim. Biophys. Acta-Bioenerg. 2007, 1767, 1252–1259. [Google Scholar] [CrossRef]
- Labs, M.; Rühle, T.; Leister, D. The antimycin A-sensitive pathway of cyclic electron flow: From 1963 to 2015. Photosynth. Res. 2016, 129, 231–238. [Google Scholar] [CrossRef]
- Shikanai, T. Central role of cyclic electron transport around photosystem I in the regulation of photosynthesis. Curr. Opin. Biotechnol. 2014, 26, 25–30. [Google Scholar] [CrossRef]
- Vermaas, W.F. Photosynthesis and Respiration in Cyanobacteria. Encycl. Life Sci. 2001. [Google Scholar] [CrossRef]
- Lea-Smith, D.J.; Bombelli, P.; Vasudevan, R.; Howe, C.J. Photosynthetic, respiratory and extracellular electron transport pathways in cyanobacteria. Biochim. Biophys. Acta-Bioenerg. 2016, 1857, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Cooley, J.W.; Vermaas, W.F.J. Succinate Dehydrogenase and Other Respiratory Pathways in Thylakoid Membranes of Synechocystis sp. Strain PCC 6803: Capacity Comparisons and Physiological Function. J. Bacteriol. 2001, 183, 4251–4258. [Google Scholar] [CrossRef] [PubMed]
- Bronstein, M.; Schütz, M.; Hauska, G.; Padan, E.; Shahak, Y. Cyanobacterial sulfide-quinone reductase: Cloning and heterologous expression. J. Bacteriol. 2000, 182, 3336–3344. [Google Scholar] [CrossRef][Green Version]
- Theissen, U.; Hoffmeister, M.; Grieshaber, M.; Martin, W. Single Eubacterial Origin of Eukaryotic Sulfide:Quinone Oxidoreductase, a Mitochondrial Enzyme Conserved from the Early Evolution of Eukaryotes During Anoxic and Sulfidic Times. Mol. Biol. Evol. 2003, 20, 1564–1574. [Google Scholar] [CrossRef]
- Berry, S.; Schneider, D.; Vermaas, W.F.J.; Rögner, M. Electron transport routes in whole cells of Synechocystis sp. Strain PCC 6803: The role of the cytochrome bd-type oxidase. Biochemistry 2002, 41, 3422–3429. [Google Scholar] [CrossRef]
- McDonald, A.E.; Ivanov, A.G.; Bode, R.; Maxwell, D.P.; Rodermel, S.R.; Hüner, N.P.A. Flexibility in photosynthetic electron transport: The physiological role of plastoquinol terminal oxidase (PTOX). Biochim. Biophys. Acta-Bioenerg. 2011, 1807, 954–967. [Google Scholar] [CrossRef]
- Pisareva, T.; Shumskaya, M.; Maddalo, G.; Ilag, L.; Norling, B. Proteomics of Synechocystis sp. PCC 6803. FEBS J. 2007, 274, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Parmryd, I.; Nilsson, F.; Persson, A.L.; Pakrasi, H.B.; Andersson, B.; Norling, B. Proteomics of Synechocystis sp. Strain PCC 6803. Mol. Cell. Proteomics 2002, 1, 956–966. [Google Scholar] [CrossRef] [PubMed]
- Feilke, K.; Ajlani, G.; Krieger-Liszkay, A. Overexpression of plastid terminal oxidase in Synechocystis sp. PCC 6803 alters cellular redox state. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160379. [Google Scholar] [CrossRef] [PubMed]
- Yeremenko, N.; Jeanjean, R.; Prommeenate, P.; Krasikov, V.; Nixon, P.J.; Vermaas, W.F.J.; Havaux, M.; Matthijs, H.C.P. Open Reading Frame ssr2016 is Required for Antimycin A-sensitive Photosystem I-driven Cyclic Electron Flow in the Cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2005, 46, 1433–1436. [Google Scholar] [CrossRef] [PubMed]
- Los, D.A.; Zorina, A.; Sinetova, M.; Kryazhov, S.; Mironov, K.; Zinchenko, V.V.; Los, D.A.; Zorina, A.; Sinetova, M.; Kryazhov, S.; et al. Stress Sensors and Signal Transducers in Cyanobacteria. Sensors 2010, 10, 2386–2415. [Google Scholar] [CrossRef] [PubMed]
- Kanesaki, Y.; Yamamoto, H.; Paithoonrangsarid, K.; Shumskaya, M.; Suzuki, I.; Hayashi, H.; Murata, N. Histidine kinases play important roles in the perception and signal transduction of hydrogen peroxide in the cyanobacterium, Synechocystis sp. PCC 6803. Plant J. 2007, 49, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Murata, N.; Los, D.A. Histidine kinase Hik33 is an important participant in cold-signal transduction in cyanobacteria. Physiol. Plant. 2006, 126, 17–27. [Google Scholar] [CrossRef]
- Ogawa, T. A gene homologous to the subunit-2 gene of NADH dehydrogenase is essential to inorganic carbon transport of Synechocystis PCC6803. Proc. Natl. Acad. Sci. USA 1991, 88, 4275–4279. [Google Scholar] [CrossRef]
- Allahverdiyeva, Y.; Mustila, H.; Ermakova, M.; Bersanini, L.; Richaud, P.; Ajlani, G.; Battchikova, N.; Cournac, L.; Aro, E.-M. Flavodiiron proteins Flv1 and Flv3 enable cyanobacterial growth and photosynthesis under fluctuating light. Proc. Natl. Acad. Sci. USA 2013, 110, 4111–4116. [Google Scholar] [CrossRef]
- Dann, M.; Leister, D. Evidence that cyanobacterial Sll1217 functions analogously to PGRL1 in enhancing PGR5-dependent cyclic electron flow. Nat. Commun. 2019, 10, 1–7. [Google Scholar] [CrossRef]
- Zer, H.; Margulis, K.; Georg, J.; Shotland, Y.; Kostova, G.; Sultan, L.D.; Hess, W.R.; Keren, N. Resequencing of a mutant bearing an iron starvation recovery phenotype defines Slr1658 as a new player in the regulatory network of a model cyanobacterium. Plant J. 2018, 93, 235–245. [Google Scholar] [CrossRef]
- Williams, J.G.K. Construction of Specific Mutations in Photosystem II Photosynthetic Reaction Center by Genetic Engineering Methods in Synechocystis 6803. Methods Enzymol. 1988, 167, 766–778. [Google Scholar] [CrossRef]
- Shcolnick, S.; Shaked, Y.; Keren, N. A role for mrgA, a DPS family protein, in the internal transport of Fe in the cyanobacterium Synechocystis sp. PCC6803. Biochim. Biophys. Acta Bioenerg. 2007, 1767, 814–819. [Google Scholar] [CrossRef]
- Hayashi, K.; Nakazawa, M.; Ishizaki, Y.; Hiraoka, N.; Obayashi, A. Regulation of inter- and intramolecular ligation with T4 DNA ligase in the presence of polyethylene glycol. Nucleic Acids Res. 1986, 14, 7617–7631. [Google Scholar] [CrossRef] [PubMed]
- Eaton-Rye, J.J. Construction of Gene Interruptions and Gene Deletions in the Cyanobacterium Synechocystis sp. Strain PCC 6803. In Methods in Molecular Biology (Clifton, N.J.); Humana Press: Totowa, NJ, USA, 2011; Volume 684, pp. 295–312. [Google Scholar]
- Satoh, S.; Ikeuchi, M.; Mimuro, M.; Tanaka, A. Chlorophyll b expressed in Cyanobacteria functions as a light-harvesting antenna in photosystem I through flexibility of the proteins. J. Biol. Chem. 2001, 276, 4293–4297. [Google Scholar] [CrossRef] [PubMed]
- Kamei, A.; Ogawa, T.; Ikeuchi, M. Photosynthesis: Mechanism and Effects; Garab, G., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1998. [Google Scholar]
- Salomon, E.; Keren, N. Manganese limitation induces changes in the activity and in the organization of photosynthetic complexes in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 2011, 155, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Gorbunov, M.Y.; Kolber, Z.S.; Falkowski, P.G. Measuring photosynthetic parameters in individual algal cells by Fast Repetition Rate fluorometry. Photosynth. Res. 1999, 62, 141–153. [Google Scholar] [CrossRef]
- Porra, R.J.; Thompson, W.A.; Kriedemann, P.E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta Bioenerg. 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Zor, T.; Selinger, Z. Linearization of the Bradford protein assay increases its sensitivity: Theoretical and experimental studies. Anal. Biochem. 1996, 236, 302–308. [Google Scholar] [CrossRef]
- De Porcellinis, A.; Frigaard, N.-U.; Sakuragi, Y. Determination of the Glycogen Content in Cyanobacteria. J. Vis. Exp. 2017, e56068. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Babraham Bioinformatics—FastQC A Quality Control tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 June 2020).
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997v2. [Google Scholar]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [PubMed]
- Zak, E.; Norling, B.; Maitra, R.; Huang, F.; Andersson, B.; Pakrasi, H.B. The initial steps of biogenesis of cyanobacterial photosystems occur in plasma membranes. Proc. Natl. Acad. Sci. USA 2001, 98, 13443–13448. [Google Scholar] [CrossRef] [PubMed]
- Ball, S.G.; Morell, M.K. From Bacterial Glycogen to Starch: Understanding the Biogenesis of the Plant Starch Granule. Annu. Rev. Plant Biol. 2003, 54, 207–233. [Google Scholar] [CrossRef] [PubMed]
- Klotz, A.; Forchhammer, K. Glycogen, a major player for bacterial survival and awakening from dormancy. Future Microbiol. 2017, 12, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Damrow, R.; Maldener, I.; Zilliges, Y. The Multiple Functions of Common Microbial Carbon Polymers, Glycogen and PHB, during Stress Responses in the Non-Diazotrophic Cyanobacterium Synechocystis sp. PCC 6803. Front. Microbiol. 2016, 7, 966. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, H.; Ikeuchi, M.; Ogawa, T.; Fukuzawa, H.; Sonoike, K. Large-Scale Analysis of Chlorophyll Fluorescence Kinetics in Synechocystis sp. PCC 6803: Identification of the Factors Involved in the Modulation of Photosystem Stoichiometry. Plant Cell Physiol. 2007, 48, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Hihara, Y.; Ikeuchi, M. Mutation in a novel gene required for photomixotrophic growth leads to enhanced photoautotrophic growth of Synechocystis sp. PCC 6803. Photosynth. Res. 1997, 53, 243–252. [Google Scholar] [CrossRef]
- Hihara, Y.; Sonoike, K.; Ikeuchi, M.; Bryant, D.A. A Novel Gene, pmgA, Specifically Regulates Photosystem Stoichiometry in the Cyanobacterium Synechocystis Species PCC 6803 in Response to High Light. Plant Physiol. 1998, 117, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Shabestary, K.; Björk, S.M.; Asplund-Samuelsson, J.; Joensson, H.N.; Jahn, M.; Hudson, E.P. Pooled CRISPRi screening of the cyanobacterium Synechocystis sp PCC 6803 for enhanced industrial phenotypes. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- de Porcellinis, A.J.; Klähn, S.; Rosgaard, L.; Kirsch, R.; Gutekunst, K.; Georg, J.; Hess, W.R.; Sakuragi, Y. The Non-Coding RNA Ncr0700/PmgR1 is Required for Photomixotrophic Growth and the Regulation of Glycogen Accumulation in the Cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2016, 57, 2091–2103. [Google Scholar] [CrossRef]
- Ohta, H.; Shibata, Y.; Haseyama, Y.; Yoshino, Y.; Suzuki, T.; Kagasawa, T.; Kamei, A.; Ikeuchi, M.; Enami, I. Identification of genes expressed in response to acid stress in Synechocystis sp. PCC 6803 using DNA microarrays. Photosynth. Res. 2005, 84, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Summerfield, T.C.; Li, H.; Sherman, L.A. The heat shock response in the cyanobacterium Synechocystis sp. Strain PCC 6803 and regulation of gene expression by HrcA and SigB. Arch. Microbiol. 2006, 186, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Ascher, D.B.; Blundell, T.L. mCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014, 30, 335–342. [Google Scholar] [CrossRef]
- Yu, J.; Mcintosh, L. Isolation and genetic characterization of pseudorevertants from site- directed PSI mutants in Synechocystis 6803. Methods Enzymol. 1998, 297, 18–26. [Google Scholar] [CrossRef]
- Vermaas, W. Molecular genetics of the cyanobacterium Synechocystis sp. PCC 6803: Principles and possible biotechnology applications. J. Appl. Phycol. 1996, 8, 263–273. [Google Scholar] [CrossRef]
- Nishijima, Y.; Kanesaki, Y.; Yoshikawa, H.; Ogawa, T.; Sonoike, K.; Nishiyama, Y.; Hihara, Y. Analysis of spontaneous suppressor mutants from the photomixotrophically grown pmgA-disrupted mutant in the cyanobacterium Synechocystis sp. PCC 6803. Photosynth. Res. 2015, 126, 465–475. [Google Scholar] [CrossRef]
- Ermakova, S.Y.; Elanskaya, I.V.; Kallies, K.U.; Weihe, A.; Börner, T.; Shestakov, S.V. Cloning and sequencing of mutant psbB genes of the cyanobacterium Synechocystis PCC 6803. Photosynth. Res. 1993, 37, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Cano, M.; Holland, S.C.; Artier, J.; Burnap, R.L.; Ghirardi, M.; Morgan, J.A.; Yu, J. Glycogen Synthesis and Metabolite Overflow Contribute to Energy Balancing in Cyanobacteria. Cell Rep. 2018, 23, 667–672. [Google Scholar] [CrossRef] [PubMed]
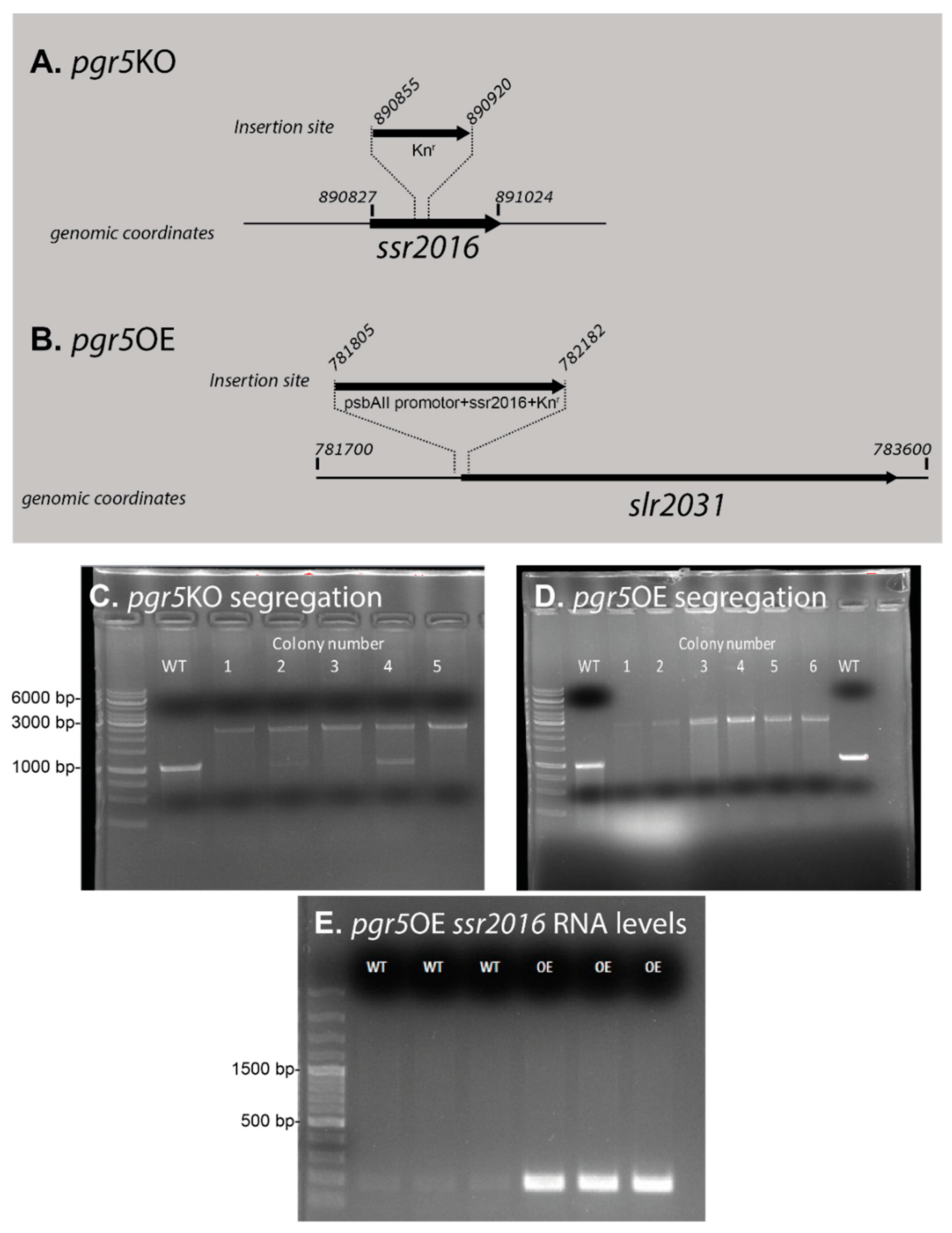
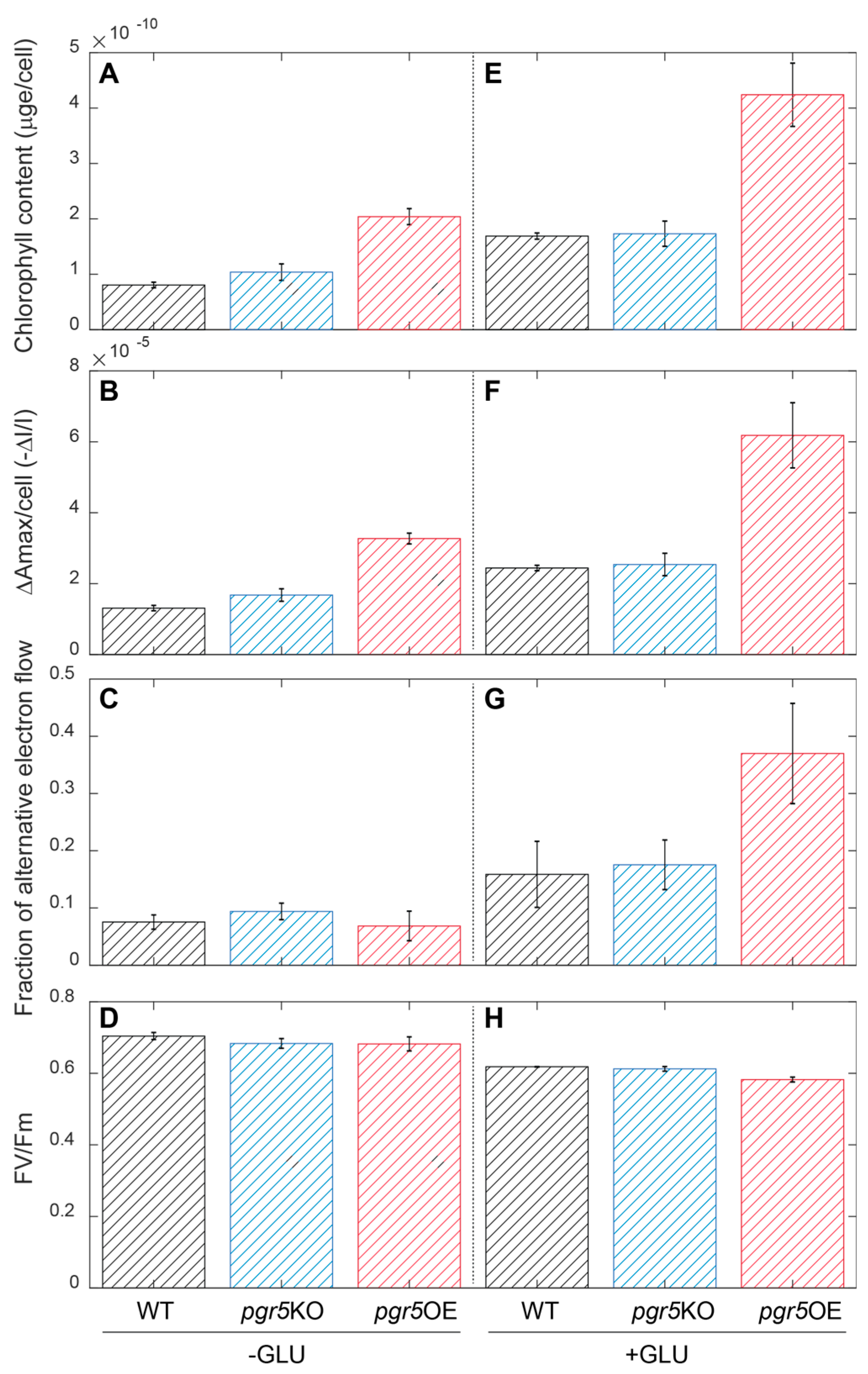
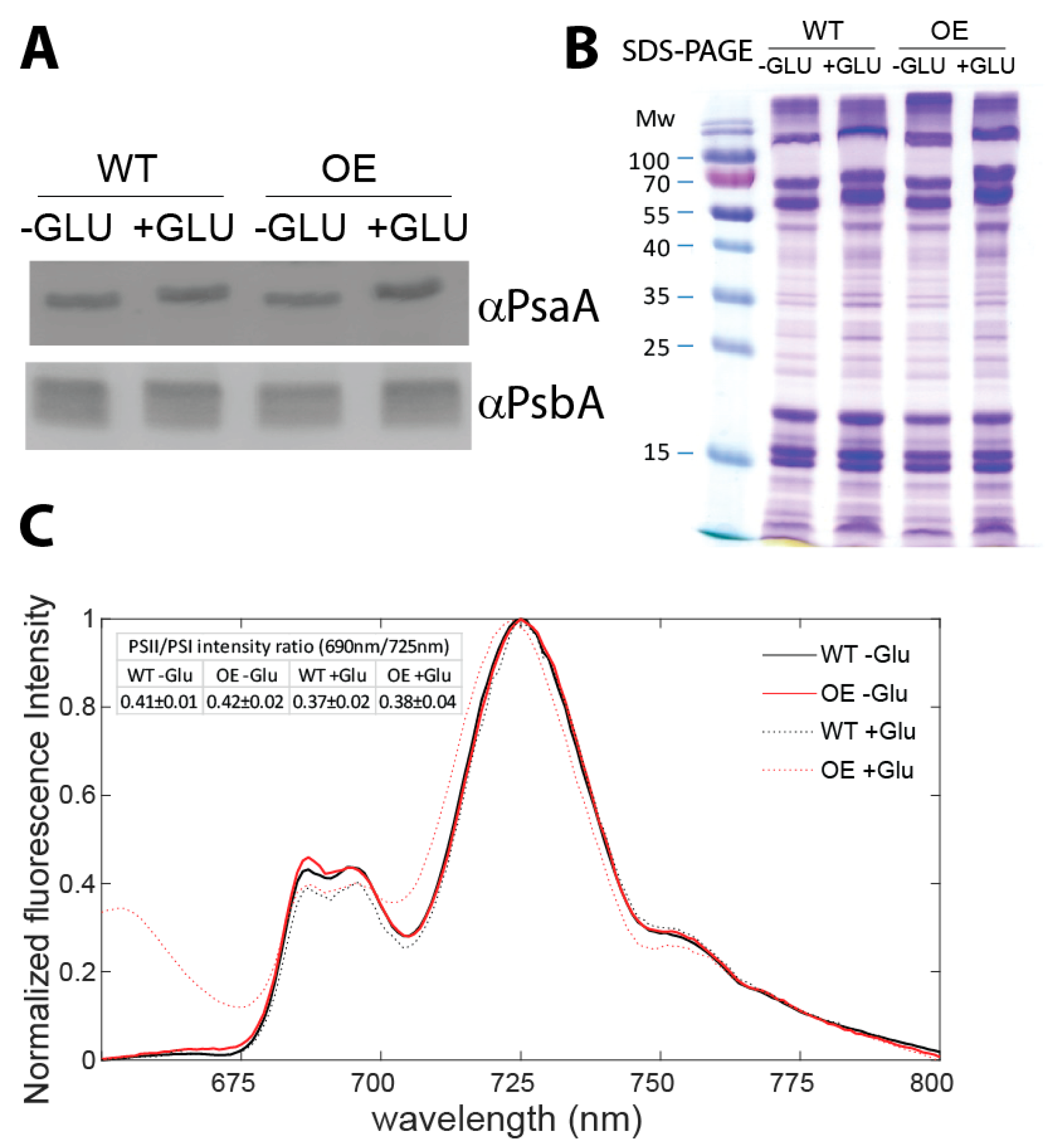
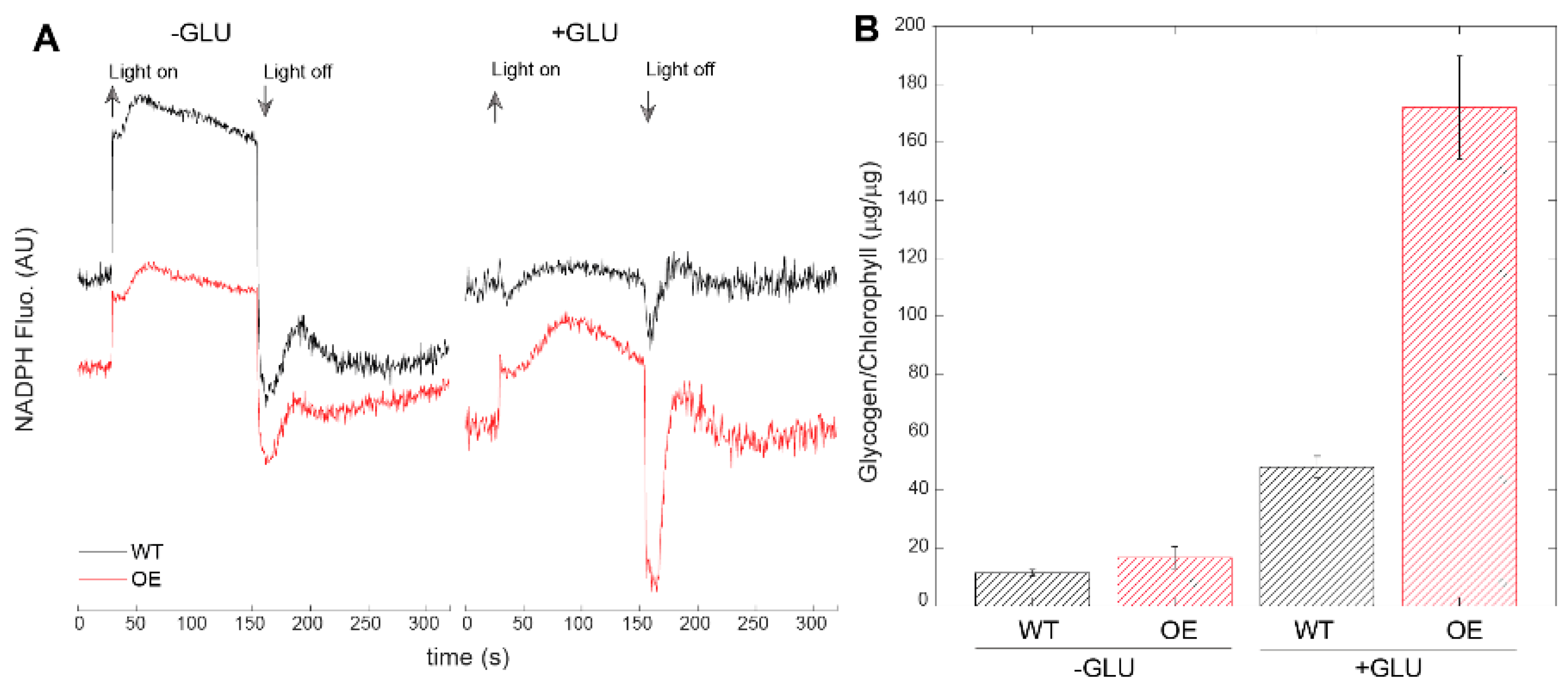
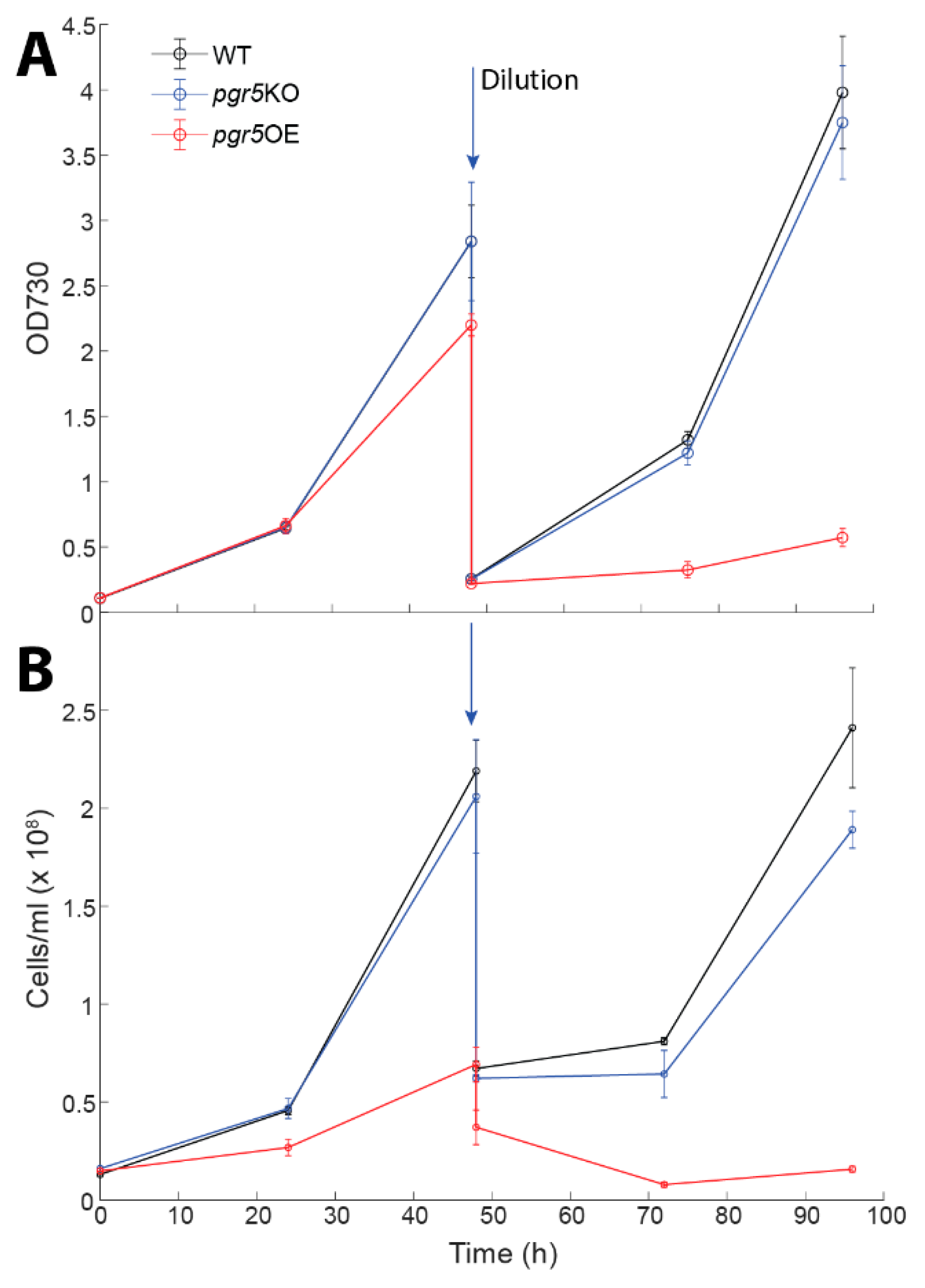
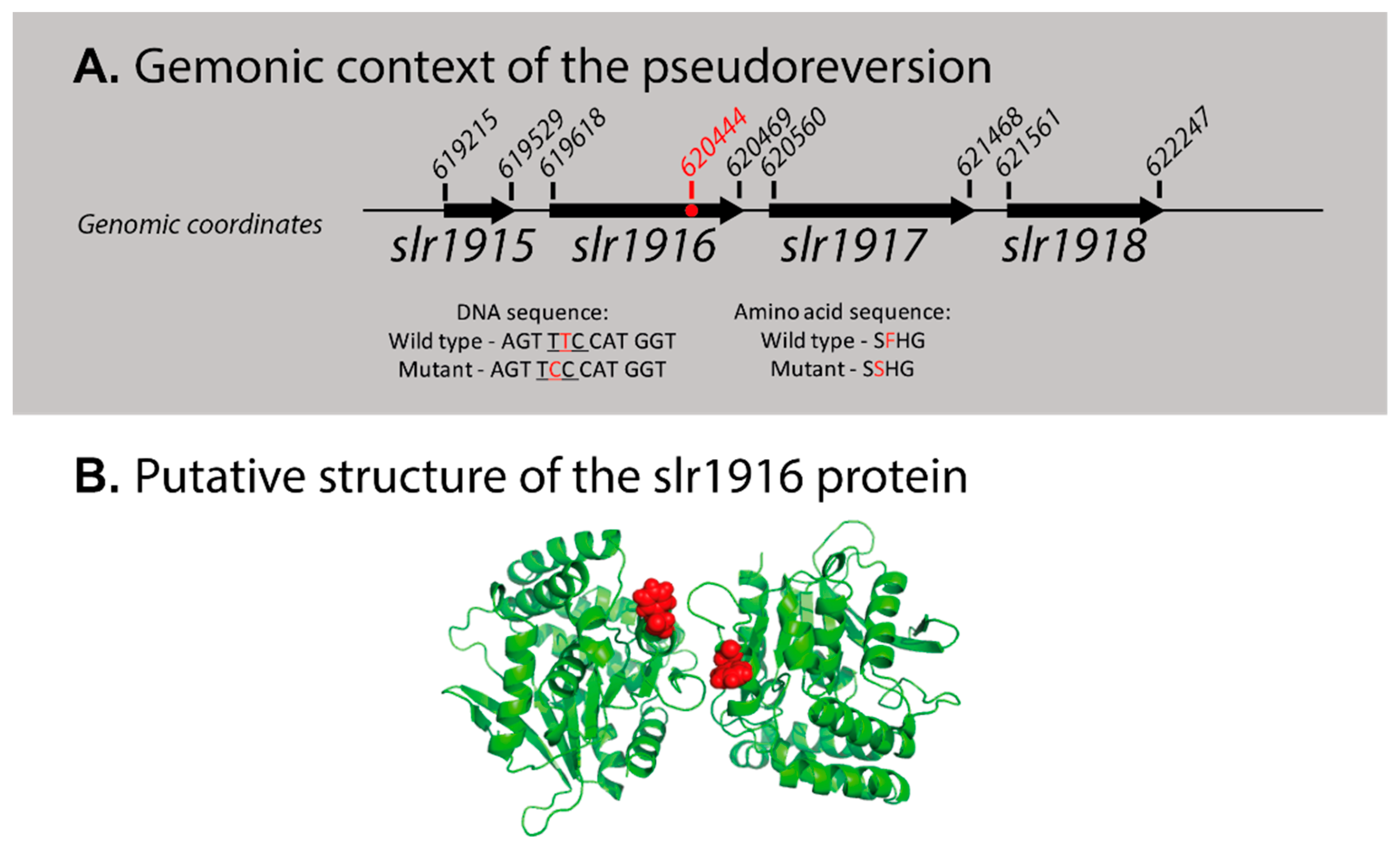
| Mutant Name | Genomic Location | Primer | |
|---|---|---|---|
| pgr5KO | 890467–890486 | F | 5′-CACCATTGGCCTGGTATTGG-3′ |
| 890848–890867 | R | 5′-TTGGTTCGTCAACAGTTAGG-3′ | |
| 890915–890938 | F | 5′-GCCAGACCATCACCAACTTTTGTA-3′ | |
| 891404–891424 | R | 5′-AAATGCCAGGTAACTAATTTG-3′ | |
| pgr5KO segregation check | 890268–890287 | F | 5′-ACGTCACGTCCTTTGAGGTC-3′ |
| 891290–891309 | R | 5′-GGATGACCAGGAAGCCAACC-3′ | |
| pgr5OE | 890816–890835 | F | 5′-GAGTCACTGCCATGTTCGCC-3′ |
| 891025–891049 | R | 5′-CTCTTCGTTTTCAATAATTCTTGCC-3′ | |
| slr2030–slr2031 segregation check | 781363–781383 | F | 5′-TGGGCACAACCATTTACCCTG-3′ |
| 782328–782348 | R | 5′-AACTATGACCAACTGCGCCAG-3′ | |
| pgr5OE RT-PCR | 890827–890852 | F | 5′-ATGTTCGCCCCCATCGTTATCTTGG-3′ |
| 891289–891314 | R | 5′-GAGGGTTTTGCCGTTGGACTTAGCT-3′ | |
| slr1916 mutation verification | 619827–619847 | F | 5′-CCCGTTCAGAATATGACCTGG-3′ |
| 620881–620902 | R | 5′-GCCGTACTTATTGGCAATTCC-3′ | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Margulis, K.; Zer, H.; Lis, H.; Schoffman, H.; Murik, O.; Shimakawa, G.; Krieger-Liszkay, A.; Keren, N. Over Expression of the Cyanobacterial Pgr5-Homologue Leads to Pseudoreversion in a Gene Coding for a Putative Esterase in Synechocystis 6803. Life 2020, 10, 174. https://doi.org/10.3390/life10090174
Margulis K, Zer H, Lis H, Schoffman H, Murik O, Shimakawa G, Krieger-Liszkay A, Keren N. Over Expression of the Cyanobacterial Pgr5-Homologue Leads to Pseudoreversion in a Gene Coding for a Putative Esterase in Synechocystis 6803. Life. 2020; 10(9):174. https://doi.org/10.3390/life10090174
Chicago/Turabian StyleMargulis, Ketty, Hagit Zer, Hagar Lis, Hanan Schoffman, Omer Murik, Ginga Shimakawa, Anja Krieger-Liszkay, and Nir Keren. 2020. "Over Expression of the Cyanobacterial Pgr5-Homologue Leads to Pseudoreversion in a Gene Coding for a Putative Esterase in Synechocystis 6803" Life 10, no. 9: 174. https://doi.org/10.3390/life10090174
APA StyleMargulis, K., Zer, H., Lis, H., Schoffman, H., Murik, O., Shimakawa, G., Krieger-Liszkay, A., & Keren, N. (2020). Over Expression of the Cyanobacterial Pgr5-Homologue Leads to Pseudoreversion in a Gene Coding for a Putative Esterase in Synechocystis 6803. Life, 10(9), 174. https://doi.org/10.3390/life10090174





